Abstract
Gata1 is a prototype transcription factor that regulates hematopoiesis, yet the molecular mechanisms by which Gata1 transactivates its target genes in vivo remain unclear. We previously showed, in transgenic zebra fish, that Gata1 autoregulates its own expression. In this study, we characterized the molecular mechanisms for this autoregulation by using mutations in the Gata1 protein which impair autoregulation. Of the tested mutations, replacement of six lysine residues with alanine (Gata1KA6), which inhibited self-association activity of Gata1, reduced the Gata1-dependent induction of reporter gene expression driven by the zebra fish gata1 hematopoietic regulatory domain (gata1 HRD). Furthermore, overexpression of wild-type Gata1 but not Gata1KA6 rescued the expression of Gata1 downstream genes in vlad tepes, a germ line gata1 mutant fish. Interestingly, both GATA sites in the double GATA motif in gata1 HRD were critical for the promoter activity and for binding of the self-associated Gata1 complex, whereas only the 3′-GATA site was required for Gata1 monomer binding. These results thus provide the first in vivo evidence that the ability of Gata1 to self-associate critically contributes to the autoregulation of the gata1 gene.
Hematopoietic development is regulated in large part by transcription factors that activate or repress certain sets of genes that are characteristic of individual lineages. The transcription factor Gata1 recognizes (T/A)GATA(A/G) sequences, which are found in the control regions of most hematopoietic genes, and activates transcription (37). The biological importance of Gata1 has been demonstrated by genetic studies with mice and zebra fish, which showed a strict requirement for Gata1 in erythroid cell development (10, 23, 43). In addition, selective loss of Gata1 expression in megakaryocytes of mutant mice results in a reduction in the number of platelets and hyperproliferation of megakaryocytes (42). Gata1 contains two zinc finger domains, which are highly conserved among different species (6, 7, 49, 61) and the other members of Gata factor family (56). The C-terminal zinc finger domain (CF) is required for DNA binding, and the N-terminal zinc finger domain (NF) modulates the DNA binding specificity of CF and stabilizes Gata1 binding to palindromic GATA sites (25, 47, 48, 58). NF is also important for the physical interaction with a transcriptional cofactor, Fog1 (51). The in vivo requirements for these zinc finger domains were analyzed by transgenic rescue assay of Gata1 knockdown mice, which demonstrated that both CF and NF are indispensable for erythropoiesis (41). In addition to the Gata1-Fog1 interaction, acetylation of Gata1 has been proposed to be an important step in the transcriptional activation (2, 13), although no direct evidence has been demonstrated so far.
In mice, the Gata1 gene is expressed in erythroid cells, megakaryocytes, and mast cells as well as in Sertoli cells in the testis (14, 26, 49, 60). Gata1 in hematopoietic cells is transcribed predominantly from the immediate-early promoter, one of two cell lineage-specific promoters (57). Reporter gene analysis exploiting the transgenic mouse system revealed that the genomic region containing the immediate-early exon, the first intron, and 3.9 kbp upstream of the transcriptional initiation site substantially recapitulates the endogenous expression profile of the Gata1 gene in erythroid cells and megakaryocytes (38). This region is referred to as the Gata1 hematopoietic regulatory domain (Gata1 HRD) (31). Importantly, transgenic expression of a wild-type Gata1 cDNA under the control of the Gata1 HRD completely rescued the Gata1 gene knockdown phenotype in mice (41, 44), indicating that this gene regulatory domain is sufficient for the function of Gata1 in hematopoietic cells. Four critical motifs for hematopoietic expression have been identified in the Gata1 HRD: the Gata1 hematopoietic enhancer, the double GATA motif, the CACCC box, and the GATA repeat in the first intron (34, 40, 50, 52). Combination of these four elements generates a vector that recapitulates the Gata1 HRD expression profile (mini-HRD vector) (36).
It is of note the GATA sites present in three out of four critical motifs in the mouse gene have also been shown to be important in human, chicken, and zebra fish Gata1 gene regulation (11, 21, 30, 33), suggesting that Gata1 gene expression is maintained by an autoregulatory mechanism during hematopoietic cell development. Along this line, through analyses of green fluorescent protein (GFP) reporter expression in transgenic zebra fish, we have previously demonstrated that Gata1 activates expression of its own promoter (21). We also showed that the double GATA motif located 6.4 kbp upstream from the translation initiation site in the zebra fish gata1 gene is crucial both for the inducible expression of GFP by ectopically expressed Gata1 and for the basal expression of gata1 in hematopoietic cells. Functional domain analyses revealed that in addition to CF, NF of Gata1 is required for ectopic GFP expression. The requirement for NF suggests that a protein-protein interaction between Gata1 and Fog1 may be important for gata1 gene autoregulation.
To understand the mechanism underlying the autoregulation of gata1, we analyzed the effects of various mutations in the Gata1 protein on ectopic GFP expression driven by the gata1 HRD in zebra fish embryos. We found that mutations of six lysine residues in the Gata1 protein (referred to as Gata1KA6) inhibited self-association of Gata1 markedly and decreased the inducible expression of GFP. Similarly, the Gata1KA6 mutant could not rescue the expression of Gata1 downstream target genes in vlad tepes (vltm651), a germ line gata1 mutant fish (23, 54). These results thus provide the first in vivo evidence that the self-association of Gata1 critically contributes to the autoregulation of the gata1 gene and the maintenance of target gene expression.
MATERIALS AND METHODS
Isolation of zebra fish Fog1 cDNA.
Total RNA was prepared from fish embryos at 24 h postfertilization (hpf) by using RNAzol (Tel-Test). A partial cDNA fragment encoding zebra fish Fog1 was prepared from this RNA sample by reverse transcription-PCR (RT-PCR) with Superscript reverse transcriptase (Roche). The primers for RT-PCR were designed on the basis of sequence data (contig identification number NA26031.1) in the zebra fish genomic DNA database (http://www.ensembl.org/Danio_rerio/) and have the sequences 5′-GGCCATGGCCATGTCTGAGATGGTGCACAG and 5′-CGTAAGAAGATGTATGAGATCTAAACTAGTCC. Expression of fog1 was analyzed by whole-mount in situ hybridization as described previously (20).
Plasmid construction.
The plasmid pCS2fog1f5-6 was constructed by subcloning partial Fog1 cDNA into pCS2+. For construction of pCS2zGATA1V239 M, a Val-to-Met mutation was introduced into pCS2zGATA1 at valine 239 of Gata1 by PCR. PCR was used to introduce Lys-to-Ala mutations (KA6 mutation) into the basic stretch motifs of zebra fish Gata1; the N-terminal basic stretch motif (LIRPKKRLIV; amino acids 275 to 284) and the C-terminal motif (SNRNKKGKKNAASE; amino acids 344 to 357) were mutated to LIRPAARLIV and SNRNAAGAANAASE, respectively. To construct pCS2zGATA1KA6, the full-length Gata1 cDNA containing the KA6 mutation was inserted into pCS2+. Constructs pCS2HAzGATA1KA6, pCS2FLzGATA1, and pCS2FLzGATA1KA6 were generated by inserting cDNA fragments for a hemagglutinin (HA) (MEYPYDVPDYAA) or FLAG (MDYKDDDDKA) peptide tag just upstream of the first ATG sites of pCS2zGATA1 (21) and pCS2zGATA1KA6, respectively. For construction of p8.1kG1-HAzGATA1 and p8.1kG1-HAzGATA1KA6, a DNA fragment of enhanced GFP (EGFP) in p8.1kG1-eGFP was replaced with that of HAzGATA1 or HAzGATA1KA6, respectively. pMALfzGATA1, pMALfzGATA1KA6, pGEXfzGATA1, and pGEXfzGATA1KA6 were constructed by inserting PCR fragments corresponding to amino acid residues 212 to 368 of wild-type Gata1 or Gata1KA6 between the EcoRI and XbaI sites of pMAL-c2 (New England Biolabs) and the EcoRI and XhoI sites of pGEX-5X-1 (Amersham Biosciences). For construction of p8.1kG1m5′G-eGFP and p8.1kG1m3′G-eGFP, either of the two GATA sites in the double GATA motif (AGATAGCTTCTTATCA) in p8.1kG1-eGFP (21) was mutated by PCR, giving AGCCAGCTTCTTATCA and AGATAGCTTCTTCCCA, respectively. All constructs were verified by DNA sequencing. Other plasmids used in this study (pCS2zGATA1, pCS2HAzGATA1, p8.1kG1-eGFP, p8.1kG1mG-eGFP, pCS2HAzGATA1dNF, and pCS2HA zGATA1dCF) were described previously (21).
Preparation of recombinant proteins.
Full-length wild-type Gata1 or Gata1KA6 protein and a Fog1 protein comprising fingers 5 and 6 were generated by in vitro translation with rabbit reticulocyte lysate (Promega). Maltose binding protein (MBP) and glutathione S-transferase (GST) fusion proteins containing zinc finger domains of wild-type Gata1 or Gata1KA6 were expressed in Escherichia coli strain BL21 codon plus RIL (Stratagene). Briefly, cells grown in Luria broth medium containing 50 μg of ampicillin per ml and 50 nM zinc acetate were treated with isopropyl-1-thio-β-d-galactopyranoside for 1 h at 37°C. Harvested cells were lysed in phosphate-buffered saline (PBS) by sonication, and the proteins were purified by separating lysates by affinity chromatography over an amylose resin (New England Biolabs) or glutathione-Sepharose beads (Amersham Biosciences). FLAG-tagged human PCAF was expressed in Sf9 cells by using a baculovirus expression system and purified as previously described (28). His6-tagged human p300 protein was purified from Sf9 cells infected with a recombinant baculovirus, which was kindly provided by T. Ito and W. L. Kraus (15, 22).
Zebra fish strains.
Zebra fish embryos were obtained by natural mating (55) and staged accordingly (18). Germ line transgenic fish were identified under a fluorescence microscope by their expression of GFP. All experiments were carried out with wild-type strain AB or vltm651 (54).
Microinjection of zebra fish embryos.
p8.1kG1m5′G-eGFP, p8.1kG1m3′G-eGFP, p8.1kG1-HAzGATA1, and p8.1kG1-HAzGATA1KA6 were linearized by digesting the vector backbone with KpnI. Digested DNA was resuspended in water and injected into the blastomere of early one-cell stage embryos. For RNA injection, synthetic capped RNA was made with the SP6 mMESSAGE mMACHINE in vitro transcription kit (Ambion), using linearized DNAs of the pCS2 derivatives described above. RNA was injected into the yolk at the one-cell stage for expression in whole bodies.
Rescue analysis.
Digested DNA was injected into early one-cell stage embryos from heterozygote intercrosses of vltm651. Embryos injected with the transgenic Gata1 constructs were fixed at 22 through 26 hpf, and the expression of Gata1 downstream genes was analyzed by in situ hybridization (20) with klfd (35) and urod (53) cDNA probes. Embryos which had more than 10 stained cells in the intermediate cell mass (ICM) were scored as rescued. cDNAs for zebra fish klfd and urod were obtained by PCR. To identify homozygous vltm651 embryos, genomic DNA was extracted from each embryo after in situ hybridization by the method described previously (55). The primers used for genotyping were 5′-AAGGATGGCATACAAACTC as a wild-type sense primer, 5′-AAGGATGGCATACAAACTT as a mutant sense primer, and 5′-AAAAATAGGACGGGATGCATTTATTGAGGG as an antisense primer for both the wild-type and mutant fish.
FACS analysis.
Embryos injected with Gata1 mRNA or uninjected embryos were homogenized in PBS at 16 hpf, and cells were collected by centrifugation at 600 × g for 5 min. After digestion with 25 mM trypsin-0.1 mM EDTA for 15 min at 32°C, the cells were washed twice with PBS, passed through a 40-μm-pore-size nylon mesh filter, and analyzed with a FACSCalibur (Becton Dickinson). Fluorescence activating cell sorting (FACS) analysis was performed with the CellQuest program (Becton Dickinson).
In vitro acetylation assays.
Acetylation assays were performed, with a minor modification, as previously described (27). Briefly, proteins were incubated with 100 ng of p300 or p/CAF in the presence of 14C-labeled acetyl coenzyme A for 1 h at 30°C in an acetylation buffer (50 mM Tris-HCl [pH 8.0], 10% glycerol, 0.5 mM dithiothreitol, 1× protease inhibitor cocktail [Roche Diagnostics], 0.1 mM EDTA, 10 mM sodium butyrate). Reaction mixtures were separated by on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and analyzed with a BAS 1600 phosphorimager (Fuji Film).
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed as previously described (19). Approximately 3 fmol of 32P-labeled dG, m5′G, m3′G, and mG were used as probes. dG (5′-CAGCAATCTAGATAGCTTCTTATCACAGTTCTG), m5′G (5′-CAGCAATCTAGCCAGCTTCTTATCACAGTTCTG), m3′G (5′-CAGCAATCTAGATAGCTTCTTCCCACAGTTCTG), and mG (5′-CAGCAATCTAGCCAGCTTCTTCCCACAGTTCTG) were prepared by annealing synthetic oligonucleotides (the GATA sites and the corresponding mutated sequence are underlined). Anti-HA antibody was purchased from Roche Diagnostics.
Immunoprecipitation and pull-down assays.
For analyzing Gata1-Fog1 interaction, MBP-Gata1 fusion proteins were mixed with FLAG-tagged Fog1 protein in a binding buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM mercaptoethanol, 1× protease inhibitor cocktail [Roche Diagnostics], 10 μM ZnSO4) and incubated with amylose resin for 1 h. For analyzing Gata1 self-association, HA-tagged Gata1 proteins were mixed with FLAG-tagged proteins in a binding buffer (33 mM Tris-HCl [pH 7.5], 100 mM NaCl, 3.3 mM EDTA, 0.67 mM dithiothreitol, 0.33% NP-40, 1× protease inhibitor cocktail) and incubated with anti-HA antibody-conjugated agarose beads (Roche Diagnostics) for 4 h. The beads were collected by centrifugation at 13,000 × g for 1 min and washed three times in binding buffer. Precipitated proteins were eluted in SDS sample buffer and resolved on SDS-10% polyacrylamide gel electrophoresis, followed by immunoblot analysis with anti-MBP (Santa Cruz), anti-HA (Roche Diagnostics), and anti-FLAG (Sigma) antibodies.
Immunoblot analysis.
Nuclear proteins of RNA-injected embryos were examined by immunoblot analysis with anti-HA antibody as described previously (21).
Nucleotide sequence accession number.
The nucleotide sequence data for zebra fish fog1 have been deposited in the DDBJ/EMBL/GenBank databases with the accession number AB112073.
RESULTS
Importance of NF and CF in Gata1 autoregulation.
Our previous analysis of stable transgenic fish lines containing the gata1 HRD-eGFP construct (8.1kG1-eGFP fish) revealed that Gata1 can autoregulate its own gene expression and that both NF and CF are required for this autoregulation (21). The functional role of NF is of interest because it is not essential for DNA binding (25, 58). Given the finding that CF was sufficient for DNA binding, further analysis of NF function was undertaken by examining the effects of various point mutations on transactivation activity. In this study, we counted GFP-positive cells in embryos of 8.1kG1-eGFP fish overexpressing wild-type, NF deletion, or CF deletion Gata1 proteins by FACS analysis in order to quantitate the effect of deleting each zinc finger (Fig. 1). Approximately 30% of the cells were GFP positive in embryos injected with wild-type Gata1 (Fig. 1C), while only 6.6 and 2.8% of cells were GFP positive in the embryos overexpressing NF deletion and CF deletion Gata1, respectively. These results confirmed our previous observation that both NF and CF are critical for Gata1 autoregulation.
FIG. 1.
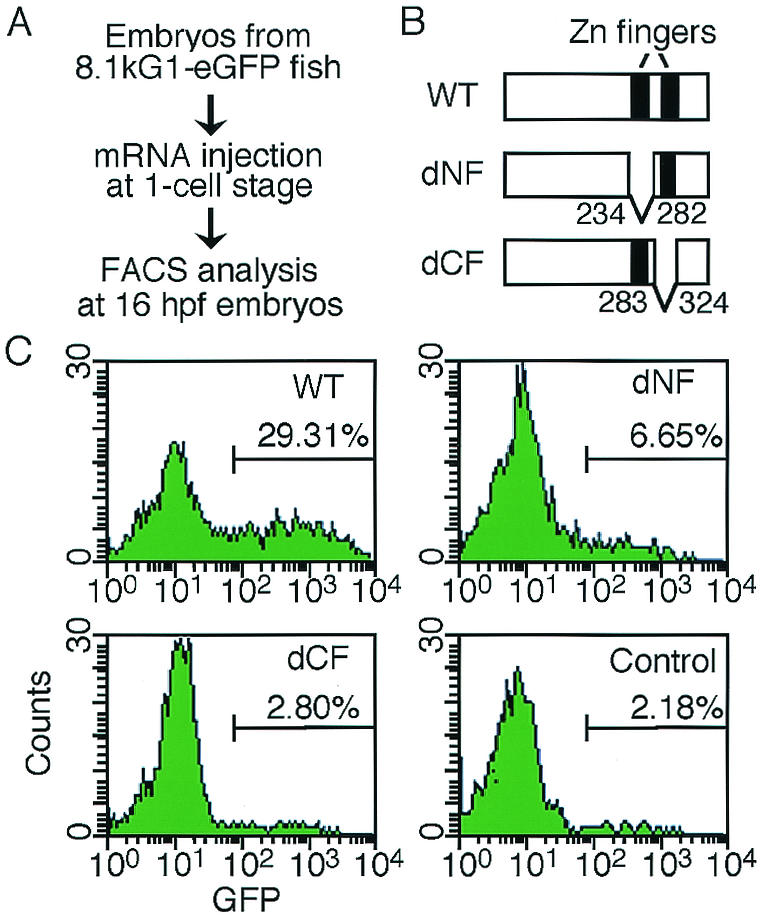
Effects of deletion of the zinc finger domains on GFP induction. (A) FACS analysis for counting GFP-positive cells in zebra fish embryos. (B) Deletion mutant proteins of Gata1 used in the analyses. WT, wild type; dNF, NF deletion mutant; dCF, CF deletion mutant. (C) FACS profiles of cells from 8.1kG1-eGFP L1 line embryos that were uninjected (control) or injected with wild-type Gata1, Gata1 dNF, or Gata1 dCF mRNA. GFP-positive cells were counted within the indicated windows. More than eight embryos were analyzed for each construct.
Gata1 cooperates with Fog1 to promote erythroid and megakaryocytic differentiation (51). Recent progress in the zebra fish genome project led us to hypothesize that Fog1 may also exist in zebra fish and play important roles in hematopoiesis through interaction with Gata1. We therefore cloned a partial cDNA which was most likely to be a zebra fish homologue of mammalian Fog1 by the RT-PCR method. Since phylogenetic analysis suggested that the isolated clone was a zebra fish homologue of Fog1 rather than Fog2 (Fig. 2A), we referred to this gene to as zebra fish fog1. The embryonic expression of fog1 was examined by whole-mount in situ hybridization and was observed in the lateral plate mesoderm at 15 hpf and in the ICM at 20 hpf (Fig. 2B), places where gata1 expression is observed and prospective hematopoietic cells develop (6). We next examined whether zebra fish Fog1 and Gata1 can interact with each other, similar to the case for mammalian proteins. MBP-fused Gata1 and FLAG-tagged truncated Fog1 comprising Gata1-binding zinc fingers 5 and 6 (8) were prepared in E. coli and by an in vitro translation system, respectively, and their interaction was tested by pull-down analysis with amylose resin, which binds MBP fusion proteins. The results clearly show that MBP-Gata1, but not MBP protein alone, binds to Fog1 (Fig. 2C, lanes 1 and 3), indicating that Gata1-Fog1 interaction is conserved in zebra fish.
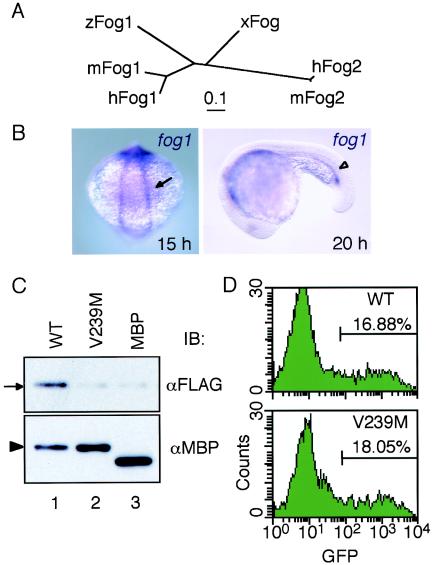
FIG. 2.
Identification of zebra fish Fog1. (A) Phylogenetic tree of Fog family proteins. Amino acid sequences in fingers 5 and 6 were analyzed. The tree was constructed by the neighbor-joining method (39) with the CLUSTAL W program (46). hFog1, human Fog1 (9); hFog2, human Fog2 (12); mFog1, mouse Fog1 (51); mFog2, mouse Fog2 (45); xFog, Xenopus Fog (5). Scale bar, genetic distance. (B) Embryonic expression pattern of fog1. Dorsal and lateral views of embryos at 15 and 20 hpf, respectively, are shown. The arrow and arrowhead indicate fog1-expressing sites in the lateral plate mesoderm and ICM, respectively. (C) Interactions between FLAG-tagged Fog1 proteins and MBP-fused proteins of wild-type Gata1 (WT) (lane 1) or Gata1V239M (lane 2) or MBP (lane 3) were examined by pull-down analysis with amylose resin and by immunoblot (IB) analysis with anti-FLAG (αFLAG) (upper panel) or anti-MBP (lower panel) antibodies. The arrow and arrowhead indicate migration positions of the Fog1 and MBP-fused Gata1 proteins, respectively. (D) FACS profiles of cells from 8.1kG1-eGFP L1 line embryos that were injected with wild-type Gata1 or Gata1V239M mRNA. GFP-positive cells were counted within the indicated windows. More than eight embryos were analyzed for each construct.
By a reverse yeast two-hybrid screen, three amino acid residues in the NF region of mouse Gata1 (203Glu, 205Val, and 222His) (Fig. 3) were previously found to contribute specifically to Fog1 binding (3). 205Val and 208Gly mutations of Gata1 in human (Fig. 3) cause X-linked thrombocytopenia and dyserythropoietic anemia (29, 32). These amino acid residues for Fog1 binding are highly conserved among vertebrate Gata1 proteins (Fig. 3). It is possible the amino acid residue corresponding to 205Val in the zebra fish Gata1 protein is critical for the fish Gata1-Fog1 interaction. To assess the contribution of Fog1 to Gata1 autoregulation, we generated a zebra fish Gata1 with a methionine replacement at valine 239 (Gata1V239M) (Fig. 3), which corresponds to 205Val of the mouse protein, and analyzed its activity on GFP induction in 8.1kG1-eGFP embryos. We chose this Val residue since its mutations caused severe defects in Fog1-dependent Gata1 function in both human and mouse.
FIG. 3.
Alignment of the zinc finger domains of the vertebrate Gata1 proteins and their adjacent sequences. z, zebra fish; m, mouse; c, chicken; h, human; x, Xenopus. Black-highlighted Cs, cysteine residues coordinated to zinc atoms. Red and green letters, acetylation sites in mouse and chicken Gata1 (2, 13) and critical residues for Fog1 interaction in mouse and human Gata1 (3, 29, 32), respectively. Open and closed circles, conserved residues among vertebrates for acetylation and interaction with Fog1, respectively. Pink-highlighted Ks and blue-highlighted V, mutation sites in Gata1KA6 and Gata1V239M, respectively. Gray-highlighted and underlined letters, mutation sites for the self-association (24) and the acetylation mutants (13), respectively. Note that these sites overlap.
We first examined the effect of the V239M mutation on binding to zebra fish Fog1 by pull-down analysis. As shown in Fig. 2C, interaction between MBP-fused Gata1 and Fog1 was significantly decreased when the V239M mutation was introduced (lane 2), suggesting that the valine residue is critical for Fog1 binding not only in mammals but also in zebra fish. We next tested the effect of this V239 M mutation on GFP induction in 8.1kG1-eGFP fish by FACS analysis. Surprisingly, overexpression of Gata1V239M induced the expression of GFP as strongly as that of wild-type Gata1 (Fig. 2D). This result suggests that an interaction between Fog1 and Gata1 may not be essential for the autoregulation of gata1 in zebra fish embryos.
Lysine residues adjacent to the zinc finger domains are important for Gata1 autoregulation.
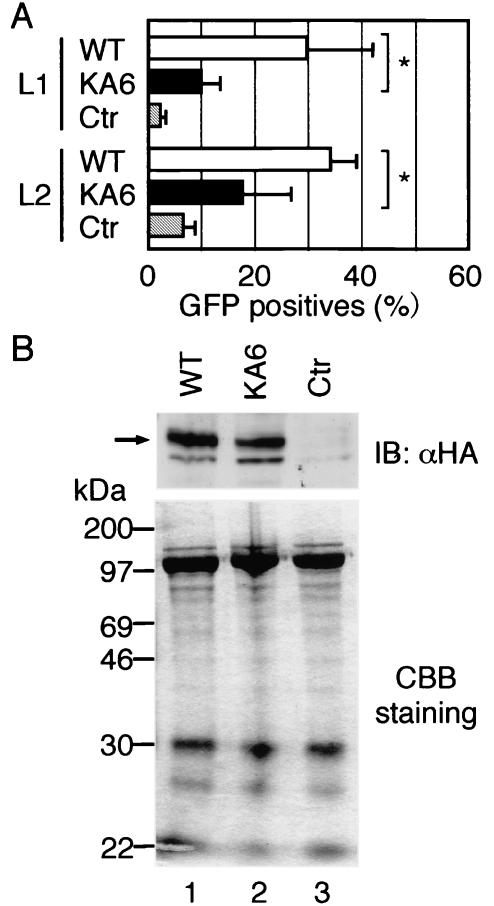
We next addressed whether acetylation of the Gata1 protein plays a role in autoregulation, since it has been demonstrated that lysine residues in the adjacent region of NF were strongly acetylated in vitro and this may be an important step for Gata1 activation (2, 13). The amino acid residues markedly acetylated in the chicken and mouse Gata1 proteins are indicated in Fig. 3 (2, 13). It is noteworthy that these lysine residues are also conserved in the zebra fish protein (Fig. 3). It has been shown that in vitro acetylation of mouse Gata1 with CREB binding protein is greatly reduced by replacement of six lysine residues with alanines (Fig. 3) (13). In order to elucidate whether acetylation of these lysine residues is important for Gata1 autoregulation, we introduced alanine substitutions at the corresponding lysine residues in zebra fish Gata1 (Gata1KA6) (Fig. 3) and examined the effect of these substitutions on GFP-inducing activity in 8.1kG1-eGFP fish by FACS analysis. Overexpression of Gata1KA6 induced GFP-positive cells threefold less than the wild-type protein did (Fig. 4A, L1). To confirm the reproducibility of this observation, we tested the effects of the KA6 mutation in a different transgenic line (L2) and obtained similar results.
FIG. 4.
Effects of the KA6 mutation on Gata1 functions. (A) Transactivation in 8.1kG1-eGFP fish. Ratios of GFP-positive cells in embryos injected with mRNA for wild-type Gata1 (WT) or Gata1KA6 or in uninjected embryos (Ctr) were determined by FACS analysis from at least three independent experiments (means ± standard deviations). More than eight embryos were analyzed in each experiment. *, P < 0.05. (B) Immunoblot (IB) analysis of nuclear proteins of uninjected embryos or embryos injected with mRNA for wild-type Gata1 or Gata1KA6. Upper panel, immunoblot analysis with anti-HA (αHA) antibodies. Lower panel, Coomassie brilliant blue (CBB) staining. Migration positions of the Gata1 proteins are indicated by an arrow.
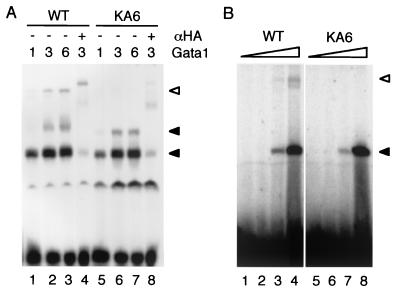
To exclude the possibility that the reduction in GFP-inducing activity was due to instability of the KA6 mutant protein, we examined expression levels of the recombinant proteins by immunoblot analysis with an antibody against HA (Fig. 4B). Immunoblot analysis of nuclear proteins from whole embryos showed that expression levels of wild-type Gata1 and Gata1KA6 proteins were similar, implying that the KA6 mutation did not affect protein stability. We also compared the abilities of wild-type Gata1 and Gata1KA6 for DNA binding to the double GATA motif by using in vitro-translated proteins (Fig. 5A) or E. coli-expressed proteins (Fig. 5B). The results indicate that Gata1KA6, as well as wild-type Gata1, binds to the double GATA motif, suggesting that the KA6 mutation did not affect the DNA binding activity of Gata1.
FIG. 5.
DNA binding activity of Gata1KA6. (A) EMSA for full-length wild-type Gata1 (WT) (lanes 1 to 4) and Gata1KA6 (lanes 5 to 8) prepared by in vitro translation with oligonucleotide dG (see Fig. 9) as a probe. Retarded complexes containing Gata1 proteins are indicated by closed arrowheads, which represent one or two Gata1 monomers bound to two GATA sites in the dG probe. The open arrowhead indicates the slow-migrating complex observed only for wild-type Gata1. Indicated volume (microliters) of rabbit reticulocyte lysate reaction product was used for each assay. The protein concentrations of wild-type Gata1 and Gata1KA6 were similar in each reaction. Anti-HA (αHA) antibody was used to detect the presence of HA-Gata1 fusion proteins (lanes 4 and 8). (B) EMSA for GST fusion proteins, which were purified from E. coli, containing zinc finger domains of wild-type Gata1 (lanes 1 to 4) and Gata1KA6 (lanes 5 to 8), using oligonucleotide dG as a probe; 0.06 (lanes 1 and 5), 0.6 (lanes 2 and 6), 6 (lanes 3 and 7), or 28 (lanes 4 and 8) pmol of each protein was used. The upper band indicated by a closed arrow in panel A is invisible in panel B, probably because of effects of spatial interference by GST.
The KA6 mutation reduces activation of Gata1 downstream genes.
The vltm651 mutant is one of the bloodless zebra fish mutants isolated through a large-scale chemical mutagenesis screen (54). Recently, it was demonstrated that vltm651 has a point mutation in the gata1 gene, resulting in a truncated Gata1 protein that cannot bind DNA (23). In vltm651 homozygotes, the expression of hematopoietic genes is impaired (23), as is the case for mice with a disruption in gata1 (10, 43). Since we have shown that the KA6 mutation reduces the ability of Gata1 to transactivate a transgenic GFP reporter gene in zebra fish embryos, we were interested in testing whether the KA6 mutation affects the endogenous expression of gata1 target genes. To answer this question, we investigated the ability of both wild-type and KA6 mutant Gata1 proteins to rescue the expression of gata1 downstream genes in vltm651 homozygous mutant embryos (Fig. 6).
FIG. 6.
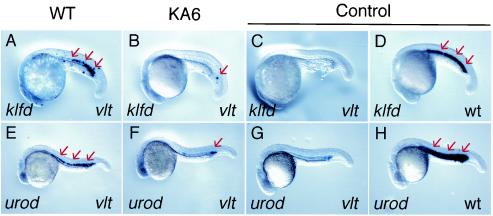
Activation of downstream genes by Gata1 overexpression. Expression of klfd (A to D) and urod (E to H) in vltm651 (vlt) and wild-type (wt) embryos was analyzed by whole-mount in situ hybridization. Plasmid constructs with gata1 HRD driving wild-type Gata1 (WT) (A and E) and Gata1KA6 (B and F) were injected into vltm651 embryos. Injected and uninjected embryos were fixed at 22 to 26 hpf, and the expression (arrows) of klfd and urod was analyzed.
We analyzed the expression of zebra fish hematopoietic genes klfd (35) and urod (53) in vltm651 embryos. The expression of both klfd and urod in prospective hematopoietic cells (Fig. 6D and H) was greatly reduced in homozygous vltm651 mutants (Fig. 6C and G). The reduction was rescued when wild-type Gata1 was expressed in these cells by transgenic expression with gata1 HRD (Fig. 6A and E). The proportions of rescued embryos among tested embryos were 36.8% (n = 19) for klfd and 61.5% (n = 13) for urod. On the other hand, rescue of klfd or urod expression was dramatically reduced when Gata1KA6 was expressed (Fig. 6B and F) (0% [n = 20] for klfd and 25% [n = 12] for urod), suggesting that the KA6 mutation impairs the ability of Gata1 to rescue the phenotype of vltm651 embryos. The results demonstrate that the KA6 mutation not only reduces the ability of Gata1 to induce ectopic expression of a GFP reporter gene in stable transgenic fish but also impairs transactivation of endogenous downstream genes.
KA6 is not a complete mutation for acetylation.
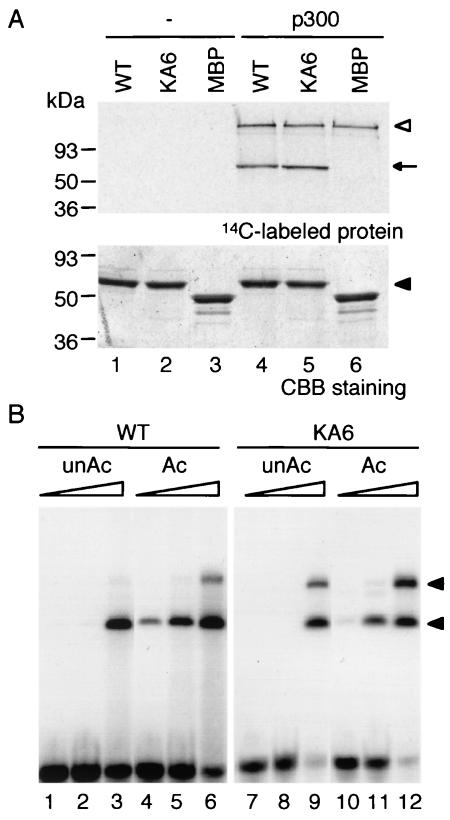
Because it is likely that acetylation of lysine residues adjacent to NF and CF is important for the activity, we examined whether zebra fish Gata1 can be acetylated in vitro by using a recombinant human p300 protein as an enzyme. We prepared zebra fish wild-type Gata1 and Gata1KA6 proteins containing both NF and CF (amino acids 212 to 368) as MBP fusion proteins by using the E. coli system. As shown in Fig. 7A, the wild-type Gata1 protein was strongly labeled by 14C-labeled acetyl coenzyme A, suggesting that zebra fish Gata1 can be acetylated by p300 as well as other vertebrate Gata1 proteins (lane 4). This reaction seemed to be specific, since recombinant p/CAF did not acetylate zebra fish Gata1 (data not shown), as is the case for mouse Gata1 (13). Surprisingly, Gata1KA6 was also acetylated by p300 (Fig. 7A, lane 5). It was previously reported that acetylation of mouse Gata1 was almost completely eliminated by introduction of mutations in the six lysine residues (13). One plausible explanation of this difference is that the lysine residues in the adjacent region of CF are not completely conserved between the mouse and zebra fish proteins (Fig. 3), and 286Lys or 342Lys (Fig. 3) or other lysine residues may be the major acetylation sites in zebra fish Gata1.
FIG. 7.
In vitro acetylation assay of Gata1 with human p300. (A) Wild-type Gata1 (WT) (lanes 1 and 4), Gata1KA6 (lanes 2 and 5), and MBP (lanes 3 and 6) were mixed with p300 (lanes 4 to 6) or left without p300 (lanes 1 to 3) and analyzed by SDS-polyacrylamide gel electrophoresis. Upper panel, 14C-labeled proteins were visualized by autoradiography. Lower panel, the relative amounts of tested fusion proteins were shown by Coomassie brilliant blue (CBB) staining. Migration positions of the Gata1 proteins are indicated by an arrow and a closed arrowhead. The high-molecular-mass bands (open arrowhead) in lanes 4 to 6 represent autoacetylated p300. (B) EMSA for acetylated (Ac) (lanes 4 to 6 and 10 to 12) and unacetylated (unAc) (lanes 1 to 3 and 7 to 9) proteins of wild-type Gata1 (lanes 1 to 6) and Gata1KA6 (lanes 7 to 12), using oligonucleotide dG as a probe; 0.3 (lanes 1, 4, 7, and 10), 0.7 (lanes 2, 5, 8, and 11), or 1.3 (lanes 3, 6, 9, and 12) pmol of each protein was used for EMSA. Retarded complexes containing Gata1 proteins are indicated by arrowheads.
Boyes et al. reported that acetylation of chicken Gata1 by p300 activates its DNA binding activity (2). To elucidate whether acetylation by p300 activates DNA binding activity of the zebra fish Gata1 protein, we performed EMSA with acetylated or unacetylated forms of Gata1 and Gata1KA6. As shown in Fig. 7B, acetylation caused a mild increase in DNA binding activity in both cases. These results suggest that acetylation enhances DNA binding by zebra fish Gata1 and that acetylation of Gata1KA6 contributes to the Gata1 function in this context. Since enhancement of DNA binding is the only function reported for Gata1 acetylation, we considered that the acetylated Gata1KA6 was functional. These results indicate that Gata1KA6 is not a complete deacetylation mutant and suggest that a defect in acetylation is not the fundamental reason for the reduction of transcriptional activity of the KA6 mutant.
Reduction of self-association by the KA6 mutation.
During the EMSA of the Gata1 proteins, we found a slowly migrating complex that was observed for wild-type Gata1 but significantly reduced for Gata1KA6 regardless of whether we used in vitro-translated proteins or E. coli-expressed proteins (Fig. 5). Supershift analysis of this complex by anti-HA antibody revealed that Gata1 is included in the complex (Fig. 5A, lane 4). We therefore hypothesized that the complex consisted of multimeric self-associated Gata1 proteins and that the KA6 mutation weakens this self-association activity. Interestingly, mutation study of mouse Gata1 showed that stretches of basic amino acid residues in the adjacent region of both NF (245Lys-246Lys-247Arg) and CF (315Lys-316Lys-317Arg) are important for self-association of Gata1 (24) (Fig. 3). Intriguingly, three out of six amino acid residues in the zebra fish Gata1 corresponding to these basic stretches were mutated in Gata1KA6 (Fig. 3).
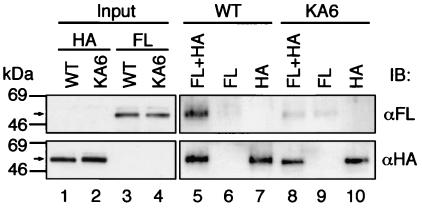
To elucidate whether wild-type zebra fish Gata1 and Gata1KA6 can self-associate, we prepared FLAG-tagged and HA-tagged Gata1 proteins by in vitro translation and tested their interaction by immunoprecipitation assay with anti-HA antibody. Immunoprecipitates were analyzed by immunoblot assay with both anti-FLAG and anti-HA antibodies. Figure 8 shows that FLAG-tagged Gata1 was efficiently immunoprecipitated when it was mixed with HA-tagged Gata1 (lane 5), indicating that zebra fish Gata1 can physically self-associate. This self-association was dramatically reduced when the KA6 mutation was introduced in both FLAG- and HA-tagged proteins (Fig. 8, lane 8). To confirm this observation, we prepared MBP and GST fusion proteins of Gata1 and Gata1KA6 by using the E. coli system and tested their self-interaction by pull-down analysis with amylose resin. Similar to the results with FLAG- and HA-tagged proteins, GST-Gata1 was recovered efficiently whereas GST-Gata1KA6 was not (data not shown). These results thus indicate that the KA6 mutation weakens the self-association activity of Gata1 and suggest that the KA6 mutation reduces the Gata1 activity in zebra fish embryos through impairing the self-association process.
FIG. 8.
Self-association of zebra fish Gata1 proteins. HA-tagged (lanes 1 and 2) and FLAG (FL)-tagged (lanes 3 and 4) proteins of wild-type Gata1 (WT) (lanes 1 and 3) and Gata1KA6 (lanes 2 and 4) were examined by immunoblot (IB) analysis with anti-FLAG (αFL) (upper panel) or anti-HA (lower panel) antibodies. Arrows indicate migration positions of the Gata1 proteins. Mixtures of FLAG-tagged and HA-tagged proteins (lanes 5 and 8), FLAG-tagged protein alone (lanes 6 and 9), HA-tagged protein alone (lanes 7 and 10) for wild-type Gata1 (lanes 5 to 7) or Gata1KA6 (lanes 8 to 10) were immunoprecipitated with anti-HA-conjugated agarose beads. Precipitated proteins were analyzed by immunoblotting.
Both GATA sites in the double GATA motif are necessary for DNA binding of self-associated Gata1 complex.
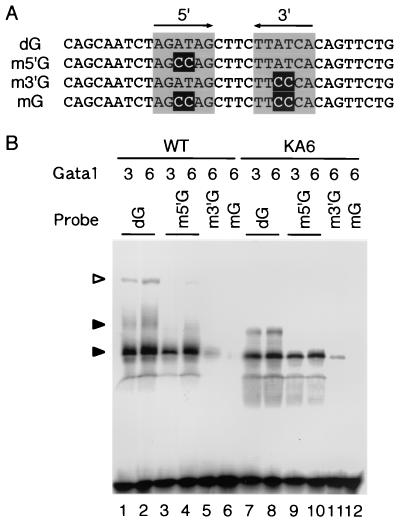
The double GATA motif in the zebra fish gata1 HRD, which is located approximately 6.4 kbp upstream from the translation initiation site, is important for the autoregulation of the gata1 gene (21). We showed in the present study that Gata1 self-association may be critical for this autoregulation. To elucidate whether the self-associated Gata1 complex requires the double GATA motif as a binding sequence, we prepared oligonucleotides containing the double GATA motif (dG), with point mutations introduced at the 5′-GATA site (m5′G), 3′-GATA site (m3′G), or both (mG) (Fig. 9A). The binding of Gata1 to these oligonucleotides was analyzed by EMSA. The intensities of retarded bands that were expected to contain Gata1 monomer (Fig. 9B) were strong when we used dG or m5′G as the probe (lanes 1 to 4) but were much fainter with m3′G (lane 5). Similar results were obtained when Gata1KA6 was used (lanes 7 to 12), suggesting that the 3′-GATA site is necessary and sufficient for the binding of Gata1 monomer. These results are consistent with results for mouse Gata1, which binds primarily to the 5′-GATA site in the double GATA motif of the mouse Gata1 gene (50).
FIG. 9.
Effect of mutations in the double GATA motif on Gata1 binding. (A) Oligonucleotides used as probes for EMSA. Arrows and gray boxes indicate the 5′ and 3′ GATA sites. Black-highlighted Cs indicate mutation sites. (B) EMSA with dG (lanes 1, 2, 7, and 8), m5′G (lanes 3, 4, 9, and 10), m3′G (lanes 5 and 11), or mG (lanes 6 and 12) as a probe and HA-tagged wild-type Gata1 (lanes 1 to 6) or HA-tagged Gata1KA6 (lanes 7 to 12) as the protein. Closed arrowheads, retarded complexes containing Gata1 monomer; open arrowheads, retarded complex containing multimeric Gata1. The indicated volume (microliters) of rabbit reticulocyte lysate reaction product was used for each assay.
On the other hand, the slowly migrating band that appears to contain a multimeric Gata1 complex (Fig. 9B) was markedly decreased by mutations at either 5′- or 3′-GATA sites in the double GATA motif (lanes 3 to 5). These results indicated that both GATA sites in the double GATA motif are critical for binding of the self-associated Gata1 complex and that this binding mode is different from that of Gata1 monomer.
Both GATA sites in the double GATA motif are necessary for Gata1 transactivation.
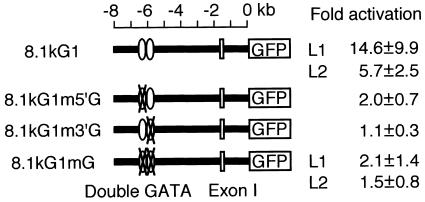
The fact that both GATA sites in the double GATA motif are important for binding of the self-associated Gata1 complex prompted us to hypothesize that these two sites should also contribute significantly to the transactivation by Gata1. To test this possibility, we introduced the mutations shown in Fig. 9A into the reporter construct p8.1kG1-eGFP and analyzed how these mutations affect the GFP-inducing activity of Gata1. Since it was not technically feasible to evaluate the transactivation activity quantitatively in the transient DNA injection system, we established stable transgenic zebra fish lines containing m5′G, m3′G, or both and named them 8.1kG1m5′G-eGFP, 8.1kG1m3′G-eGFP, and 8.1kG1mG-eGFP fish, respectively.
In order to examine whether Gata1 transactivates these mutant reporter genes, Gata1 was overexpressed in embryos of each line by mRNA injection and the GFP-inducing activity was quantitated by FACS analysis, comparing the numbers of GFP-expressing cells in Gata1 mRNA-injected and uninjected embryos (Fig. 10). In 8.1kG1-eGFP fish, which contain an intact double GATA motif, overexpression of Gata1 resulted in 14.6- and 5.7-fold accumulation of GFP-positive cells compared to that in uninjected embryos in two independent lines (L1 and L2, respectively), whereas the accumulation was less than 2-fold higher in 8.1kG1m5′G-eGFP, 8.1kG1m3′G-eGFP, and 8.1kG1mG-eGFP embryos. These results indicate that both GATA sites in the double GATA motif are indeed necessary for the transactivation activity of zebra fish Gata1. Although it is still possible to assume that the double GATA motif has functions in addition to its interaction with self-associated Gata1 complex, our results strongly argue that the self-association of Gata1 is critical for the autoregulation.
FIG. 10.
Effect of mutations in the double GATA motif on Gata1 transactivation. Gata1 mRNA was injected into embryos of 8.1kG1-eGFP, 8.1kG1m5′G-eGFP, 8.1kG1m3′G-eGFP, or 8.1kG1mG-eGFP fish, and the numbers of GFP-positive cells in 16-hpf embryos were counted by FACS analysis. Values indicate ratios of GFP-positive cells in embryos injected with Gata1 mRNA to those in uninjected embryos from at least three independent experiments (means ± standard deviations). More than eight embryos were analyzed in each experiment.
DISCUSSION
In this study, we demonstrated the in vivo functional significance of zebra fish Gata1 self-association. Although the presence of Gata1 self-association has been reported previously, the physiological roles of the Gata1-Gata1 interaction remained to be elucidated. We showed here that mutations of Gata1 that decreased self-association greatly reduced the transactivation activity of Gata1. The mutations decreased expression of both the gata1 HRD-driven GFP reporter gene and Gata1-target genes. These results are consistent with the previous finding that mouse Gata1 self-associates in vitro, which can stimulate transcription of a reporter gene containing GATA sites in the regulatory regions (4). Chicken Gata1 has also been reported to interact homotypically and can superactivate Gata1-mediated transcription (59). Our present study further revealed that the self-association of Gata1 is an important mechanism underlying autoregulation of gata1 gene expression.
Some Gata1 downstream genes, such as klfd and urod, can be directly regulated by self-associated Gata1. We could find double GATA motif-related sequences in the upstream regions of klfd and urod when we searched the zebra fish genome database (http://www.ensembl.org/Danio_rerio/) (AGATAAAATAATCA [klfd, −2910 from the 5′ end of the published cDNA sequence], GGATAAAATATCC [klfd, −300], and TGATAAATCAAGTTATCA [urod, −5600]). Therefore, it seems possible that self-associated Gata1 transactivates these genes. However, it is also possible that the KA6 mutation affects activation of these genes indirectly through the lack of amplification of exogenous Gata1 proteins, whose amount depends on the activity of the transgenic gata1 promoter. Regulation of Gata1 downstream genes in the context of Gata1 self-association remains to be clarified.
How does the self-association enhance transcriptional activity of Gata1? One possible explanation is that it can elevate the local concentration of histone acetylases such as CREB binding protein/p300, which are known to interact with Gata1 (1), around the regulatory region of Gata1 target genes, making RNA polymerase or other basal transcription factors easier to access to the promoter (16). Recruitment of chromatin-remodeling factors such as SWI/SNF, which also forms a complex with Gata1 and enhances the transcriptional activity of the chromatin-assembled promoter, may also become accelerated (17).
An alternative explanation is that a multimeric Gata1 complex may keep the regulatory regions more effectively unwrapped from the nucleosome structure through multiple interactions between low-affinity GATA sites and each constituent Gata1 protein. In the present study, we showed that double GATA motifs are critical for DNA binding of the self-associated Gata1 complex and for the transactivation ability of Gata1. Since more than 20 GATA sites exist in the zebra fish gata1 HRD in addition to the two GATA sites in the double GATA motif, there is a possibility that a multimeric Gata1 complex targets the double GATA motif first and successively interacts with these other GATA sites.
The following results further support the hypothesis that the self-association of Gata1 is crucial for the transcriptional activity. One study showed that GFP expression driven by a 979-bp artificial minigene composed of the core region of the Gata1 hematopoietic enhancer, the double GATA motif, GATA-GACT repeat regions in the first intron, and the two CACCC boxes recapitulated the Gata1 gene expression in primitive and definitive erythroid cells in the mouse (36). Each of the first three regions contains multiple GATA sites. GFP expression in mouse embryos was dramatically reduced when one of these regions was deleted, suggesting that multiple GATA sites are required for the expression of mouse Gata1. Mackay et al. have demonstrated that the effect on Gata1 transactivation activity of mutations impairing its self-association was much greater from a promoter containing six GATA sites than from one containing a single GATA site (24). We tried to identify a GATA site in the zebra fish gata1 HRD that is essential for the activation of the gene but outside the double GATA motif by exploiting various deletion and mutation constructs. All tested constructs, however, showed activity comparable to that of the wild-type gata1 HRD (data not shown), suggesting that those GATA sites are inactive or functionally redundant.
An intriguing finding is that while amino acid residues needed for self-association overlap heavily with those for acetylation in mouse Gata1 (Fig. 3), amino acid residues for self-association differ substantially from those for acetylation in zebra fish Gata1. We could not find a difference in the level of acetylation between wild-type Gata1 and Gata1KA6, although this does not necessarily exclude the possibility that acetylation of these six lysine residues, which should be abolished in Gata1KA6, plays important roles in Gata1 activation. In this regard, it is important to note that acetylation of lysine residues results in neutralization of their positive charge. This is particularly important since the lysine residues adjacent to the zinc finger domains appear to be critical for Gata1 self-association, and therefore it is possible that acetylation and neutralization of these residues weaken the Gata1-Gata1 interaction. To test this possibility, we performed a pull-down analysis using MBP and GST fusion proteins of wild-type Gata1 under acetylated or unacetylated conditions. No difference, however, was observed in the pull-down results under the acetylated and unacetylated conditions (data not shown). Thus, the relationship between self-association and acetylation of Gata1 remains to be elucidated.
Basic amino acid residues in the zebra fish Gata1 protein, which are important for self-association, are conserved among vertebrate hematopoietic Gata factors Gata1, Gata2, and Gata3. The conservation implies that Gata2 and Gata3 are also able to self-associate homotypically or heterotypically. Indeed, mouse Gata2 and Gata3 have been demonstrated to interact with Gata1 in vitro (4), and chicken Gata2, but not Gata5, was shown to be able to superactivate Gata1-mediated transcription in avian fibroblasts (59). Interestingly, most of these basic amino acid residues are not conserved among cardiac Gata factors (i.e., Gata4, Gata5, and Gata6), suggesting that one of the critical differences between hematopoietic and cardiac Gata factors is the ability to self-associate. Since we have shown that transgenic expression of Gata2 or Gata3 proteins could rescue the embryonic lethal phenotype of Gata1 knockdown mutant mice (44), it is important to clarify whether cardiac Gata factors possess similar rescue activity.
Erythropoiesis is a highly coordinated process that is regulated by a finely tuned combination of transcription factors in a stage-specific and context-dependent manner. These transcription factors, including Gata1, may play multiple roles at different developmental stages with different modes of action. In this paper, we demonstrate that self-association of Gata1 participates in the regulation of erythroid gene expression and in autoregulation of gata1. Our findings shed new light on the complex nature of transcriptional regulation of erythropoiesis.
Acknowledgments
We thank Toshiko Arai, Ayako Hayashi, and Toyoko Kinoshita for help with fish maintenance and Norio Suzuki, Yaeko Takagi, Hitoshi Osanai, and Takafumi Suzuki for help and discussion. We also thank Igor B. Dawid, Kazuhiko Igarashi, Kinuko Ohneda, and Ritsuko Shimizu for critical reading of the manuscript and Kyosuke Nagata and Yasutake Katoh for the gift of E. coli strains.
This work was supported by grants-in-aid from the NOVARTIS Foundation (Japan) for the Promotion of Science, the Japan Society for Promotion of Sciences (JSPS-RFTF), the Japan Science and Technology Corporation (ERATO), the Naito Foundation, and the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 3.Crispino, J. D., M. B. Lodish, J. P. MacKay, and S. H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219-228. [DOI] [PubMed] [Google Scholar]
- 4.Crossley, M., M. Merika, and S. H. Orkin. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deconinck, A. E., P. E. Mead, S. G. Tevosian, J. D. Crispino, S. G. Katz, L. I. Zon, and S. H. Orkin. 2000. FOG acts as a repressor of red blood cell development in Xenopus. Development 127:2031-2040. [DOI] [PubMed] [Google Scholar]
- 6.Detrich, H. W., III, M. W. Kieran, F. Y. Chan, L. M. Barone, K. Yee, J. A. Rundstadler, S. Pratt, D. Ransom, and L. I. Zon. 1995. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA 92:10713-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, T., and G. Felsenfeld. 1989. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell 58:877-885. [DOI] [PubMed] [Google Scholar]
- 8.Fox, A. H., C. Liew, M. Holmes, K. Kowalski, J. Mackay, and M. Crossley. 1999. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18:2812-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freson, K., C. Thys, C. Wittewrongel, J. Vermylen, M. F. Hoylaerts, and C. Van Geet. 2003. Molecular cloning and characterization of the GATA1 cofactor human FOG1 and assessment of its binding to GATA1 proteins carrying D218 substitutions. Hum. Genet. 112:42-49. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara, Y., C. P. Browne, K. Cunniff, S. C. Goff, and S. H. Orkin. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 93:12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannon, R., T. Evans, G. Felsenfeld, and H. Gould. 1991. Structure and promoter activity of the gene for the erythroid transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 88:3004-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes, M., J. Turner, A. Fox, O. Chisholm, M. Crossley, and B. Chong. 1999. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J. Biol. Chem. 274:23491-23498. [DOI] [PubMed] [Google Scholar]
- 13.Hung, H. L., J. Lau, A. Y. Kim, M. J. Weiss, and G. A. Blobel. 1999. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, E., T. Toki, H. Ishihara, H. Ohtani, L. Gu, M. Yokoyama, J. D. Engel, and M. Yamamoto. 1993. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature 362:466-468. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899-1907. [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, K. D., J. A. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimmel, C. B., W. W. Ballard, S. R. Kimmel, B. Ullmann, and T. F. Schilling. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253-310. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, M., and K. Kawakami. 1995. ATF-1CREB heterodimer is involved in constitutive expression of the housekeeping Na, K-ATPase α1 subunit gene. Nucleic Acids Res. 23:2848-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi, M., K. Nishikawa, T. Suzuki, and M. Yamamoto. 2001. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev. Biol. 232:315-326. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, M., K. Nishikawa, and M. Yamamoto. 2001. Hematopoietic regulatory domain of gata1 gene is positively regulated by GATA1 protein in zebrafish embryos. Development 128:2341-2350. [DOI] [PubMed] [Google Scholar]
- 22.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons, S. E., N. D. Lawson, L. Lei, P. E. Bennett, B. M. Weinstein, and P. P. Liu. 2002. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc. Natl. Acad. Sci. USA 99:5454-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay, J. P., K. Kowalski, A. H. Fox, R. Czolij, G. F. King, and M. Crossley. 1998. Involvement of the N-finger in the self-association of GATA-1. J. Biol. Chem. 273:30560-30567. [DOI] [PubMed] [Google Scholar]
- 25.Martin, D. I. K., and S. H. Orkin. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886-1898. [DOI] [PubMed] [Google Scholar]
- 26.Martin, D. I. K., L. I. Zon, G. Mutter, and S. H. Orkin. 1990. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature 344:444-447. [DOI] [PubMed] [Google Scholar]
- 27.Masumi, A., and K. Ozato. 2001. Coactivator p300 acetylates the interferon regulatory factor-2 in U937 cells following phorbol ester treatment. J. Biol. Chem. 276:20973-20980. [DOI] [PubMed] [Google Scholar]
- 28.Masumi, A., I.-M. Wang, B. Lefebvre, X.-J. Yang, Y. Nakatani, and K. Ozato. 1999. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol. Cell. Biol. 19:1810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehaffey, M. G., A. L. Newton, M. J. Gandhi, M. Crossley, and J. G. Drachman. 2001. X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood 98:2681-2688. [DOI] [PubMed] [Google Scholar]
- 30.Meng, A., H. Tang, B. Yuan, B. A. Ong, Q. Long, and S. Lin. 1999. Positive and negative cis-acting elements are required for hematopoietic expression of zebrafish GATA-1. Blood 93:500-508. [PubMed] [Google Scholar]
- 31.Motohashi, H., F. Katsuoka, J. A. Shavit, J. D. Engel, and M. Yamamoto. 2000. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell 103:865-875. [DOI] [PubMed] [Google Scholar]
- 32.Nichols, K. E., J. D. Crispino, M. Poncz, J. G. White, S. H. Orkin, J. M. Maris, and M. J. Weiss. 2000. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat. Genet. 24:266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolis, S., C. Bertini, A. Ronchi, S. Crotta, L. Lanfranco, E. Moroni, B. Giglioni, and S. Ottolenghi. 1991. An erythroid specific enhancer upstream to the gene encoding the cell-type specific transcription factor GATA-1. Nucleic Acids Res. 19:5285-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura, S., S. Takahashi, T. Kuroha, N. Suwabe, T. Nagasawa, C. Trainor, and M. Yamamoto. 2000. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol. Cell. Biol. 20:713-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oates, A. C., S. J. Pratt, B. Vail, Y. Yan, R. K. Ho, S. L. Johnson, J. H. Postlethwait, and L. I. Zon. 2001. The zebrafish klf gene family. Blood 98:1792-1801. [DOI] [PubMed] [Google Scholar]
- 36.Ohneda, K., R. Shimizu, S. Nishimura, Y. Muraosa, S. Takahashi, J. D. Engel, and M. Yamamoto. 2002. A minigene containing four discrete cis elements recapitulates GATA-1 gene expression in vivo. Genes Cells 7:1243-1254. [DOI] [PubMed] [Google Scholar]
- 37.Ohneda, K., and M. Yamamoto. 2002. Roles of hematopoietic transcription factors GATA-1 and GATA-2 in the development of red blood cell lineage. Acta Haematol. 108:237-245. [DOI] [PubMed] [Google Scholar]
- 38.Onodera, K., S. Takahashi, S. Nishimura, J. Ohta, H. Motohashi, K. Yomogida, N. Hayashi, J. D. Engel, and M. Yamamoto. 1997. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl. Acad. Sci. USA 94:4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 40.Seshasayee, D., J. N. Geiger, P. Gaines, and D. M. Wojchowski. 2000. Intron 1 elements promote erythroid-specific GATA-1 gene expression. J. Biol. Chem. 275:22969-22977. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu, R., S. Takahashi, K. Ohneda, J. D. Engel, and M. Yamamoto. 2001. In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. EMBO J. 20:5250-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivdasani, R. A., Y. Fujiwara, M. A. McDevitt, and S. H. Orkin. 1997. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 16:3965-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi, S., K. Onodera, H. Motohashi, N. Suwabe, N. Hayashi, N. Yanai, Y. Nabeshima, and M. Yamamoto. 1997. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J. Biol. Chem. 272:12611-12615. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi, S., R. Shimizu, N. Suwabe, T. Kuroha, K. Yoh, J. Ohta, S. Nishimura, K.-C. Lim, J. D. Engel, and M. Yamamoto. 2000. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood 96:910-916. [PubMed] [Google Scholar]
- 45.Tevosian, S. G., A. E. Deconinck, A. B. Cantor, H. I. Rieff, Y. Fujiwara, G. Corfas, and S. H. Orkin. 1999. FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl. Acad. Sci. USA 96:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trainor, C. D., R. Ghirlando, and M. A. Simpson. 2000. GATA zinc finger interactions modulate DNA binding and transactivation. J. Biol. Chem. 275:28157-28166. [DOI] [PubMed] [Google Scholar]
- 48.Trainor, C. D., J. G. Omichinski, T. L. Vandergon, A. M. Gronenborn, G. M. Clore, and G. Felsenfeld. 1996. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol. Cell. Biol. 16:2238-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai, S.-F., D. I. K. Martin, L. I. Zon, A. D. D'Andrea, G. G. Wong, and S. H. Orkin. 1989. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature 339:446-451. [DOI] [PubMed] [Google Scholar]
- 50.Tsai, S.-F., E. Strauss, and S. H. Orkin. 1991. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 5:919-931. [DOI] [PubMed] [Google Scholar]
- 51.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 52.Vyas, P., M. A. McDevitt, A. B. Cantor, S. G. Katz, Y. Fujiwara, and S. H. Orkin. 1999. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development 126:2799-2811. [DOI] [PubMed] [Google Scholar]
- 53.Wang, H., Q. Long, S. D. Marty, S. Sassa, and S. Lin. 1998. A zebrafish model for hepatoerythropoietic porphyria. Nat. Genet. 20:239-243. [DOI] [PubMed] [Google Scholar]
- 54.Weinstein, B. M., A. F. Schier, S. Abdelilah, J. Malicki, L. Solnica-Krezel, D. L. Stemple, D. Y. R. Stainier, F. Zwartkruis, W. Driever, and M. C. Fishman. 1996. Hematopoietic mutations in the zebrafish. Development 123:303-309. [DOI] [PubMed] [Google Scholar]
- 55.Westerfield, M. 1995. The zebrafish book, 3rd ed. University of Oregon Press, Eugene.
- 56.Yamamoto, M., L. J. Ko, M. W. Leonard, H. Beug, S. H. Orkin, and J. D. Engel. 1990. Activity and tissue-specific expression of the transcription factor NF-E1 (Gata) multigene family. Genes Dev. 4:1650-1662. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto, M., S. Takahashi, K. Onodera, Y. Muraosa, and J. D. Engel. 1997. Upstream and downstream of erythroid transcription factor GATA-1. Genes Cells 2:107-115. [DOI] [PubMed] [Google Scholar]
- 58.Yang, H.-Y., and T. Evans. 1992. Distinct roles for the two cGATA-1 finger domains. Mol. Cell. Biol. 12:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, H.-Y., and T. Evans. 1995. Homotypic interactions of chicken GATA-1 can mediate transcriptional activation. Mol. Cell. Biol. 15:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yomogida, K., H. Ohtani, H. Harigae, E. Ito, Y. Nishimune, J. D. Engel, and M. Yamamoto. 1994. Developmental stage- and spermatogenic cycle-specific expression of transcription factor GATA-1 in mouse Sertoli cells. Development 120:1759-1766. [DOI] [PubMed] [Google Scholar]
- 61.Zon, L. I., C. Mather, S. Burgess, M. E. Bolce, R. M. Harland, and S. H. Orkin. 1991. Expression of GATA-binding proteins during embryonic development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 88:10642-10646. [DOI] [PMC free article] [PubMed] [Google Scholar]