Abstract
Enrichment of nicotinic acetylcholine receptors (nAChR) on the tip of the subjunctional folds of the postsynaptic membrane is a central event in the development of the vertebrate neuromuscular junction. This is attained, in part, through a selective transcription in the subsynaptic nuclei, and it has recently been shown that the GA binding protein (GABP) plays an important role in this compartmentalized expression. The neural factor heregulin (HRG) activates nAChR transcription in cultured cells by stimulating a signaling cascade of protein kinases. Hence, it is speculated that GABP becomes activated by phosphorylation, but the mechanism has remained elusive. To fully understand the consequences of GABP phosphorylation, we examined the effect of heregulin-elicited GABP phosphorylation on cellular localization, DNA binding, transcription, and mobility. We demonstrate that HRG-elicited phosphorylation dramatically changes the transcriptional activity and mobility of GABP. While phosphorylation of GABPβ seems to be dispensable for these changes, phosphorylation of GABPα is crucial. Using fluorescence resonance energy transfer, we furthermore showed that phosphorylation of threonine 280 in GABPα triggers reorganizations of the quaternary structure of GABP. Taken together, these results support a model in which phosphorylation-elicited structural changes of GABP enable engagement in certain interactions leading to transcriptional activation.
The neuromuscular junction (NMJ) is a specialized structure involved in communication between the motor neurons and skeletal muscle cells. As the NMJ forms, signals from the motor nerve terminal initiate a complex sequence of processes in the myonuclei, which result in the accumulation of a specific set of gene products in the postsynaptic membrane, such as the nicotinic acetylcholine receptors (nAChR), acetylcholine esterase, and utrophin (13, 44). However, in the absence of innervation, the muscle seems to be somewhat prepatterned (31, 60).
The present models for postsynaptic differentiation suggest that at least three different mechanisms operate in the polynucleated muscle cell. On the one hand, nerve-evoked electrical activity represses transcription of synaptic genes in extrasynaptic nuclei but on the other hand, several mechanisms may possibly contribute to the increased local concentration of nAChR at the NMJ. Two nerve-derived signaling molecules enhance both transcriptional and posttranslational mechanisms that govern clustering and targeting of the synaptic proteins directly underneath the motor nerve: (i) agrin released from the motor nerve terminal into the basal lamina of the synaptic cleft stimulates redistribution of previously unlocalized cell surface nAChR to the postsynaptic membrane domain (for reviews, see references 22 and 42), and (ii) another neurotrophic factor selectively augments transcription of nAChR genes in the subsynaptic nuclei (for reviews, see references 44 and 47). At present, the most likely candidate for the latter activity is the so-called acetylcholine receptor-inducing activity, also known as heregulin β (HRG) or neuregulin β1 (10, 25, 43).
Given that HRG plays a fundamental role in driving synaptic expression for a defined set of genes, the HRG receptor and its localization were examined. It was demonstrated that HRG binds to various dimers of the ErbB family and that these receptors are concentrated at the NMJ of innervated muscles (3, 25). Furthermore, stimulation of ErbB receptors by HRG leads to the activation of the mitogen-activated protein (MAP) kinases, Erk and Jnk, which suggested that substrates of MAP kinases play a pivotal role in regulating nAChR gene expression in the subsynaptic nuclei (2, 51, 55).
Using a systematic mutagenesis approach, efforts in our laboratory led to the identification of the minimal motif, the N box, required for the HRG-stimulated synaptic expression of the nAChRδ and -ɛ subunit genes (15, 28). Interestingly, further studies have shown that the N box is also essential for HRG-elicited transcription of the utrophin and acetylcholine esterase genes (7, 20, 26), suggesting that the N box is a general promoter element directing synaptic expression. Alternative, N-box-independent mechanisms, which regulate the expression of other synaptic genes, exist in parallel (11). Yet, the N box remains a critical element in targeting transcription of several key synaptic genes to the subjunctional domain.
Using N-box-containing oligonucleotides as bait, the factor that binds the N box was identified as an Ets-related transcription factor, the so-called GA binding protein (GABP) (18, 48). GABP is a heteromeric DNA binding protein composed of an α subunit and a β subunit (29). The α subunit is a member of the Ets family of transcription factors and harbors the DNA-binding elements of the complex (56). The β subunit contains a number of ankyrin motifs that mediate the highly specific dimerization to GABPα (4, 56). GABPβ does not contain DNA-binding activity but dramatically enhances the DNA-binding capacity of GABPα and contains the nuclear localization signal that translocates GABPα to the nucleus (45). Furthermore, whereas the α subunit is unable to stimulate transcription by itself, the C-terminal part of GABPβ is required for GABP-dependent transcriptional activation (45, 46). In solution, GABP exists exclusively as an αβ dimer, but upon binding to DNA with two Ets-binding sites, GABP forms an α2β2 heterotetramer (8). Tetramerization depends on a C-terminal leucine zipper-like domain (12, 45, 46). However, the mechanism and the significance of GABP tetramer assembly are still not clear, even though it seems to be important for the transactivation activity of GABP.
GABP becomes phosphorylated following HRG treatment of cells in culture (48). As observed with other Ets factors, in vitro studies have demonstrated that both subunits of GABP can be phosphorylated directly by MAP kinases (16, 17, 57). Threonine at position 280 of GABPα as well as serine 170 and threonine 180 of GABPβ were identified as the major phosphorylation sites in vitro and in vivo (17). Since the transcriptional activities of other Ets factors have been shown to be augmented by phosphorylation (34, 57), it has been speculated that the transcriptional activity of GABP is also increased by HRG-elicited phosphorylation. However, the consequences of GABP phosphorylation in transcriptional initiation have remained speculative, and the mechanism whereby phosphorylation would mediate transcriptional activation of GABP has remained unresolved. In fact, the mechanism whereby phosphorylation results in protein activation has been elucidated for only a few proteins (30, 53), but it is likely that phosphorylation mediates protein activation through conformational allosteric changes within the tertiary and quaternary structures of the protein.
In this study, we have addressed the role of HRG-elicited phosphorylation for the competence of the transcription factor GABP in localization, transcription, DNA binding, and mobility. Furthermore, we explored the intramolecular organization of phosphorylated and nonphosphorylated GABP. We find that phosphorylation of threonine 280 of GABPα has major impact on the transcriptional competence and the intramolecular structure of GABP, as well as on the mobility of the complex in the nucleus. These data suggest that phosphorylation stabilizes a protein conformation that renders GABP competent as a transcriptional activator as part of a larger complex.
MATERIALS AND METHODS
Cell lines and transient-transfection assays.
C2C12 cells were routinely grown in high-glucose Dulbecco's modified Eagle's medium (Gibco-BRL) with 20% fetal bovine serum (Gibco-BRL) and antibiotics. For transfection experiments, 2.5 × 105 C2C12 cells were plated on 35-mm-diameter dishes coated with collagen (Becton Dickinson). The following day, the cultures were transfected by using the Lipofectamine plus kit (Invitrogen) according to the manufacturer's recommendations. The cells were induced to differentiate into myotubes by replacing fetal bovine serum with 5% horse serum for 2 days. For the experiments with HRG (Euromedex), cells were treated with 2.5 nM HRG in differentiation medium for the indicated times starting at day 4 after seeding.
Treatments with PD98,059 (PD) (Sigma) were initiated at the indicated times before HRG addition at 5 nM.
Plasmid construction and oligonucleotides.
The oligonucleotides used in this study were all purchased from Eurogentec, Seraing, Belgium. GABPα and -β constructs were inserted in frame into the pE(C/Y/G)FP-N1 or -C1 vector (Invitrogen) by PCR amplification. All inserted segments were verified by sequencing (Genome Express). To generate site-specific mutants of GABPα and -β, standard procedures for Quickchange (Stratagene) were followed as recommended by the vendor. GABPα(T280A) denotes a site-specific mutant in which the threonine at position 280 of GABPα was replaced by an alanine. GABPβ(S170A-T180A) denotes a mutant in which serine 170 and threonine 180 were replaced by alanines. All plasmid DNA was prepared by using Qiagen kits. To verify the expression of the expected proteins, their molecular weights were validated in conventional Western blotting.
Luciferase and β-galactosidase assays.
C2C12 cells were cotransfected with wild-type or mutant GABP together with a luciferase reporter plasmid regulated by a 2,200-bp fragment of the nAChRɛ promoter (48). To normalize for transfection efficiency, the pSG5-βGal plasmid was included and the resulting β-galactosidase activity was monitored. In brief, the cells were collected by trypsination at day 4 to 6 after induction of differentiation and divided into two aliquots. Luciferase activity and β-galactosidase expression were then measured by previously published procedures (48). Treatments with HRG (Euromedex) were initiated 48 h before trypsination.
Cell extracts and antibodies.
Lysates from cultured C2C12 myotubes were obtained as described previously (2). HRG-treated cells were lysed 15 min after addition of 2.5 nM HRG. Fluorescence-tagged GABPα and/or -β was detected in Western blotting with JL-8 mouse monoclonal antibodies (Clontech). Actin was detected with I-19 goat polyclonal antibodies (Santa Cruz) and served as an internal control for equal protein loading.
Electrophoretic mobility shift assays (EMSAs).
All procedures were performed according to previously published protocols (48). In brief, approximately 2 ng of labeled probe was incubated for 30 min with cell lysate (30 μg) at room temperature before separation by 5% nondenaturing polyacrylamide gel electrophoresis. The resulting bands were visualized and quantitated by phosphorimager analysis (Molecular Dynamics) after the gel had been dried.
Insoluble-protein fractionation and protein extraction.
Insoluble proteins were isolated according to the protocol of Miura and Sasaki (37). In brief, 15 min after initiation of HRG incubation, treated and mock-treated C2C12 cells were lysed on ice in buffer I (10 mM Tris-HCl [pH 7.2], 2.5 mM MgCl2, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride). The cell lysates were centrifuged at 735 × g (Eppendorf 5415C centrifuge), and the soluble proteins (supernatant) were removed. The pellet containing the detergent-insoluble protein fraction (IPF) was lysed in buffer II (20 mM NaxHxPO4, pH 8.0, 0.5 M NaCl, 1 mM EDTA, 5 mM MgCl2, 0.75% Triton X-100, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride). The proteins were recovered by centrifugation at 11,750 × g. The obtained protein samples were sonicated, and the protein concentration was determined with a Bradford protein assay kit (Bio-Rad). Aliquots of 25 μg of IPF were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4 to 15% polyacrylamide) (Bio-Rad), transferred onto nitrocellulose membrane (Amersham Pharmacia Biotech), and analyzed by standard Western blotting.
FRAP.
Imaging and measurements of fluorescence recovery after photobleaching (FRAP) were done with a Zeiss LSM 510 inverted confocal laser scanning microscope. The temperature in the culture chamber was kept at 37°C by using a heated cell chamber (Biotechs Inc.). Individual images were taken at 0.3-s intervals before (n = 5) and after (n = 120) bleaching of a circular area at 90% of the maximal laser capacity of the system with five iterations. Imaging scans (512 × 512 pixels) were recorded at 0.1% of the bleaching laser power. For quantitative analysis of FRAP, the fluorescence intensities of the bleached region, the entire cell, and the background were measured at each time point. The relative fluorescence intensity was calculated as previously described (40).
The computer software Prism (Graphpad Software Inc.) was used for nonlinear regression analysis. Kinetic modeling was performed as previously described (9). Briefly, kinetic models assuming the coexistence of one, two, or three individual enzyme fractions with different mobilities were tested. In all cases, best fits (according to r2 values and F test significance) were obtained by assuming the coexistence of two enzyme fractions with different mobilities. Values of maximal recovery derived from nonlinear regression were used to calculate the proportion of the two enzyme fractions with different mobility.
FRAP of GABP in HRG-treated C2C12 cells was measured between 10 and 30 min after HRG addition. PD was added 4 h prior to FRAP determination.
FRET microscopy (acceptor photobleaching FRET).
Fluorescent images were captured with a Zeiss Axiovert fluorescence microscope that was fitted with a Sensicam charge-coupled device camera (12 bit) and controlled by the MetaVue software package (Universal Imaging Corporation). The temperature in the culture dish was kept at 37°C by using a heated cell chamber (Biotechs Inc.). For fluorescence resonance energy transfer (FRET) microscopy, images were taken with the donor filter set for cyan fluorescent protein (CFP) (excitation, 440 nm; dichroic mirror, 455 nm; emission, 480 nm) (Chroma), and a FRET filter set (XF88; Omega Opticals) with excitation of the donor (at 440 nm), a 455-nm dichroic mirror, and an emission filter for the acceptor (at 535 nm). Images mapping FRET between GABPα and -β subunits were analyzed with the NIH Image J software and calculated as the increase in donor (enhanced CFP [ECFP]) fluorescence intensity (I) after photobleaching of the acceptor (enhanced yellow fluorescent protein [EYFP]) fluorophore. After subtraction of background fluorescence, the FRET (dequenching) efficiency (E) for the region of interest is expressed by E (%) × 100 = [(IECFPpostbleach − IECFPprebleach)/IECFPpostbleach]. Photobleaching was performed with maximal output from a 100-W mercury lamp and the EYFP filter set (500 nm) for 5 min, where the ECFP bleaches minimally but EYFP bleaches more than 95%. Samples changing shape or position during the bleach time were disregarded.
Treatment with PD was initiated 4 h prior to acquisition of FRET images, which was followed by a 1-h intermediate wash period before HRG stimulation.
RESULTS
We have previously found that the level of GABP phosphorylation increases approximately twofold upon HRG treatment of skeletal muscle cells in culture (48). Following these observations, the major phosphorylation sites of GABPα and -β were identified as threonine 280 on the α subunit and serine 170 and threonine 180 on the β subunit (17). To determine the functional properties of the phosphorylation sites of GABP, we constructed a series of site-specific mutants of both GABPα and GABPβ cloned into vectors encoding a fluorescent tag, either N terminally or C terminally (Fig. 1).
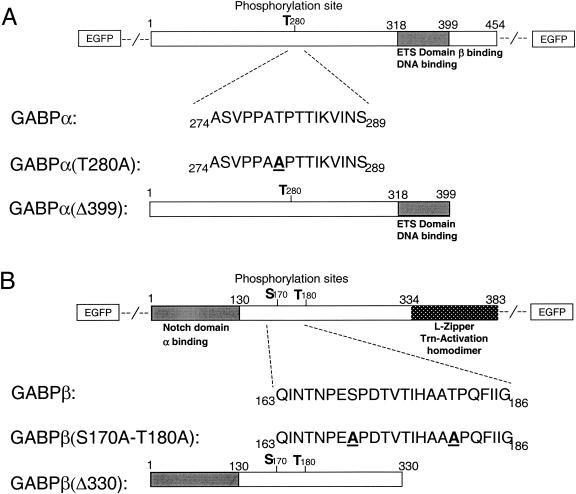
FIG. 1.
Schematic presentation of the fluorescently tagged GABP subunits α and β. (A) GABPα contains a threonine phosphorylation site at amino acid 280, which is replaced with an alanine in GABPα(T280A). Introduction of a stop codon after position 399 deletes the dimerization domain in the GABPα(Δ399) protein. The fluorescent tag is positioned either N or C terminally. (B) By using site-directed mutagenesis, two putative phosphorylation sites of GABPβ are replaced with alanines [GABPβ(S170A-T180A)]. GABPβ(Δ330) contains a truncation mutation at amino acid 330 that deletes the homodimerization domain. The fluorescent tag is positioned either N or C terminally.
Localization of fluorescently tagged GABP in C2C12 cells.
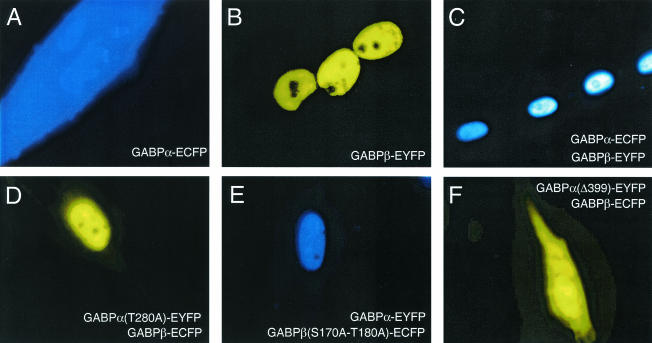
As seen in Fig. 2, the presence of the fluorescent tag does not disturb the localization properties of the GABP subunits. In agreement with previously reported observations obtained by others using immunocytochemical staining (6, 45), we noticed that in living cells, GABPα is not able to direct nuclear localization efficiently when transfected alone (Fig. 2A). In contrast, when GABPβ is transfected alone it localizes exclusively in the nucleus (Fig. 2B), and when it is cotransfected with GABPα, it is able to efficiently transport GABPα to the nucleus (Fig. 2C). Interestingly, with the fluorescently tagged proteins, it is clear that the GABPαβ complex is diffusely distributed in the nucleus, with evidence of some localized accumulations. However, GABP is systematically excluded from the nucleoli.
FIG. 2.
Localization of ECFP- or EYFP-tagged GABP subunits in living C2C12 myotubes. Epifluorescence microscope images were taken with the filter set given by the color in the panel. Images were taken 4 to 6 days after induction of differentiation. C2C12 myoblasts were transfected with GABPα-N1-ECFP (A) or GABPβ-C1-EYFP (B) or cotransfected with GABPα-N1-ECFP and GABPβ-C1-EYFP (ECFP filter set) (C), GABPα(T280A)-N1-EYFP and GABPβ-C1-ECFP (EYFP filter set) (D), GABPα-N1-EYFP and GABPβ(S170A-T180A)-C1-ECFP (ECFP filter set) (E), or GABPα(Δ399)-N1-EYFP and GABPβ-C1-ECFP (EYFP filter set) (F).
Furthermore, a site-specific mutation in the phosphorylation site, which substitutes an alanine for an threonine [GABPα(T280A)], does not alter the localization properties of the GABP complex (Fig. 2D). Similarly, the double mutation of the phosphorylation sites of GABPβ [GABPβ(S170A-T180A)] did not affect nuclear localization of the GABP complex (Fig. 2E).
Two C-terminal truncation mutants, GABPα(Δ399) and GABPβ(Δ330), have previously been shown to behave as dominant negative mutants of transcriptional activation (45, 48, 56). GABPα(Δ399) contains a deletion of the C-terminal dimerization domain, and when cotransfected with wild-type GABPβ, it was no longer transported efficiently to the nucleus and was again detected in the cytoplasm (Fig. 2F). In contrast, a deletion of the homodimerization domain of GABPβ [GABPβ(Δ330)] did not affect its nuclear localization or ability to transport GABPα to the nucleus (data not shown).
We also examined the in vitro localization pattern of GABP upon HRG-elicited phosphorylation, but we were not able to detect any significant changes for any of our constructs (data not shown).
These results present for the first time the localization of the GABP subunits in living cells. We demonstrate that neither HRG-elicited phosphorylation nor the phosphorylation sites are important for proper localization of the GABP complex in the nucleus and, very importantly, that the fluorescent tag does not disturb the localization properties of the individual GABP subunits.
HRG-elicited transcriptional activation of the nAChRɛ promoter.
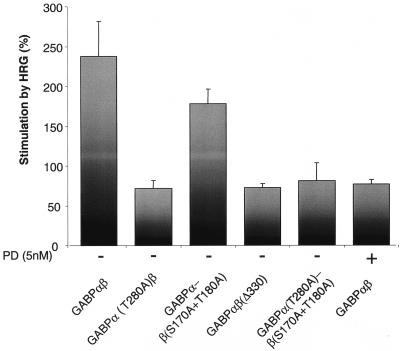
It has been speculated that the observed GABP phosphorylation following HRG treatment of cells in culture might stimulate the transcriptional activity of GABP (17, 18, 20, 26, 48). We thus sought to investigate this question directly in a luciferase-based reporter system. For this purpose, a reporter construct harboring the mouse nAChRɛ promoter was placed upstream of the luciferase gene (14) and cotransfected with the different GABP subunits. As seen in Fig. 3, quantifications of the luciferase activities revealed that cotransfections of the GABPα and -β wild-type subunits cause a ∼2.5-fold increase in luciferase activity after HRG treatment, whereas the point mutation in GABPα(T280A) dramatically hampers the HRG-induced augmentation of luciferase activity (∼0.7-fold induction). Also, we found that HRG treatment of the GABPβ(S170A-T180A) mutant did not have any significant impact on the luciferase activity. When GABPα(T280A) was cotransfected with GABPβ(S170A-T180A), the luciferase activity level was comparable to that obtained with GABPα(T280A). This indicates that phosphorylation of GABPβ in transcriptional activation is dispensable. As previously observed (48), the GABPβ dominant negative mutant caused only a very modest activation of luciferase expression (Fig. 3).
FIG. 3.
nAChRɛ promoter-driven luciferase reporter gene activation. C2C12 cells were cotransfected with the indicated alleles of GABPα and -β and a luciferase gene controlled by a 2,200-bp fragment of the mouse nAChRɛ promoter, which harbors the N-box response element. HRG was added when myoblasts were in the middle of differentiating into myotubes. PD was added 30 min prior to HRG. Measurements of luciferase activity were performed 48 h after addition of HRG. The results are presented as the average ratio of luciferase activities from HRG-treated and nontreated cells (corrected for transfection efficiency) for ≥3 independent experiments done in triplicate. The error bars correspond to the standard errors of the means.
We and others have previously found that GABP is a substrate for MAP kinase phosphorylation in vitro (17, 48). When we added PD (an inhibitor of MEK and downstream kinases [55]) 30 min prior to HRG addition, we found that the HRG-elicited luciferase induction by GABPαβ was greatly reduced [∼0.75-fold induction, which is comparable to the induction obtained with GABPα(T280A)β]. This observation directly demonstrates that MAP kinases also play a fundamental role in vitro in activating the GABP complex by phosphorylation.
Importantly, these data reveal that the fluorescent tag does not interfere with the functional activity of GABP, which maintains its ability to be activated by HRG treatment. Collectively, these results establish that MAP kinase phosphorylation of the T280 residue of GABPα is essential for HRG-elicited stimulation of nAChRɛ promoter-driven luciferase expression, whereas the phosphorylation of GABPβ seems to be less pivotal for transcriptional activation.
DNA binding by GABP is not changed by phosphorylation.
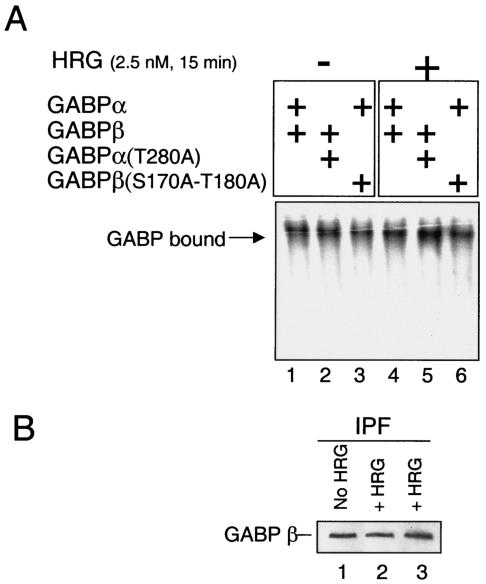
In order to understand the mechanisms leading towards higher expression of the nAChRɛ promoter-driven reporter gene after stimulation with HRG, we next tested the possibility that higher expression was reached through higher DNA-binding capabilities of phosphorylated GABP. We thus examined the ability of wild-type and mutant GABPs to bind to a DNA duplex containing three consecutive N boxes in EMSAs with whole-cell lysates of transfected C2C12 cells. As illustrated in Fig. 4A, no significant differences in the DNA-binding capacities were detected whether the cells were stimulated with HRG or not (compare lanes 1 to 3 with lanes 4 to 6). Thus, phosphorylation of GABP does not influence the DNA-binding properties of the complex. Equal loading (30 μg) was verified by Western blotting with antibodies against the fluorescent tag (data not shown).
FIG. 4.
DNA-binding properties of GABP in EMSA and protein fractionation. (A) EMSA comparison of the DNA-binding efficiencies of GABP wild-type and mutant proteins. C2C12 myotubes transfected with GABPαβ (lanes 1 and 4), GABPα(T280A)β (lanes 2 and 5), or GABPαβ(S170A-T180A) (lanes 3 and 6) were mock treated (−) or treated (+) with HRG 15 min prior to preparation of lysates. C2C12 lysates (30 μg/lane) were incubated with 32P-labeled N-box-containing oligoduplex DNA for 30 min at room temperature. (B) C2C12 myotubes transfected with GABPβ-ECFP were either not treated (lane 1) or treated (lanes 2 and 3) with HRG for 15 min before preparation of the IPFs. Twenty-five micrograms of IPF was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (4 to 15% polyacrylamide), blotted onto a polyvinylidene difluoride membrane, and reacted with anti-EGFP (JL-8).
Since the EMSAs described above were done with naked oligoduplex DNA, we wanted to eliminate the possibility that different DNA-binding capacities of phosphorylated or nonphosphorylated GABP could be found on endogenous DNA. In order to quantify the portion of GABP that is bound to endogenous DNA in the cells, we made a simple extraction of insoluble proteins and analyzed this fraction by Western blotting. This method has previously been utilized to demonstrate the soluble-nonsoluble distribution of other DNA-metabolizing factors (37). Due to the fact that GABPα has some intrinsic DNA-binding affinity on its own (26, 54), we would like to exclude this nonfunctional monomer fraction and exclusively expose the functional GABP tetrameric complexes residing in the IPF. Hence, we transfected C2C12 cells with nontagged GABPα and fluorescent-tagged GABPβ. As seen in Fig. 4B, we were not able to identify any differences in the quantity of GABPβ detectable in the IPF from HRG-treated or nontreated C2C12 cells.
These data solidly confirm and expand our and previously presented EMSA data (18), showing that the phosphorylation state of GABP is not an important parameter for the DNA-binding activity of the complex to naked or endogenous DNA sequences.
HRG stimulation causes GABPα T280-dependent mobility changes.
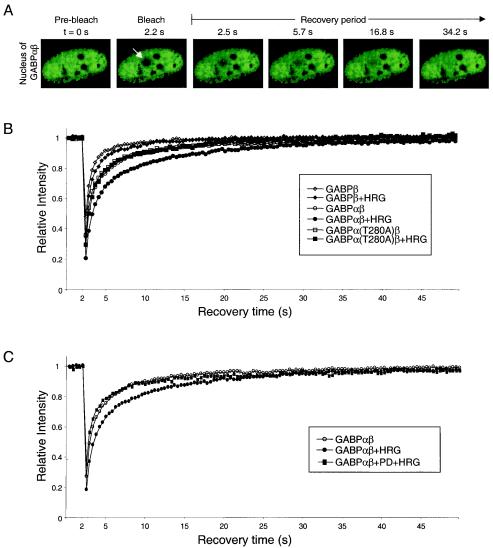
Knowing that the DNA-binding properties of GABP remain unchanged upon HRG treatment, we became interested in investigating whether the HRG-elicited transcriptional activation is attained through the formation of interactions of phosphorylated GABP and other cellular complexes. Since the formation of larger complexes or the transient interaction with less mobile cellular components, such as chromatin, will result in a decrease in the mobility of GABP, we undertook an examination of the mobility of GABP by using a photobleaching technique called FRAP. In this technique, fluorescent GABP molecules are irreversibly bleached in a small area of the nucleus by a high-power laser beam (Fig. 5A, arrow [diameter of bleach spot, ∼2 μm]). The subsequent fluorescent recovery by nonbleached GABP molecules from the surrounding areas moving into the bleached region is recorded at low laser power (Fig. 5A) (for reviews, see references 32 and 58).
FIG. 5.
FRAP analysis of the mobilities of GABP proteins upon HRG stimulation. (A) C2C12 myotubes expressing EGFP-GABPα and nontagged GABPβ were bleached at 2.2 s in the region indicated by the arrow. Images were taken before bleaching and after bleaching with an interval of 0.3-s. Only selected images from the recovery period are shown. (B) FRAP analysis of living cells expressing GABPβ, GABPαβ, and GABPα(T280A)β either mock treated or treated with HRG. (C) FRAP analysis of living cells expressing GABPαβ either mock treated, treated with HRG, or treated with HRG in the presence of PD. Quantitative data of fluorescence recovery kinetics are plotted. Fluorescence intensities in the bleached region were measured and expressed as the relative recovery over time after the bleach pulse. The relative intensity is shown as the average from ≥3 independent experiments with ≥6 individual cells per experiment. Standard deviations were in each case less than 5% of the average value (not shown).
Since GABPβ does not possess any DNA-binding activities when transfected alone (45), we used it as a mobility control for the nonbound fraction. In order to minimize the contribution of GABPα or -β monomers to the mobility of the GABP complex, we exploited the feature of GABPα being transported to the nucleus only in the presence of GABPβ (45). Therefore, GABPβ was tagged only in the experiments addressing the mobility of GABPβ alone; otherwise, enhanced green fluorescent protein (EGFP)-tagged GABPα was cotransfected with nontagged GABPβ.
It is readily apparent from time-lapse fluorescent images (Fig. 5A) and quantitative plots of recovery kinetics (Fig. 5B) that FRAP was fast and complete for all constructs. As evident in Fig. 5B, the recovery of GABPβ is significantly faster than that of GABPαβ in untreated cells, which indicates that a fraction of GABPαβ interacts with DNA. Interestingly, we find that the mobility of GABPαβ is dramatically decreased upon treatment with HRG (Fig. 5B), whereas GABPβ is only slightly retarded in its mobility. Indeed, it is likely that the HRG-elicited mobility shift of GABPβ reflects that a small part of the fluorescently tagged GABPβ interacts with endogenous GABP. To test whether the HRG-elicited mobility change is dependent on the GABPα T280 phosphorylation site, we assessed the mobility of GABPα(T280A) with and without HRG stimulation (Fig. 5B). Importantly, we were not able to discern any kinetic difference between nonstimulated wild-type GABPαβ and the mutant, whether it was HRG stimulated or not.
Since we found that the HRG-elicited transcriptional activity induction of GABP is MAP kinase dependent (Fig. 3), we tested whether the mobility change of GABP following HRG treatment was equally dependent. As evident in Fig. 5C, treatment of transfected C2C12 myotubes with the MAP kinase inhibitor PD completely reverted the fluorescently tagged GABP to the nonstimulated, more mobile form. Together, these results demonstrate that the drop in mobility observed after HRG treatment is dependent on MAP kinase phosphorylations, which render the complex more active in N-box-mediated transcriptional activation. This suggests that the lower mobility is caused by engagement in transcriptional processes on DNA.
Nonlinear regression of the data from Fig. 5B and C indicated with significance (P < 0.0001) that in all cases two different mobility states of the fluorescent enzymes contributed to the apparent FRAP kinetics (Table 1). A major fraction moved with fast kinetics, whereas a smaller fraction moved with slow kinetics. Significantly, we found that the slow population of wild-type GABPαβ was increased from 27.9% ± 1.9% to 33.6% ± 1.1% after HRG treatment, whereas the slow population of GABPα(T280A)β did not change (from 26.5% ± 1.4% to 25.9% ± 1.7%). The increase of the slow population is accompanied by a lower mobility of the wild-type GABP complex after HRG treatment; the half-time needed for fluorescent recovery in the bleached spot increased from 4.59 ± 0.4 to 7.12 ± 0.4 s. Also, analysis of the data obtained with the PD-pretreated cells revealed a pronounced similarity to nontreated wild-type GABPαβ.
TABLE 1.
Nonlinear regression of FRAP data
| Protein (treatment) | Slow population
|
Fast population
|
||
|---|---|---|---|---|
| % of total population (mean ± SEM) | t1/2 (s) (mean ± SEM) | % of total population | t1/2 (s) | |
| GABPβ (none) | 15.4 ± 1.3 | 3.46 ± 0.3 | 84.9 ± 1.3 | 0.37 ± 0.01 |
| GABPβ (HRG) | 19.3 ± 1.9 | 3.96 ± 0.4 | 81.3 ± 2.0 | 0.47 ± 0.02 |
| GABPαβ (none) | 27.9 ± 1.9 | 4.59 ± 0.4 | 70.7 ± 2.0 | 0.55 ± 0.03 |
| GABPαβ (HRG) | 33.6 ± 1.1 | 7.12 ± 0.4 | 64.6 ± 1.3 | 0.72 ± 0.03 |
| GABPαβ (PD + HRG) | 21.1 ± 1.7 | 5.54 ± 0.7 | 76.0 ± 1.8 | 0.45 ± 0.03 |
| GABPα(T280A)β (none) | 26.5 ± 1.4 | 4.98 ± 0.4 | 72.6 ± 1.5 | 0.47 ± 0.02 |
| GABPα(T280A)β (HRG) | 25.9 ± 1.7 | 6.01 ± 0.6 | 75.5 ± 1.9 | 0.66 ± 0.03 |
These data clearly indicate that phosphorylation of T280 in GABPα renders the complex able to engage in certain activities on DNA that keeps the complex immobile for longer periods of time than without HRG-induced phosphorylation.
Efficient FRET is phosphorylation dependent.
Besides increasing the negative charge of the protein, it can be hypothesized that phosphorylation-specific alterations of the quaternary structure of the GABP complex might prime it for further protein-protein interactions. One way of monitoring such structural changes of GABP in living cells is to measure the changes in energy transfer between the subunits upon phosphorylation, the so-called FRET signal (53). This involves the measurement of transfer of energy from the excited state of a donor molecule (ECFP) to an appropriate acceptor molecule (EYFP) (for a review, see reference 32). Experimentally, we applied the acceptor photobleaching technique to detect FRET (for a review, see reference 33). In this technique, energy transfer is measured as an increase in donor fluorescence (dequenching) upon photobleaching of the acceptor fluorophore (see Materials and Methods), which allows FRET determinations with higher reproducibility (59).
First, we tested the dependence of dequenching on donor and acceptor positions in the α and β subunits, as summarized in Table 2. We found that the maximal dequenching (11.7% ± 2.1%) was obtained when GABPα was tagged at the C terminus while GABPβ was N-terminally tagged. As controls, we measured FRET of ECFP and EYFP cotransfections (1.02% ± 0.9%) and from an ECFP-EYFP tandem construct (29.0% ± 3.5%), and the results were comparable to those previously obtained (49).
TABLE 2.
Calculated dequenching values after acceptor bleaching of transiently transfected C2C12 myotubes
| GABPα construct | % Dequenching (mean ± SEM) with the following GABPβ construct:
|
||
|---|---|---|---|
| pEC/YFP-N1-GABPβ | pEC/YFP-C1-GABPβ | pEC/YFP-C1-GABPβ (S170A-T180A) | |
| pEC/YFP-N1-GABPα | 5.10 ± 1.0 | 11.7 ± 2.1 | NDa |
| pEC/YFP-C1-GABPα | 1.52 ± 0.7 | 8.2 ± 1.1 | 9.3 ± 0.8 |
| pEC/YFP-N1-GABα(T280A) | ND | 1.28 ± 0.3 | ND |
ND, not determined.
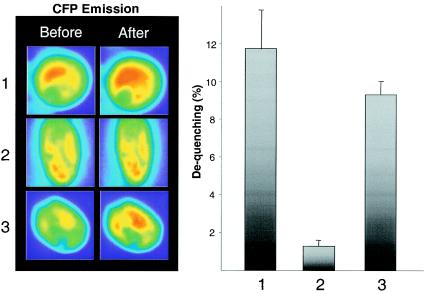
Next, we examined the consequences of site-directed mutations in GABPα and -β for FRET efficiencies by using the same experimental setup. As seen in Fig. 6, we found that ECFP-GABPβ emission was dequenched only 1.3% ± 0.3% after complete photobleaching of EYFP-GABPα(T280A), whereas dequenching of the ECFP-GABPβ(S170A-T180A)-EYFP-GABPα pair reached a level only slightly different from that obtained with the wild-type subunits. These observations suggest that the phosphorylation status of GABP is important for maintaining the correct intermolecular structure of the GABP complex and that the GABPα(T280) site is important in this aspect, whereas the putative phosphorylation sites on GABPβ are less critical.
FIG. 6.
FRET analysis of the GABPαβ heterodimer after acceptor photobleaching. C2C12 cells were transfected with equal amounts of vectors encoding EYFP- and ECFP-GABP fusion proteins. Images were taken before and after the bleach pulse, using both the CFP and FRET filter sets. Left panels, after the cells were differentiated into myotubes, representative images (before and after photobleaching) of the following GABP wild-type and mutant protein combinations were obtained with the ECFP filter set (λ = 480 nm): 1, EYFP-N1-GABPα plus ECFP-C1-GABPβ; 2, EYFP-N1-GABPα(T280A) plus ECFP-C1-GABPβ; 3, EYFP-N1-GABPα plus ECFP-C1-GABPβ(S170A-T180A). Right panel, dequenching calculated as the increase in fluorescent intensity of the donor fluorophore ECFP after acceptor bleaching. Bars, 1, EYFP-N1-GABPα plus ECFP-C1-GABPβ; 2, EYFP-N1-GABPα(T280A) plus ECFP-C1-GABPβ; 3, EYFP-N1-GABPα plus ECFP-C1-GABPβ(S170A-T180A). Data are represented as the averages from ≥3 independent experiments with ≥3 individual cells. Error bars correspond to the standard errors of the means.
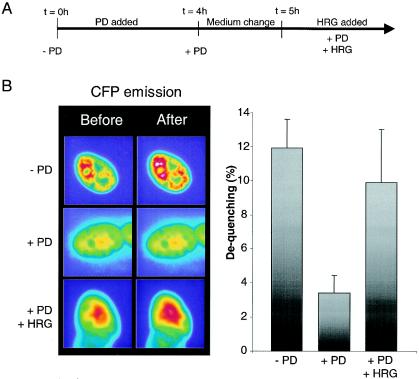
In order to test this directly, we measured the FRET signal between wild-type GABPα and -β after HRG treatment, but we were unable to detect any significant augmentation of the FRET signal at any time point after HRG addition (data not shown). The phosphorylation level is increased maximally twofold upon HRG treatment (48), and this may be below the detection threshold of our FRET system. On the other hand, when we applied a stimulation protocol as outlined in Fig. 7A, we found that preincubation with PD 4 h before FRET determination led to a significantly reduced dequenching of the wild-type GABP subunit combination (from 11.9% ± 1.7% to 3.4% ± 1.3%) (Fig. 7B). Furthermore, after removing PD from the medium, we were able to restore the dequenching levels to near-normal levels with a 15-min HRG treatment (Fig. 7B). These observations show that the overall phosphorylation level is an important determinant in the intermolecular folding pattern.
FIG. 7.
FRET analysis after phosphorylation modifications. (A) Stimulation protocol. At time zero, PD was added and incubated for 4 h. The cells were then washed and reincubated in differentiation medium for 1 h. The medium was again changed, and stimulation with HRG was initiated. (B) In order to determine the impact of phosphorylation on FRET efficiencies, dequenching experiments similar to those presented in Fig. 6 were performed. Left panel, images obtained before and after acceptor bleaching from C2C12 cells cotransfected with EYFP-N1-GABPα plusECFP-C1-GABPβ before (− PD) or after (+ PD) PD treatment or after medium change and reincubation with HRG (+PD + HRG). Right panel, dequenching calculations as described in the legend to Fig. 6.
In conclusion, our experiments demonstrate that the phosphorylation status of GABP is an important parameter for the correct quaternary architecture of a complex, which seems to be apt for interactions governing transcriptional initiation.
DISCUSSION
The N box is a crucial element for the highly compartmentalized expression of nAChRδ and -ɛ subunits in subsynaptic nuclei (15, 28), and the Ets-related transcriptional activator GABP is required for the restricted expression and for the HRG-elicited transcriptional activation (18, 48). Furthermore, the level of GABPα phosphorylation is increased ∼2-fold upon treatment with HRG (48). In this study, we examined the mechanism of GABP phosphorylation. We found that phosphorylation is crucial for transcriptional activation, that phosphorylation significantly changes the mobility of the GABP complex in the nuclei of living cells, that phosphorylation causes important intermolecular conformational changes, and that the threonine 280 phosphorylation site in GABPα is pivotal in these activities.
In agreement with previous reports, we show that the GABP complex is located exclusively in the nucleus (6, 45). However, our in vitro approach with living C2C12 cells allowed us to determine that fluorescent GABP is entirely excluded from the nucleoli.
The results in this study clearly show that the T280A substitution in GABPα dramatically hampers the ability of the protein to support HRG-elicited transcriptional activation. In fact, the stimulation is comparable to what is obtained with GABPβ(Δ330), a dominant negative deletion mutant of GABPβ. On the other hand, a double substitution of phosphorylation sites S170 and T180 of GABPβ had only minor significance in our reporter system assay, indicating that T280 of GABPα is the major functional phosphorylation site in GABP in transcriptional activation in vitro. This finding is a direct corroboration of the phosphorylation data presented by Fromm and Burden showing that T280 is the major phosphorylation acceptor site in vitro (17).
Reversible phosphorylation of transcription factors is a versatile way to accomplish short-term regulation of gene expression (for a review, see reference 23). Phosphorylation of transcription factors has been shown to regulate transcriptional activity by modulating nuclear and cytoplasmic localization (1, 24), modifying DNA-binding affinity (41, 52), or regulating protein-protein interactions (19). These possibilities are not mutually exclusive, and phosphorylation and dephosphorylation of multiple sites can result in dynamic regulation of a transcription factor at various levels. Since neither the localization nor the DNA-binding affinity of GABP was altered upon phosphorylation, we sought to address whether GABP engaged in complex formation upon HRG treatment by using the FRAP approach.
Our findings, which demonstrate that the mobility of GABP is dramatically changed upon phosphorylation, have important implications for its function in transcriptional activation. Recent FRAP experiments revealed that only a few nuclear components can be considered to be immobile, whereas the rest rapidly diffuse in the nucleoplasm (5, 27, 35, 40). On the other hand, GABP is by no means freely diffusible, since EGFP alone, which is thought to be freely diffusible, exhibited recovery rates that were even too high to be detected by our experimental settings. It has been proposed that a decrease in mobility is caused by the interaction with less mobile cellular constituents; however, the nature of such a constituent (chromatin, the cytoskeleton, or a large complex) remains an unsolved issue (36, 50). Nonlinear regression analysis revealed that GABP exists in two different mobility states, a fast and a slow state. Interestingly, after HRG-elicited phosphorylation of threonine 280 on GABPα, an increased portion of the complex was in the slow population. Since the luciferase-based transcriptional assay demonstrated that phosphorylation of this residue augments the transcriptional activation of GABP, we speculate that the fraction exhibiting slow kinetics represents promoter-bound GABP. Importantly, no immobile GABP fraction was detected in either the HRG-treated or the untreated cells. This is consistent with recent mobility studies, which demonstrated that only RNA polymerase II is transiently immobile, whereas transcriptional activators are in a more dynamic equilibrium with a rapid on-off rate at their cognate regulatory elements (for a review, see reference 21).
The mechanism by which phosphorylation of GABPα(T280) activates the complex in transcription is not known; however, the present study points out the importance of this site. Using the FRET approach, we have been able to show that conformational changes between two subunits of a transcription factor may be responsible for its activation upon phosphorylation. As such, phosphorylation can be thought of as contributing to a molecular switch that modifies the transcriptional activity of GABP for as long as it remains phosphorylated, and in this context GABPα(T280) seems to be very critical. From the PD-repressed FRET state, the GABPα(T280)-dependent dequenching peaked at ∼15 min after HRG stimulation, which correlates with kinetics obtained for MAP kinase activity after HRG stimulation of C2C12 cells (17, 51). This report further supports the notion that GABP is phosphorylated in vitro by MAP kinases, since we found that the luciferase activity and the obtained FRET signals were severely hampered by treatment of the cells with the specific MAP kinase inhibitor PD. However, a minor stimulation of the nAChRɛ promoter-driven reporter gene could be observed even in the presence of PD. We suggest that this background is caused either by a leaky promoter or by N-box-independent mechanisms (11).
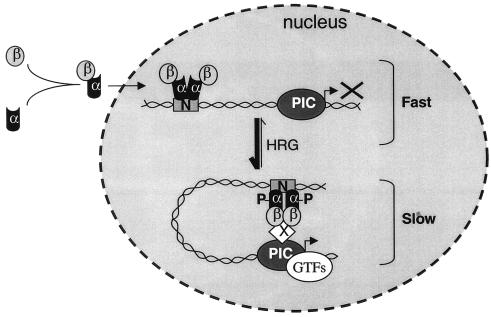
On the basis of our data, we propose a model of GABP transcriptional activation that involves a phosphorylation-dependent mobility shift of GABP (Fig. 8). Monomers of GABPα and -β dimerize in the cytoplasm, and as a complex they are transported into the nucleus. Since it is able to exert DNA-binding activities, we propose that the heterodimer engages in a scanning mode, where it frequently but only very transiently associates with the relatively immobile binding sites in the genomic DNA. Concomitant with DNA binding, a heterotetrameric GABPα2β2 complex is formed, and if this complex becomes phosphorylated, the structural organization is changed in a manner that allows interactions with certain factors of the preinitiation complex. In this way, we speculate that the GABP subunits exist in three cellular modes: a monomeric cytoplasmic mode, a fast DNA scanning mode, and a slow transcriptional activation mode (Fig. 8). Ongoing studies are aimed at clarifying with which factors GABP may be interacting in the transcriptional activation mode.
FIG. 8.
Speculative model of the mechanism of action of GABP in transcriptional activation. The subunits of GABP hardly exist as monomers in solution; heterodimers are quickly formed and transported into the nucleus (54). Once GABPα interacts with GABPβ, a high-affinity DNA-binding complex is formed. Binding to DNA promotes the formation of a GABPα2β2 heterotetrameric complex (8), which is able to scan DNA for N-box sites but not to initiate transcription. HRG-induced phosphorylation generates structural changes of the heterotetrameric GABP complex, which allow interaction with other cellular components of the preinitiation complex (PIC). This will ultimately lead to the assembly of the general transcription factors (GTFs) around the start site, and RNA polymerase II transcription of an N-box-containing gene can be initiated. Fast and slow refer to the mobility of GABP as determined from Fig. 5.
The importance of N-box-dependent transcription has recently been highlighted by the finding of point mutations in the N boxes of the nAChRɛ subunits from patients with a particular form of congenital myasthenia syndrome (38, 39). The point mutation was found to decrease the affinity of GABP for the N box, resulting in lower mRNA and protein levels of the nAChRɛ subunit. Thus, failure to activate transcription may cause an impairment of neuromuscular transmission. A better understanding of the mechanisms by which GABP initiates transcription may provide a novel molecular approach for designing strategies against myasthenic syndromes.
Acknowledgments
We thank H. H. Sitte for the tandem construct. We appreciate the technical support of S. Pons, L. Strochlic, and R. Grailhe. We thank L. Schaeffer and N. Mechawar for critically reading the manuscript.
M.S. was supported by the Carlsberg Foundation, The Danish Research Council, and The Lundbeck Foundation. M.O.C. was supported by the Deutsche Forschungsgemeinschaft (Bo 910/3-1 and Bo 910/4-1).
REFERENCES
- 1.Agarwal, R., and O. Cohen-Fix. 2002. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 16:1371-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altiok, N., S. Altiok, and J. P. Changeux. 1997. Heregulin-stimulated acetylcholine receptor gene expression in muscle: requirement for MAP kinase and evidence for a parallel inhibitory pathway independent of electrical activity. EMBO J. 16:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altiok, N., J. L. Bessereau, and J. P. Changeux. 1995. ErbB3 and ErbB2/neu mediate the effect of heregulin on acetylcholine receptor gene expression in muscle: differential expression at the endplate. EMBO J. 14:4258-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batchelor, A. H., D. E. Piper, F. C. de la Brousse, S. L. McKnight, and C. Wolberger. 1998. The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science 279:1037-1041. [DOI] [PubMed] [Google Scholar]
- 5.Becker, M., C. Baumann, S. John, D. A. Walker, M. Vigneron, J. G. McNally, and G. L. Hager. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3:1188-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briguet, A., and M. A. Ruegg. 2000. The Ets transcription factor GABP is required for postsynaptic differentiation in vivo. J. Neurosci. 20:5989-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, R. Y., C. Boudreau-Lariviere, L. M. Angus, F. A. Mankal, and B. J. Jasmin. 1999. An intronic enhancer containing an N-box motif is required for synapse- and tissue-specific expression of the acetylcholinesterase gene in skeletal muscle fibers. Proc. Natl. Acad. Sci. USA 96:4627-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinenov, Y., M. Henzl, and M. E. Martin. 2000. The alpha and beta subunits of the GA-binding protein form a stable heterodimer in solution. Revised model of heterotetrameric complex assembly. J. Biol. Chem. 275:7749-7756. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, M. O., H. U. Barthelmes, S. Feineis, B. R. Knudsen, A. H. Andersen, F. Boege, and C. Mielke. 2002. Changes in mobility account for camptothecin-induced subnuclear relocation of topoisomerase I. J. Biol. Chem. 277:15661-15665. [DOI] [PubMed] [Google Scholar]
- 10.Chu, G. C., L. M. Moscoso, M. X. Sliwkowski, and J. P. Merlie. 1995. Regulation of the acetylcholine receptor epsilon subunit gene by recombinant ARIA: an in vitro model for transynaptic gene regulation. Neuron 14:329-339. [DOI] [PubMed] [Google Scholar]
- 11.De Kerchove D'Exaerde, A., J. Cartaud, A. Ravel-Chapuis, T. Seroz, F. Pasteau, L. M. Angus, B. J. Jasmin, J. P. Changeux, and L. Schaeffer. 2002. Expression of mutant Ets protein at the neuromuscular synapse causes alterations in morphology and gene expression. EMBO Rep. 3:1075-1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Brousse, F. C., E. H. Birkenmeier, D. S. King, L. B. Rowe, and S. L. McKnight. 1994. Molecular and genetic characterization of GABP beta. Genes Dev. 8:1853-1865. [DOI] [PubMed] [Google Scholar]
- 13.Duclert, A., and J. P. Changeux. 1995. Acetylcholine receptor gene expression at the developing neuromuscular junction. Physiol. Rev. 75:339-368. [DOI] [PubMed] [Google Scholar]
- 14.Duclert, A., N. Savatier, and J. P. Changeux. 1993. An 83-nucleotide promoter of the acetylcholine receptor epsilon-subunit gene confers preferential synaptic expression in mouse muscle. Proc. Natl. Acad. Sci. USA 90:3043-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duclert, A., N. Savatier, L. Schaeffer, and J. P. Changeux. 1996. Identification of an element crucial for the sub-synaptic expression of the acetylcholine receptor epsilon-subunit gene. J. Biol. Chem. 271:17433-17438. [DOI] [PubMed] [Google Scholar]
- 16.Flory, E., A. Hoffmeyer, U. Smola, U. R. Rapp, and J. T. Bruder. 1996. Raf-1 kinase targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J. Virol. 70:2260-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromm, L., and S. J. Burden. 2001. Neuregulin-1-stimulated phosphorylation of gabp in skeletal muscle cells. Biochemistry 40:5306-5312. [DOI] [PubMed] [Google Scholar]
- 18.Fromm, L., and S. J. Burden. 1998. Synapse-specific and neuregulin-induced transcription require an ets site that binds GABPalpha/GABPbeta. Genes Dev. 12:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallagher, E. D., S. Xu, C. Moomaw, C. A. Slaughter, and M. H. Cobb. 2002. Binding of JNK/SAPK to MEKK1 is regulated by phosphorylation. J. Biol. Chem. 277:45785-45792. [DOI] [PubMed] [Google Scholar]
- 20.Gramolini, A. O., L. M. Angus, L. Schaeffer, E. A. Burton, J. M. Tinsley, K. E. Davies, J. P. Changeux, and B. J. Jasmin. 1999. Induction of utrophin gene expression by heregulin in skeletal muscle cells: role of the N-box motif and GA binding protein. Proc. Natl. Acad. Sci. USA 96:3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hager, G. L., C. Elbi, and M. Becker. 2002. Protein dynamics in the nuclear compartment. Curr. Opin. Genet. Dev. 12:137-141. [DOI] [PubMed] [Google Scholar]
- 22.Huh, K. H., and C. Fuhrer. 2002. Clustering of nicotinic acetylcholine receptors: from the neuromuscular junction to interneuronal synapses. Mol Neurobiol. 25:79-112. [DOI] [PubMed] [Google Scholar]
- 23.Hunter, T., and M. Karin. 1992. The regulation of transcription by phosphorylation. Cell 70:375-387. [DOI] [PubMed] [Google Scholar]
- 24.Ihle, J. N. 1996. STATs: signal transducers and activators of transcription. Cell 84:331-334. [DOI] [PubMed] [Google Scholar]
- 25.Jo, S. A., X. Zhu, M. A. Marchionni, and S. J. Burden. 1995. Neuregulins are concentrated at nerve-muscle synapses and activate ACh-receptor gene expression. Nature 373:158-161. [DOI] [PubMed] [Google Scholar]
- 26.Khurana, T. S., A. G. Rosmarin, J. Shang, T. O. Krag, S. Das, and S. Gammeltoft. 1999. Activation of utrophin promoter by heregulin via the ets-related transcription factor complex GA-binding protein alpha/beta. Mol. Biol. Cell 10:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura, H., K. Sugaya, and P. R. Cook. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koike, S., L. Schaeffer, and J. P. Changeux. 1995. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor delta-subunit gene. Proc. Natl. Acad. Sci. USA 92:10624-10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaMarco, K., C. C. Thompson, B. P. Byers, E. M. Walton, and S. L. McKnight. 1991. Identification of Ets- and notch-related subunits in GA binding protein. Science 253:789-792. [DOI] [PubMed] [Google Scholar]
- 30.Lin, K., V. L. Rath, S. C. Dai, R. J. Fletterick, and P. K. Hwang. 1996. A protein phosphorylation switch at the conserved allosteric site in GP. Science 273:1539-1542. [DOI] [PubMed] [Google Scholar]
- 31.Lin, W., R. W. Burgess, B. Dominguez, S. L. Pfaff, J. R. Sanes, and K. F. Lee. 2001. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature 410:1057-1064. [DOI] [PubMed] [Google Scholar]
- 32.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 33.Majoul, I., M. Straub, R. Duden, S. W. Hell, and H. D. Soling. 2002. Fluorescence resonance energy transfer analysis of protein-protein interactions in single living cells by multifocal multiphoton microscopy. J. Biotechnol. 82:267-277. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy, S. A., D. Chen, B. S. Yang, J. J. Garcia Ramirez, H. Cherwinski, X. R. Chen, M. Klagsbrun, C. A. Hauser, M. C. Ostrowski, and M. McMahon. 1997. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol. Cell. Biol. 17:2401-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNally, J. G., W. G. Muller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 36.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 37.Miura, M., and T. Sasaki. 1999. Detection of chromatin-bound PCNA in cultured cells following exposure to DNA-damaging agents. Methods Mol. Biol. 113:577-582. [DOI] [PubMed] [Google Scholar]
- 38.Nichols, P., R. Croxen, A. Vincent, R. Rutter, M. Hutchinson, J. Newsom-Davis, and D. Beeson. 1999. Mutation of the acetylcholine receptor epsilon-subunit promoter in congenital myasthenic syndrome. Ann. Neurol. 45:439-443. [PubMed] [Google Scholar]
- 39.Ohno, K., B. Anlar, and A. G. Engel. 1999. Congenital myasthenic syndrome caused by a mutation in the Ets-binding site of the promoter region of the acetylcholine receptor epsilon subunit gene. Neuromuscul. Disord. 9:131-135. [DOI] [PubMed] [Google Scholar]
- 40.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 41.Piwien-Pilipuk, G., O. MacDougald, and J. Schwartz. 2002. Dual regulation of phosphorylation and dephosphorylation of C/EBPbeta modulate its transcriptional activation and DNA binding in response to growth hormone. J. Biol. Chem. 277:44557-44565. [DOI] [PubMed] [Google Scholar]
- 42.Ruegg, M. A., and J. L. Bixby. 1998. Agrin orchestrates synaptic differentiation at the vertebrate neuromuscular junction. Trends Neurosci. 21:22-27. [DOI] [PubMed] [Google Scholar]
- 43.Sandrock, A. W., S. E. Dryer, K. M. Rosen, S. N. Gozani, R. Kramer, L. E. Theill, and G. D. Fischbach. 1997. Maintenance of acetylcholine receptor number by neuregulins at the neuromuscular junction in vivo. Science 276:599-603. [DOI] [PubMed] [Google Scholar]
- 44.Sanes, J. R., and J. W. Lichtman. 1999. Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22:389-442. [DOI] [PubMed] [Google Scholar]
- 45.Sawa, C., M. Goto, F. Suzuki, H. Watanabe, J. Sawada, and H. Handa. 1996. Functional domains of transcription factor hGABP beta1/E4TF1-53 required for nuclear localization and transcription activation. Nucleic Acids Res. 24:4954-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawada, J., M. Goto, C. Sawa, H. Watanabe, and H. Handa. 1994. Transcriptional activation through the tetrameric complex formation of E4TF1 subunits. EMBO J. 13:1396-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaeffer, L., A. de Kerchove d'Exaerde, and J. P. Changeux. 2001. Targeting transcription to the neuromuscular synapse. Neuron 31:15-22. [DOI] [PubMed] [Google Scholar]
- 48.Schaeffer, L., N. Duclert, M. Huchet-Dymanus, and J. P. Changeux. 1998. Implication of a multisubunit Ets-related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 17:3078-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid, J. A., P. Scholze, O. Kudlacek, M. Freissmuth, E. A. Singer, and H. H. Sitte. 2001. Oligomerization of the human serotonin transporter and of the rat GABA transporter 1 visualized by fluorescence resonance energy transfer microscopy in living cells. J. Biol. Chem. 276:3805-3810. [DOI] [PubMed] [Google Scholar]
- 50.Shopland, L. S., and J. B. Lawrence. 2000. Seeking common ground in nuclear complexity. J. Cell Biol. 150:F1-F4. [DOI] [PubMed] [Google Scholar]
- 51.Si, J., Q. Wang, and L. Mei. 1999. Essential roles of c-JUN and c-JUN N-terminal kinase (JNK) in neuregulin-increased expression of the acetylcholine receptor epsilon-subunit. J. Neurosci. 19:8498-8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stefanovsky, V. Y., G. Pelletier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063-1073. [DOI] [PubMed] [Google Scholar]
- 53.Stultz, C. M., A. D. Levin, and E. R. Edelman. 2002. Phosphorylation-induced conformational changes in a mitogen-activated protein kinase substrate. Implications for tyrosine hydroxylase activation. J. Biol. Chem. 277:47653-47661. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki, F., M. Goto, C. Sawa, S. Ito, H. Watanabe, J. Sawada, and H. Handa. 1998. Functional interactions of transcription factor human GA-binding protein subunits. J. Biol. Chem. 273:29302-29308. [DOI] [PubMed] [Google Scholar]
- 55.Tansey, M. G., G. C. Chu, and J. P. Merlie. 1996. ARIA/HRG regulates AChR epsilon subunit gene expression at the neuromuscular synapse via activation of phosphatidylinositol 3-kinase and Ras/MAPK pathway. J. Cell Biol. 134:465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, C. C., T. A. Brown, and S. L. McKnight. 1991. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science 253:762-768. [DOI] [PubMed] [Google Scholar]
- 57.Wasylyk, B., J. Hagman, and A. Gutierrez-Hartmann. 1998. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23:213-216. [DOI] [PubMed] [Google Scholar]
- 58.White, J., and E. Stelzer. 1999. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 9:61-65. [DOI] [PubMed] [Google Scholar]
- 59.Wouters, F. S., and P. I. Bastiaens. 1999. Fluorescence lifetime imaging of receptor tyrosine kinase activity in cells. Curr. Biol. 9:1127-1130. [DOI] [PubMed] [Google Scholar]
- 60.Yang, X., S. Arber, C. William, L. Li, Y. Tanabe, T. M. Jessell, C. Birchmeier, and S. J. Burden. 2001. Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron 30:399-410. [DOI] [PubMed] [Google Scholar]