Abstract
Abasic (AP) sites are among the most frequent endogenous lesions in DNA and present a strong block to replication. In Saccharomyces cerevisiae, an apn1 apn2 rad1 triple mutant is inviable because of its incapacity to repair AP sites and related 3′-blocked single-strand breaks (M. Guillet and S. Boiteux, EMBO J. 21:2833, 2002). Here, we investigated the origin of endogenous AP sites in yeast. Our results show that the deletion of the UNG1 gene encoding the uracil DNA glycosylase suppresses the lethality of the apn1 apn2 rad1 mutant. In contrast, inactivation of the MAG1, OGG1, or NTG1 and NTG2 genes encoding DNA glycosylases involved in the repair of alkylation or oxidation damages does not suppress lethality. Although viable, the apn1 apn2 rad1 ung1 mutant presents growth delay due to a G2/M checkpoint. These results point to uracil as a critical source of the formation of endogenous AP sites in DNA. Uracil can arise in DNA by cytosine deamination or by the incorporation of dUMP during replication. Here, we show that the overexpression of the DUT1 gene encoding the dUTP pyrophosphatase (Dut1) suppresses the lethality of the apn1 apn2 rad1 mutant. Therefore, this result points to the dUTP pool as an important source of the formation of endogenous AP sites in eukaryotes.
Endogenous abasic (AP) sites in DNA may arise from the hydrolytic loss of normal and damaged bases or from the removal of damaged or inappropriate bases by DNA glycosylases in the course of the base excision repair (BER) process (11, 26). The measured spontaneous depurination rate in double-stranded DNA translates to the loss of 10,000 purines per human cell per day (26, 27). Damaged DNA bases can arise in several ways, most importantly from the methylation, oxidation, and deamination of normal bases, yielding a variety of lesions, such as N7-methylguanine (N7-meG), 8-oxo-7,8-dihydroguanine (8-oxoG), 5,6-dihydroxy-5,6-dihydrothymine (Tg), or uracil (5, 45, 49). Inappropriate bases such as uracil can be incorporated into DNA during replication (54). The vast majority of damaged and inappropriate bases in DNA are removed by specific DNA glycosylases, thus yielding AP sites (23, 28, 47). Recent information indicates that normal human liver cells present a steady-state level of about 50,000 AP sites per genome (33, 34). Thus, AP sites are likely to represent the most common spontaneous lesions in DNA. In addition to being abundant, AP sites may be cytotoxic, blocking DNA replication and transcription (16, 60). Furthermore, the bypass of AP sites is primarily mutagenic and results in base substitutions and frameshift mutations (16, 30). Moreover, AP sites can be converted into single-strand breaks (SSBs) after cleavage by AP endonucleases or DNA glycosylases/AP lyases (23, 47). Finally, 3′- or 5′-blocked SSBs can be converted into highly toxic double-strand breaks during replication (24).
In Saccharomyces cerevisiae several DNA repair pathways are involved in the elimination of AP sites and related 3′-blocked SSBs (15). BER is the major pathway, and it is initiated by an AP endonuclease, Apn1 or Apn2 (18, 28). Apn1, which provides the major AP endonuclease activity, shares homology with Escherichia coli Nfo (endonuclease IV) (42, 43). Apn2 accounts for less than 5% of total AP endonuclease activity, and it shares homology with E. coli Xth (exonuclease III) (3, 20). Apn1 and Apn2 catalyze the hydrolytic cleavage of the phosphodiester backbone at the 5′-side of AP sites in DNA, yielding SSBs with 3′-OH and 5′-deoxyribose-phosphate (5′-dRP) ends (7). Apn1 and Apn2 are also endowed with a 3′-phosphodiesterase activity, removing 3′-blocking groups such as 3′-phosphate, 3′-phosphoglycolate, or 3′-dRP (42, 43, 56, 57). Besides BER, nucleotide excision repair (NER) acts as a backup activity in the repair of AP sites in yeast (51, 55). However, an apn1 apn2 rad14 triple mutant is viable and only presents a modest spontaneous mutator phenotype (55). Finally, our recent results have suggested that the Rad1/Rad10 and Mus81/Mms4 3′-flap endonucleases play a critical role in the repair of AP sites by removing 3′-blocking lesions (3′-dRP) generated after the cleavage of AP sites by DNA glycosylases/AP lyases (15). Indeed, apn1 apn2 rad1 triple mutants and apn1 apn2 rad1 mus81 quadruple mutants are not viable, forming only microcolonies of about 300 cells or about 20 cells, respectively (15). The synthetic lethality of mutations in APN1, APN2, and RAD1 led us to conclude that, in the absence of DNA repair, endogenous AP sites are formed at a rate that causes cell death. This finding is in agreement with the proposal that AP sites are the most common lesions that are formed in DNA under physiological conditions.
In this study, we attempted to identify the origins of spontaneous AP sites in DNA in S. cerevisiae. Four sources of the formation of AP sites have been investigated: the removal of normal and alkylated purine by the 3-methyladenine DNA glycosylase 1 (Mag1) (4, 6), the removal of oxidatively damaged guanine by the 8-oxoG DNA glycosylase 1 (Ogg1) (58), the removal of oxidatively damaged pyrimidines by the thymine glycol DNA glycosylases 1 and 2 (Ntg1 and Ntg2) (48), and the removal of uracil by the uracil DNA glycosylase 1 (Ung1) (41). The inactivation of Mag1, Ogg1, Ntg1, and Ntg2 or Ung1 should prevent the formation of AP sites from lesions such as N7-meG, 8-oxoG, Tg, or uracil residues in DNA. Therefore, we deleted the MAG1, OGG1, NTG1, and NTG2 or UNG1 genes in a yeast strain that cannot support the burden of spontaneous AP sites, such as the apn1 apn2 rad1 triple mutant (15). The results show that the deletion of the UNG1 gene suppresses the lethality of the apn1 apn2 rad1 mutant, whereas deletion of the MAG1, OGG1, or NTG1 and NTG2 genes does not. Furthermore, the overexpression of the dUTP pyrophosphatase Dut1 (12) also suppresses the lethality of the apn1 apn2 rad1 triple mutant. These results point to the incorporation of uracil from dUTP during replication and to cellular metabolism as primary sources of endogenous AP sites in yeast.
MATERIALS AND METHODS
Yeast culture and genetic procedures.
Yeast strains were grown at 30°C in YP or YNB medium supplemented with appropriate amino acids and bases and 2% glucose (YPD or YNBD medium) or 2% galactose (YPGal or YNBGal medium). All media including agar were from Difco. Presporulation and sporulation procedures were performed as previously described (44). Micromanipulation and dissection of asci were performed with a Singer MSM system as previously described (50).
Yeast strains and plasmids.
S. cerevisiae strains used in this study are listed in Table 1. All yeast strains are isogenic to wild-type (WT) strain FF18733 (MATa leu2-3,112 trp1-289 his7-2 ura3-52 lys1-1). The APN2, MAG1, MUS81, and UNG1 gene deletions were produced by a PCR-mediated one-step replacement technique. All disruptions were confirmed by PCR and crossing and/or auxotrophy. APN1, NTG1, and NTG2 disruptions were produced as previously described (13, 15). RAD1 and RAD14 gene disruptions were performed by using plasmids pWJ163 (from R. Rothstein) and pBM190 (2). To construct plasmid p414GAL1-TDG, the TDG cDNA was amplified by PCR from pETZ8a TDG (T.R. O'Connor, City of Hope, Duarte, Calif.) with TDG Bam5 (5′-CGGATCCATGGAAGCGGAGAACGCGGGCAGC-3′) and TDG EcoIII (5′-CGAATTCTTAAGCATGGCTTTCTTCTTCCTG-3′) as primers. To construct p424GAL1-DUT1, the DUT1 gene was amplified from genomic DNA with DUT1 Bam5 (5′-ATCGGATCCATGACTGCTACTAGCGAC-3′) and DUT1 EcoIII (5′-CTAGAATTCTCAAGGGTCCTTTGATTCTGAC-3′) as primers. The PCR products were digested by EcoRI and BamHI (TDG and DUT1) and cloned in p414GAL1 (TDG) and p424GAL1 (DUT1) (31) previously digested with EcoRI and BamHI. Yeast strains were transformed with plasmids p414GAL1, p424GAL1, p414GAL1-TDG, or p424GAL1-DUT1 by the lithium acetate method as previously described (14). The haploid strain BG157/p414GAL1-TDG was isolated by tetrad dissection after sporulation of the diploid BG211 strain which had been previously transformed with p414GAL1-TDG.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| FF18733 | MATa leu2-3,112 trp1-289 his7-2 ura3-52 lys1-1 | F. Fabre |
| FF18734 | MATα leu2-3,112 trp1-289 his7-2 ura3-52 lys1-1 | F. Fabre |
| FF181482 | FF18734 with rad1Δ::LEU2 | F. Fabre |
| BG4 | FF18734 with apn1Δ::URA3 apn2Δ::kanMX6 | S. Boiteux |
| BG15 | FF18734 with apn2Δ::kanMX6 | S. Boiteux |
| BG17 | FF18734 with apn1Δ::URA3 rad1Δ::LEU2 | This study |
| BG40 | FF18733 with apn1Δ::URA3 apn2Δ::kanMX6 rad14Δ::LEU2 | This study |
| BG135 | FF18733 with ung1Δ::URA3 | This study |
| BG136 | FF18733 with mag1Δ::URA3 | This study |
| BG154 | FF18733 with mus81Δ::kanMX6 rad1Δ::LEU2 | F. Fabre |
| BG4 | FF18734 with apn1Δ::URA3 apn2Δ::kanMX6 | S. Boiteux |
| BG17 | FF18734 with apn1Δ::URA3 rad1Δ::LEU2 | This study |
| BG110 | FF18733 with apn2Δ::kanMX6 mag1Δ::URA3 | This study |
| BG24 | FF18733 with apn1Δ::URA3 apn2Δ::kanMX6 ogg1Δ::TRP1 | This study |
| BG163 | FF18733 with apn1Δ::URA3 rad1Δ::LEU2 ung1Δ::URA3 | This study |
| BG157 | FF18733 with apn1Δ::URA3 apn2Δ::kanMX6 rad1Δ::LEU2 ung1Δ::URA3 | S. Boiteux |
| BG40 | FF18733 with apn1Δ::URA3 apn2Δ::kanMX6 rad14::LEU2 | This study |
| BG159 | FF18733 with apn1Δ::URA3 apn2Δ::kanMX6 ung1Δ::URA3 | S. Boiteux |
| BG211 | Diploid apn1Δ::URA3/APN1 apn2Δ::kanMX6/APN2 rad1Δ::LEU2/RAD1 ung1Δ::URA3/UNG1 | This study |
| BG211 + p414GAL1-TDG | Diploid apn1Δ::URA3/APN1 apn2Δ::kanMX6/APN2 rad1Δ::LEU2/RAD1 ung1Δ::URA3/UNG1 with p414GAL1-TDG | This study |
| BG157 + p414GAL1-TDG | FF18733 with apn1Δ::URA3 apn2Δ::kanMX6 rad1Δ::LEU2 ung1Δ::URA3; with p414GAL1-TDG | This study |
| BG159 + p414GAL1-TDG | FF18733 with apn1Δ::URA3 apn2Δ::kanMX6 ung1Δ::URA3; with p414GAL1-TDG | This study |
| BG83 + p424GAL1-DUT1 | Diploid apn1Δ::URA3/APN1 apn2Δ::kanMX6/APN2 rad1Δ::LEU2/RAD1; with p424GAL1-DUT1 | This study |
| CC892 | FF18733 with ntg1Δ::URA3 ntg2Δ::TRP1 rad1Δ::LEU2 | S. Boiteux |
FACS analysis and DAPI staining.
Flow cytometry analysis was done with a FACScalibur (Beckton-Dickinson) cell sorter. For fluorescence-activated cell sorting (FACS) analysis, yeast cells were grown to exponential phase. Ten million cells were harvested, fixed in 70% ethanol, washed in phosphate-buffered saline, incubated with 1 mg of RNase/ml, centrifuged, and resuspended in 50 μg of propidium iodide per ml. For DAPI (4′, 6′-diamidino-2-phenylindole) staining, 107 cells were fixed in 70% ethanol, washed, and resuspended in 1 μg of DAPI per ml. Cells were analyzed on a Zeiss Axiophot 2 microscope.
Determination of the genotypes.
The genotypes of inviable spores were inferred from the segregation patterns of the three viable spores. The genotypes of viable strains were inferred from the segregation patterns and PCR when two genes possessed the same markers. The genomic DNA of the different spores was extracted with a Dneasy tissue kit (Qiagen). PCRs were performed to determine the disruptions of APN1, APN2, MAG1, NTG1, and UNG1 by using the following primers: APN232 (5′-AGGATCCTTATTCTTTCTTAGTCTTCCTC-3′) and APN25 (5′-GGGGATGCCTCGACACCTAGC-3′), KanR606 (5′-ACGGAATTTATGCCTCTTCCG-3′) and APN13 (5′-AGGATCCTTAAACCCACTGAAAAAACCC-3′), 5URA392 (5′-GCGGTTTGAAGCAGGCGGCGGAGAAG-3′) and MAG1flank3 (5′-GACAGTATACTCGCTTTTCCGC-3′), NTG1flank5 (5′-GCAGTTACAGTCACAGTCACAGCC-3′) and NTG1lfank3 (5′-GGCTCTGATTGGTGTCGTGATG-3′), and 5URA392 and UNG1flank3 (5′-CTTCTCCTTACTCTCTCAATTTGG-3′), respectively.
Analysis of mutant strain colonies.
The number of cells per colony or microcolony was counted by microscopic observation. Alternatively, the number of cells per colony was estimated after suspension of the cells in sterile water and counting with a hemocytometer. For all crosses, cells were counted 4 days after dissection.
MMS sensitivity.
To estimate methyl methanesulfonate (MMS) sensitivity, yeast cells were grown in YPD medium at 30°C to an optical density at 600 nm (OD600) of 1.0 and resuspended in sterile water. Appropriate dilutions of MMS (Sigma) were added for 20 min at 30°C with agitation. The reaction was terminated by adding 1 volume of 10% sodium thiosulfate. To assess cell viability after treatment, appropriate dilutions of cell suspensions were plated on YPD medium and allowed to grow for 2 days.
Spontaneous mutation frequencies.
Yeast strains were grown in 2 ml of YNBD or YNBGal medium with the appropriate amino acids at 30°C for 3 days to a cell density of approximately 5 × 107 cells/ml. Cultures were inoculated at a starting density of 103 cells/ml. Cell density was measured by plating dilutions on YNBD or YNBGal plates and counting the colonies after 3 days. The quantification of canavanine-resistant mutants (Canr) was determined after plating 0.1 ml of undiluted culture on YNBD or YNBGal plates containing 60 μg of canavanine sulfate (Research Organics, Inc.) per ml. The plates were incubated for 5 days before cells were counted.
Preparation of cell extracts and assay for dUTPase activity.
Yeast strains (WT and apn1 apn2 rad1/p424GAL1-DUT1) were grown at 30°C in 200 ml of YNBGal medium to reach an OD600 of 1.0. The yeast cells were centrifuged, washed, and stored at −80°C. Cell-free protein extracts were prepared as previously described (52). After centrifugation, the proteins in the supernatant fraction were precipitated in the presence of ammonium sulfate (500 mg/ml), resuspended in 0.5 ml of 50 mM KH2PO4/K2HPO4 (pH 6.8), and dialyzed overnight at 2°C against the same buffer yielding the cell extract. The protein concentration in cell extract was measured according to the Bradford method. The dUTPase activity assay (100-μl final volume) contained 50 mM KH2PO4/K2HPO4 (pH 6.8), 100 pmol of dUTP (Sigma-Aldrich), and cell extract. The reactions were performed at 37°C for increasing lengths of time and/or increasing amounts of cell extract. The conversion of dUTP into dUMP was monitored after separation by high-pressure liquid chromatography (HPLC) with a C18 μBondapack column (Waters). The reaction mixture was directly injected in the system. The column was isocratically developed (1 ml/min) at room temperature with 50 mM NH4H2PO4, pH 4.5, as the mobile phase (39). The retention times of dUTP and dUMP (Sigma-Aldrich) were 4.1 min and 9.5 min, respectively. Products eluted from the HPLC column were detected by monitoring UV absorption at 254 nm. The amount of dUMP formed was calculated by using a calibration curve and an authentic dUMP marker molecule. One unit of dUTPase activity releases 1 pmol of dUMP per min at 37°C.
RESULTS
Deletion of the UNG1 gene suppresses lethality in cells lacking APN1, APN2, and RAD1, whereas inactivation of the MAG1, OGG1, or NTG1 and NTG2 genes does not.
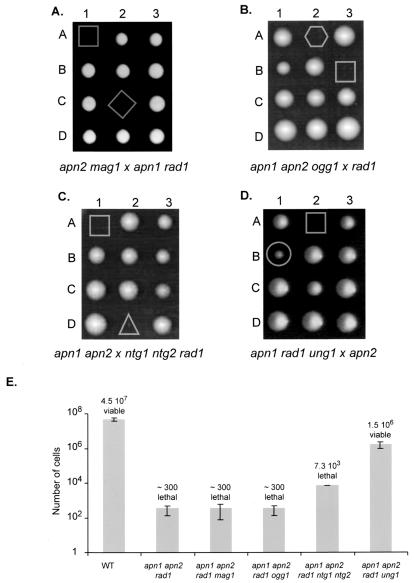
The synthetic lethality of mutations in the APN1, APN2, and RAD1 genes strongly suggests that the burden of endogenous AP sites in DNA causes cell death in S. cerevisiae (15). Although not viable, an apn1 apn2 rad1 triple mutant can form microcolonies of about 300 cells (15). The origins of spontaneous AP sites in DNA are presumably multiple; however, the involvement of alkylated or oxidized purines, oxidized pyrimidines, and uracil can be investigated by using apn1 apn2 rad1 strains where the MAG1, OGG1, NTG1, and NTG2 or UNG1 genes have been deleted. To test the roles of Mag1 and Ogg1, an apn2 mag1 strain was crossed with an apn1 rad1 mutant, and an apn1 apn2 ogg1 strain was crossed with a rad1 mutant (Fig. 1A and B). A high degree of spore lethality was observed in both cases, and no apn1 apn2 rad1 mag1 or apn1 apn2 rad1 ogg1 quadruple mutant was obtained (36 tetrads dissected, with 9 quadruple mutants expected and 48 tetrads dissected, with 12 quadruple mutants expected, for the crossing of mag1 [Fig. 1A] and ogg1 [Fig. 1B], respectively). Microscopic analysis does not reveal significant differences among the microcolonies of apn1 apn2 rad1, apn1 apn2 rad1 mag1, and apn1 apn2 rad1 ogg1 (Fig. 1E). To examine the role of Ntg1 and Ntg2, an apn1 apn2 strain was crossed with an ntg1 ntg2 rad1 mutant (Fig. 1C). A high degree of spore inviability was observed, and no apn1 apn2 rad1 ntg1 ntg2 mutant was obtained (24 tetrads dissected with three apn1 apn2 rad1 ntg1 ntg2 mutants expected [Fig. 1C]). However, 4 days after dissection, two types of microcolonies composed of an average of 300 cells and of 7 · 103 cells were observed (Fig. 1E). By replica plating and PCR analysis, the microcolonies were genotyped as apn1 apn2 rad1 and apn1 apn2 rad1 ntg1 ntg2 mutants, respectively. This result shows that the absence of the two DNA glycosylases/AP lyases, Ntg1 and Ntg2, allows the apn1 apn2 rad1 ntg1 ntg2 mutant to form a larger colony but does not suppress lethality. To evaluate the role of Ung1, the apn1 rad1 ung1 strain was crossed with the apn2 mutant (Fig. 1D). Four days after dissection, two types of unusual colonies were observed: 17 microcolonies and 16 visible colonies, which were clearly smaller than those of the WT (71 tetrads dissected) (Fig. 1D). By replica plating and PCR analysis, the microcolonies were genotyped as apn1 apn2 rad1 triple mutants, and all the small colonies were genotyped as apn1 apn2 rad1 ung1 quadruple mutants. The apn1 apn2 rad1 ung1 colonies were composed of an average of 2 × 106 cells at 4 days after dissection, whereas WT colonies were composed of about 4 ×107 cells (Fig. 1E). These results show that the deletion of the UNG1 gene suppresses the lethality of the apn1 apn2 rad1 triple mutant, whereas the deletion of MAG1, OGG1, or NTG1 and NTG2 does not. Therefore, uracil is a critical source of the formation of spontaneous AP sites in DNA.
FIG. 1.
Effect of inactivation of MAG1, OGG1, NTG1, and NTG2, or UNG1 upon viability of an apn1 apn2 rad1 triple mutant. (A) Inactivation of MAG1. The BG110 (apn2 mag1) mutant was crossed with the BG17 (apn1 rad1) strain. After sporulation of the diploids, tetrads were dissected on YPD plates. Spore genotypes were inferred by replica plating on appropriate media and by PCR. A selection of tetrads containing an apn1 apn2 rad1 (A1) and an apn1 apn2 rad1 mag1 (C2) mutant is presented. (B) Inactivation of OGG1. Strain BG24 (apn1 apn2 ogg1) was crossed with FF181482 (rad1). The cross was analyzed as previously described. A selection of tetrads containing an apn1 apn2 rad1 (B3) and an apn1 apn2 rad1 ogg1 (A2) mutant is presented. (C) Inactivation of NTG1 and NTG2. Strain BG4 (apn1 apn2) was crossed with CC892 (ntg1 ntg2 rad1). The cross was analyzed as previously described. A selection of tetrads containing an apn1 apn2 rad1 (A1) and an apn1 apn2 rad1 ntg1 ntg2 (D2) mutant is presented. (D) Inactivation of UNG1. Strain BG163 (apn1 rad1 ung1) was crossed with BG15 (apn2). The cross was analyzed as previously described. A selection of tetrads containing an apn1 apn2 rad1 (A2) and an apn1 apn2 rad1 ung1 (B1) mutant is presented. (E) The average numbers of cells per microcolony or colony were estimated 4 days after dissection. The mutants are characterized as viable or lethal when growth in liquid medium (YPD) was observed or not, respectively, 3 days after inoculation of the microcolony (the microcolonies were inoculated 4 days after dissection). All numbers of cells are the means ± standard deviations from at least three independent colonies.
The apn1 apn2 rad1 ung1 mutant presents a growth delay due to a G2/M checkpoint and possesses synthetic lethality with mus81.
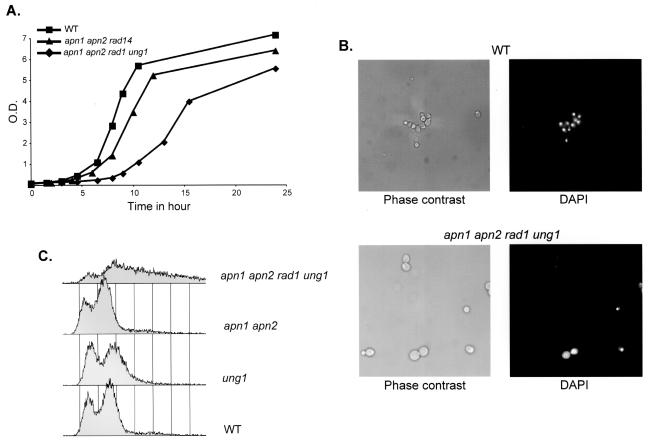
Although viable, the apn1 apn2 rad1 ung1 quadruple mutant generates colonies smaller than those of the WT, suggesting a growth defect. Growth curves in YPD medium show that the apn1 apn2 rad1 ung1 mutant has a doubling time of 150 min compared to 76 min for the WT strain (Fig. 2A). Exponential phase cultures of the apn1 apn2 rad1 ung1 mutant contain larger cells and a higher number of large-budded cells than cultures of the WT strain (55% and 40%, respectively) (Fig. 2B). DAPI staining shows that most of these large-budded cells have the nucleus at the bud neck: 52% of the large-budded cells of the apn1 apn2 rad1 ung1 mutant compared to 22% of those of the WT strain (Fig. 2B and data not shown). This localization of the nucleus is characteristic of cells arrested at the G2/M checkpoint, more specifically, at the preanaphase stage of mitosis (53). FACS analysis of apn1 apn2 rad1 ung1 shows a decrease in the number of cells in all phases of the cell cycle and an accumulation of cells at the right of the G2 peak that could represent dead cells (Fig. 2C). The presence of dead cells in exponentially growing cultures of apn1 apn2 rad1 ung1 is also suggested by a reduced plating efficiency (52%) compared to that of the WT (≥80%). Taken together, these results suggest that the growth delay of the apn1 apn2 rad1 ung1 mutant is due to a G2/M checkpoint that causes a transient or irreversible arrest at the G2/M transition.
FIG. 2.
Properties of an apn1 apn2 rad1 ung1 quadruple mutant strain. (A) Growth curves at 30°C in YPD medium of the WT, apn1 apn2 rad14, and apn1 apn2 rad1 ung1 strains. Cells were grown in complete medium and diluted to an OD600 of 0.05 at time zero; the growth of the three cultures was measured as a function of time at 30°C with agitation. (B) Microscopic analysis of the WT (top panels) and apn1 apn2 rad1 ung1 (bottom panels) strains. Cells were grown to exponential phase in YPD medium and observed by microscopy in phase contrast (left panels) and after DAPI staining (right panels). The same magnification (×100) was used for both strains. (C) FACS analysis of exponential phase cultures in YPD medium of the WT, ung1, apn1 apn2, and apn1 apn2 rad1 ung1 strains.
The reduced viability of the apn1 apn2 rad1 ung1 mutant points to the formation of spontaneous AP sites and 3′-blocked SSBs which are not generated by Ung1. To test this hypothesis the apn1 apn2 rad1 ung1 mutant was crossed with the mus81 rad1 strain. After dissection of the spores, a high degree of spore lethality was observed, and no apn1 apn2 rad1 ung1 mus81 colony was obtained (47 tetrads were dissected, with 12 quintuple mutants expected). Microscopic analysis allowed us to identify three classes of microcolonies, one composed of an average of 20 cells, one composed of an average of 300 cells, and one composed of an average of 104 cells (13, 12, and 8 spores, respectively). By replica plating and segregation patterns, we identified the two first classes as apn1 apn2 rad1 mus81 and apn1 apn2 rad1 mutants, respectively (15), and the third one as an apn1 apn2 rad1 ung1 mus81 quintuple mutant. Therefore, the presence of the Mus81/Mms4 complex is essential for the viability of the apn1 apn2 rad1 ung1 mutant. However, the inactivation of UNG1 allows a substantial increase in the number of generations that an apn1 apn2 rad1 mus81 mutant can undergo after dissection. These results confirm the critical role of AP sites generated by Ung1, but they also show that other endogenous sources of AP sites exist and generate a burden of lesions that cause cell death and mutations in the absence of the relevant repair systems.
NER-independent role of the Rad1/Rad10 heterodimer in the repair of AP sites in DNA.
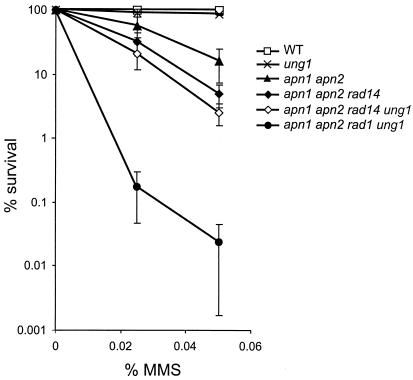
The viability of the apn1 apn2 rad1 ung1 mutant allowed us to investigate the specific role of Rad1/Rad10 in the repair of DNA damages induced by MMS. The lethal effect of MMS is generally attributed to the formation of AP sites resulting from the chemical or enzymatic release of N3-methyladenine and N7-meG (59). The deletion of UNG1 does not enhance MMS sensitivity in WT and apn1 apn2 rad14 backgrounds (Fig. 3). Figure 3 also shows that the apn1 apn2 rad14 triple mutant is more sensitive than the apn1 apn2 double mutant to MMS, confirming the involvement of NER in the removal of AP sites (51, 55). Furthermore, the apn1 apn2 rad1 ung1 mutant exhibits an extreme sensitivity to MMS compared to the sensitivity of the apn1 apn2 rad14 ung1 mutant (Fig. 3). These results point to a role of Rad1/Rad10, independent of NER, in the repair of MMS-induced AP sites. These results are consistent with previous studies showing that mutations in APN1, APN2, and RAD1 are synthetic lethal mutations, whereas mutations in APN1, APN2, and RAD14 are not (15, 51, 55).
FIG. 3.
Effect of inactivation of RAD14 and RAD1 genes upon MMS sensitivity. The different mutants were grown at 30°C in YPD medium to exponential phase and exposed to MMS. Experimental points are the averages of at least three independent experiments.
Origin of uracil in DNA: role of cytosine deamination and expression of the human TDG protein.
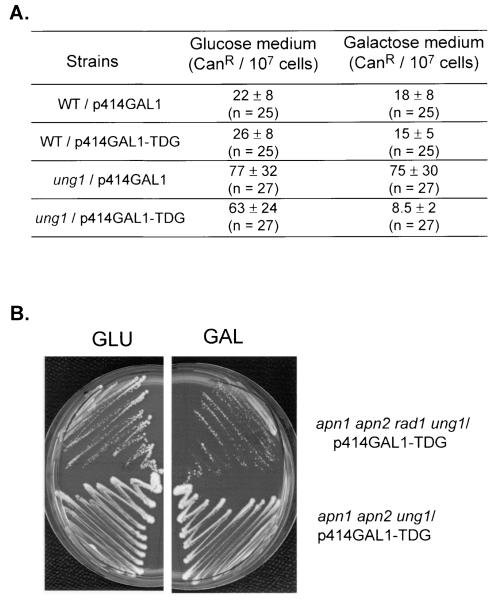
In double-stranded DNA, cytosine deamination occurs spontaneously, leading to the formation of U · G mismatches that are responsible for the CG-to-TA mutator phenotype of ung1 mutants of E. coli and S. cerevisiae (8, 19). The role of cytosine deamination on the formation of spontaneous AP sites can be specifically investigated in apn1 apn2 rad1 ung1 cells expressing a DNA glycosylase that excises uracil in U · G mismatches but not in U · A pairs, such as the human uracil or thymine DNA glycosylase TDG (17, 29, 35). If U · G is a critical source of the formation of AP sites, the expression of TDG in the apn1 apn2 rad1 ung1 mutant should mimic the expression of Ung1 and cause cell death. Therefore, we cloned the human cDNA encoding TDG under the control of a galactose-inducible promoter in the p414GAL1 centromeric plasmid yielding p414GAL1-TDG. To assess the expression of a functional human TDG in yeast, we transformed an ung1 mutant with the empty p414GAL1 plasmid and with plasmid p414GAL1-TDG. The spontaneous mutation frequencies of the resulting strains were measured by using the CAN1 gene as a reporter. Figure 4A shows that the ung1 mutant exhibits a fourfold-higher frequency of the spontaneous canavanine-resistant (Canr) mutant than the WT. Figure 4A also shows that the spontaneous mutator phenotype of the ung1 mutant is suppressed in the ung1/p414GAL1-TDG strain grown in the presence of galactose. In contrast, the ung1/p414GAL1-TDG strain grown in the presence of glucose exhibits a spontaneous mutator phenotype (Fig. 4A). These results demonstrate that the human TDG expressed in yeast is functional and regulated by the growth medium. To explore the role of U · G mismatches in the formation of endogenous AP sites, we transformed an apn1 apn2 rad1 ung1 mutant with plasmid p414GAL1-TDG. Figure 4B shows that the apn1 apn2 rad1 ung1/p414GAL1-TDG strain, expressing (GAL) or not (GLU) TDG can form colonies on plates. These results show that the removal of uracil in U · G mismatches does not lead to cell death and, in turn, strongly suggest that cytosine deamination is not a critical cause of the formation of spontaneous AP sites in DNA.
FIG. 4.
Expression of human TDG in ung1 and apn1 apn2 rad1 ung1 mutant strains. (A) Effect of TDG on spontaneous mutagenesis. The WT and ung1 strains harboring p414GAL1 or p414GAL1-TDG were grown at 30°C in YNBD (Glucose) or YNBGal (Galactose) medium to saturation and plated onto canavanine-containing plates. Mutation frequencies were calculated by using the method of the medians (n = number of independent cultures). (B) Effect of TDG on survival. Mutant strains, apn1 apn2 rad1 ung1/p414GAL1-TDG and apn1 apn2 ung1/p414GAL1-TDG, were grown in YNBD medium with appropriate amino acids to an OD600 of 1.0. Cells were washed and plated on YNBD (GLU) or YNBGal (GAL) plates.
Origin of uracil in DNA: role of dUMP incorporation during replication and expression of the dUTP pyrophosphatase Dut1.
In addition to cytosine deamination, uracil can arise in DNA after the incorporation of dUMP by DNA polymerases by using dUTP during replication or repair (9, 12). The incorporation of dUMP in DNA leads to the formation of the U · A pair that is not mutagenic by itself; however, it can be at the origin of the AP sites after the removal of uracil by a DNA glycosylase, such as the yeast Ung1. To assess the role of dUMP incorporated in DNA as a significant source of the formation of AP sites, we overexpressed the DUT1 gene in an apn1 apn2 rad1 triple mutant. In S. cerevisiae, the DUT1 gene encodes a dUTP pyrophosphatase that hydrolyses dUTP to dUMP and PPi (12). Therefore, overexpression of DUT1 should decrease the intracellular dUTP/dTTP ratio and thereby prevent the incorporation of uracil into DNA. The DUT1 gene was cloned under the control of a galactose-inducible promoter in the 2μm plasmid p424GAL1 yielding p424GAL1-DUT1. Control experiments showed that p424GAL1-DUT1 expressed a functional Dut1 that can rescue the lethality of the dut1 deletion mutant (data not shown). Therefore, the diploid strain BG83 (apn1/APN1 apn2/APN2 rad1/RAD1) was transformed with p424GAL1-DUT1. Four days after dissection of the spores on galactose plates, we observed microcolonies and slowly growing colonies (Fig. 5A). The tetrads were analyzed, and the genotypes of the spores were inferred from segregation patterns after replica plating. All microcolonies were genotyped as apn1 apn2 rad1 mutants, whereas slowly growing colonies were apn1 apn2 rad1/p424GAL1-DUT1 (Fig. 5A). The apn1 apn2 rad1/p424GAL1-DUT1 strain is viable; it forms colonies and grows to saturation in YNBGal medium. To confirm the overexpression of Dut1, the dUTPase activity was measured in cell-free protein extracts. Figure 5B shows that the dUTPase activity of the apn1 apn2 rad1 strain hosting p424GAL1-DUT1 is four- to fivefold higher than that of the WT. Therefore, the overexpression of Dut1 can suppress the lethality of an apn1 apn2 rad1 triple mutant. These results strongly suggest that the incorporation of dUMP from the dUTP pool and, consequently, the U · A pairs in DNA, are at the origin of an important part of endogenous AP sites in yeast.
FIG. 5.
Overexpression of DUT1 suppresses the lethality of an apn1 apn2 rad1 mutant. (A) The diploid strain BG83 (apn1/APN1 apn2/APN2 rad1/RAD1) was transformed with p424GAL1-DUT1 and dissected after sporulation on YPGal plates. Spore genotypes were inferred by replica plating on selective media containing galactose. A selection of tetrads containing an apn1 apn2 rad1/p424GAL1-DUT1 (C2) and an apn1 apn2 rad1 (C3) mutant is presented. (B) The dUTP pyrophosphatase (dUTPase) activity was measured in cell extracts of the WT and apn1 apn2 rad1/p424GAL1-DUT1 strains. Cell extract preparations and the dUTPase assay are described in Materials and Methods. Reactions were performed for 15 min at 37°C with increasing amounts of total cell proteins. The assays measured the conversion of dUTP into dUMP, which was identified and quantified after separation of the products of the reaction by HPLC. One unit produces 1 pmol of dUMP per min at 37°C.
DISCUSSION
AP sites are currently thought to be among the most frequent spontaneous lesions that occur in DNA; they are potentially lethal or mutagenic. Consistent with the expected abundance and toxicity, the burden of the spontaneous AP sites is lethal in the absence of Apn1, Apn2, and Rad1/Rad10 in S. cerevisiae (15). To explain the synthetic lethality of the apn1 apn2 rad1 triple mutant, we proposed that AP endonucleases or 3′-phosphodiesterases (Apn1 and Apn2) cooperate with 3′-flap endonucleases (Rad1/Rad10 and Mus81/Mms4) to repair AP sites and 3′-blocked SSBs resulting from the chemical or enzymatic (AP lyase) cleavage of AP sites in DNA (15). In the present study, we explored the origin of endogenous AP sites in yeast. The strategy used was to suppress the synthetic lethality of an apn1 apn2 rad1 triple mutant by overexpressing or inactivating cellular function(s) that can modulate the number of AP sites in DNA. Therefore, we inactivated Mag1, Ogg1, Ntg1, and Ntg2 or Ung1 to assess the impact of endogenous DNA base damage, such as N7-meG, 8-oxoG, Tg, and uracil lesions, respectively, on the formation of AP sites. Here, we show that the deletion of MAG1 or OGG1 in the apn1 apn2 rad1 triple mutant does not suppress lethality. Furthermore, the number of generations that an apn1 apn2 rad1 mag1 or apn1 apn2 rad1 ogg1 quadruple mutant can undergo is not significantly different from that of an apn1 apn2 rad1 triple mutant. These results strongly suggest that N7-meG and 8-oxoG are not major sources of the formation of AP sites in the DNA of dividing cells. Although we cannot exclude the action of other repair pathways, it should be noted that both BER and NER are inactivated in these mutants. The deletion of NTG1 and NTG2 in an apn1 apn2 rad1 background allows the cells to undergo several more generations. This phenotype might be due to a reduced number of AP sites that result from the inactivation of the DNA glycosylase activity of Ntg1 and Ntg2 (48) or to an inefficient cleavage of AP sites by the AP lyase activity of Ogg1, which is only able to cleave AP sites opposite a cytosine (58). The second mechanism would result in a reduced number of 3′-blocked SSBs which have been suggested to be more toxic than AP sites alone (15). Together, these mechanisms can contribute to enhance the survival of the apn1 apn2 rad1 ntg1 ntg2 mutant versus the apn1 apn2 rad1 mutant. In contrast, the deletion of the UNG1 gene suppresses the lethality of the apn1 apn2 rad1 triple mutant. This result points to uracil as a critical source of the formation of AP sites in DNA. These data can be compared with those showing that E. coli cells deficient in AP endonuclease (lacking xth and nfo) and NER (lacking uvrA) are not viable, but they can be rescued by the inactivation of the ung1 gene (10). Although viable, the apn1 apn2 rad1 ung1 quadruple mutant presents growth defects characterized by increased doubling time, cellular abnormalities, and altered FACS profiles. Indeed, cells at the right of the G2 peak visible by FACS analysis are probably dead cells that have accumulated too much DNA damage. Furthermore, the deletion of UNG1 cannot suppress lethality in absence of Apn1, Apn2, Rad1/Rad10, and Mus81/Mms4. This finding is consistent with the occurrence, independently of uracil, of endogenous sources of AP sites, which can have a significant impact on cell viability and, more importantly, on genetic stability. Indeed, the hydrolytic depurination of DNA and the endogenous oxidative stress are probably involved in the lethality of the apn1 apn2 rad1 mus81 ung1 mutant. The deleterious action of the spontaneous oxidative stress is unambiguously revealed by the mutator phenotype of Ogg1-deficient strains (52).
Our data strongly suggest that the removal of uracil in DNA by the Ung1 DNA glycosylase is a critical cause of the formation of spontaneous AP sites. However, the data do not say much about the origin of uracil in DNA. The role of U · G mismatches was investigated with yeast strains that express human TDG, which specifically excises uracil in U · G mismatches (36). The results show that expression of TDG suppresses the mutator phenotype of ung1 strains but does not affect the viability of the apn1 apn2 rad1 ung1 mutant. Therefore, cytosine deamination is not the major endogenous cause of uracil in DNA as expected from the modest spontaneous mutator phenotype of Ung1-deficient strains (19). Finally, our results point to U · A pairs formed by use of dUTP during replication as a critical source of AP sites in DNA. The role of dUTP was strongly suggested by our data showing that the overproduction of the Dut1 activity suppresses the lethality of the apn1 apn2 rad1 triple mutant. Although viable, the apn1 apn2 rad1/p424GAL1-DUT1 strain presents severe growth defects (unpublished data). This result is presumably due to the fact that the dUTP pool is rather low in WT strains, and a fourfold increase in dUTPase activity is not sufficient to completely deplete the dUTP pool in the cell. Taken together, our results show that Dut1 and Ung1 play a critical role in the formation of endogenous AP sites in DNA. This finding is in agreement with the fact that dUTP pyrophosphatase and uracil DNA glycosylase activities are found in all living organisms with dTMP in their DNA and also in viruses (1, 46). Mammalian cells possess several uracil DNA glycosylases; the major activity is associated with UNG proteins, whereas minor activities are associated with SMUG1, TDG, or MBD4 (36). The UNG gene has apparently evolved a specialized role in the repair of U · A pairs formed by use of dUMP from the dUTP pool during replication (21, 32, 40). In fact, UNG is associated with DNA replication foci in mammalian cells, probably due to direct interaction with PCNA and RPA (40). Moreover, both UNG and DUT proteins in mammalian cells have two isoforms, one in the nucleus and another in mitochondria (25, 37). Thus, it seems that mammalian cells have efficient systems to avoid the persistence of uracil in nuclear and mitochondrial DNA that comes from the incorporation of dUMP. Recent studies showed that the cells of ung−/− mice exhibit a steady-state level of ∼2,000 uracils per genome, which is most likely due to the incorporation of dUMP, but this number is probably an underestimation of the level of uracil in DNA since SMUG1 is present in these cells (38). In contrast, a steady-state level of only 400 8-oxoG residues per genome was measured in the cells of ogg1−/− mice (22). These results in mammalian cells are in agreement with our data in yeast and point to the incorporation of dUMP during replication and its removal by uracil DNA glycosylases as a critical source of endogenous AP sites in DNA. These results led us to conclude that a critical threat to DNA in dividing cells is due to DNA metabolism itself, since dUTP is a physiological intermediate in the course of dTTP biosynthesis in all organisms (11).
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique and by the Commissariat à l'Energie Atomique. This work was also supported by the Comité de Radioprotection of Electricité de France. Marie Guillet was supported by a fellowship from the Association pour la Recherche contre le Cancer.
We thank Patricia Auffret van der Kemp for her kind assistance in the measurement of the dUTPase activity. We thank T. R. O'Connor for the kind gift of the TDG cDNA and Marcelo de Padula and J. Pablo Radicella for their support.
REFERENCES
- 1.Baldo, Angela M., and Marcella A. McClure. 1999. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J. Virol. 73:7710-7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bankmann, M., L. Prakash, and S. Prakash. 1992. Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature 355:555-558. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, Richard A. O. 1999. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol. 19:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berdal, K. G., R. F. Johansen, and E. Seeberg. 1998. Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J. 17:363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadet, J., M. Berger, T. Douki, and J. L. Ravanat. 1997. Oxidative damage to DNA: formation, measurement, and biological significance. Rev. Physiol. Biochem. Pharmacol. 131:1-87. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., B. Derfler, A. Maskati, and L. Samson. 1989. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc. Natl. Acad. Sci. USA 86:7961-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 8.Duncan, Bruce K., and Bernard Weiss. 1982. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J. Bacteriol. 151:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.el-Hajj, Hiyam H., Linghua Wang, and Bernard Weiss. 1992. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J. Bacteriol. 174:4450-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, Patricia L. 1990. Escherichia coli strains with multiple DNA repair defects are hyperinduced for the SOS response. J. Bacteriol. 172:4719-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg, E., G. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 12.Gadsden, M. H., E. M. McIntosh, J. C. Game, P. J. Wilson, and R. H. Haynes. 1993. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 12:4425-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellon, L., R. Barbey, P. Auffret van der Kemp, D. Thomas, and S. Boiteux. 2001. Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2, and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:1087-1096. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillet, M., and S. Boiteux. 2002. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 21:2833-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardeland, U., M. Bentele, T. Lettieri, R. Steinacher, J. Jiricny, and P. Schar. 2001. Thymine DNA glycosylase. Prog. Nucleic Acid Res. Mol. Biol. 68:235-253. [DOI] [PubMed] [Google Scholar]
- 18.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 19.Impellizzeri, Kimberly J., Blake Anderson, and Peter M. J. Burgers. 1991. The spectrum of spontaneous mutations in a Saccharomyces cerevisiae uracil-DNA-glycosylase mutant limits the function of this enzyme to cytosine deamination repair. J. Bacteriol. 173:6807-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, Robert E., Carlos A. Torres-Ramos, Tadahide Izumi, Sankar Mitra, Satya Prakash, and Louise Prakash. 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavli, B., O. Sundheim, M. Akbari, M. Otterlei, H. Nilsen, F. Skorpen, P. A. Aas, L. Hagen, H. E. Krokan, and G. Slupphaug. 2002. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 277:39926-39936. [DOI] [PubMed] [Google Scholar]
- 22.Klungland, A., I. Rosewell, S. Hollenbach, E. Larsen, G. Daly, B. Epe, E. Seeberg, T. Lindahl, and D. E. Barnes. 1999. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA 96:13300-13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krokan, H. E., R. Standal, and G. Slupphaug. 1997. DNA glycosylases in the base excision repair of DNA. Biochem J. 325:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzminov, A. 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 98:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ladner, R. D., D. E. McNulty, S. A. Carr, G. D. Roberts, and S. J. Caradonna. 1996. Characterization of distinct nuclear and mitochondrial forms of human deoxyuridine triphosphate nucleotidohydrolase. J. Biol. Chem. 271:7745-7751. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 27.Lindahl, T., and B. Nyberg. 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11:3610-3618. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 29.Liu, P., A. Burdzy, and L. C. Sowers. 2002. Substrate recognition by a family of uracil-DNA glycosylases: UNG, MUG, and TDG. Chem. Res. Toxicol. 15:1001-1009. [DOI] [PubMed] [Google Scholar]
- 30.Loeb, L. A. 1985. Apurinic sites as mutagenic intermediates. Cell 40:483-484. [DOI] [PubMed] [Google Scholar]
- 31.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagelhus, T. A., T. Haug, K. K. Singh, K. F. Keshav, F. Skorpen, M. Otterlei, S. Bharati, T. Lindmo, S. Benichou, R. Benarous, and H. E. Krokan. 1997. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 272:6561-6566. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, J., and J. A. Swenberg. 1999. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 59:2522-2526. [PubMed] [Google Scholar]
- 34.Nakamura, J., V. E. Walker, P. B. Upton, S. Y. Chiang, Y. W. Kow, and J. A. Swenberg. 1998. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 58:222-225. [PubMed] [Google Scholar]
- 35.Neddermann, P., P. Gallinari, T. Lettieri, D. Schmid, O. Truong, J. J. Hsuan, K. Wiebauer, and J. Jiricny. 1996. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 271:12767-12774. [DOI] [PubMed] [Google Scholar]
- 36.Nilsen, H., K. A. Haushalter, P. Robins, D. E. Barnes, G. L. Verdine, and T. Lindahl. 2001. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 20:4278-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsen, H., M. Otterlei, T. Haug, K. Solum, T. A. Nagelhus, F. Skorpen, and H. E. Krokan. 1997. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 25:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsen, H., I. Rosewell, P. Robins, C. F. Skjelbred, S. Andersen, G. Slupphaug, G. Daly, H. E. Krokan, T. Lindahl, and D. E. Barnes. 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell 5:1059-1065. [DOI] [PubMed] [Google Scholar]
- 39.O'Connor, T. R., S. Boiteux, and J. Laval. 1988. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 16:5879-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otterlei, M., E. Warbrick, T. A. Nagelhus, T. Haug, G. Slupphaug, M. Akbari, P. A. Aas, K. Steinsbekk, O. Bakke, and H. E. Krokan. 1999. Post-replicative base excision repair in replication foci. EMBO J. 18:3834-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Percival, K. J., M. B. Klein, and P. M. Burgers. 1989. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J. Biol. Chem. 264:2593-2598. [PubMed] [Google Scholar]
- 42.Popoff, S. C., A. I. Spira, A. W. Johnson, and B. Demple. 1990. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl. Acad. Sci. USA 87:4193-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramotar, Dindial, Sonya C. Popoff, Edith B. Gralla, and Bruce Demple. 1991. Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol. 11:4537-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Resnick, M. A., J. C. Game, and S. Stasiewicz. 1983. Genetic effects of UV irradiation on excision-proficient and -deficient yeast during meiosis. Genetics 104:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rydberg, B., and T. Lindahl. 1982. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sartori, A. A., S. Fitz-Gibbon, H. Yang, J. H. Miller, and J. Jiricny. 2002. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 21:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scharer, O. D., and J. Jiricny. 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays 23:270-281. [DOI] [PubMed] [Google Scholar]
- 48.Senturker, S., P. Auffret van der Kemp, H. J. You, P. W. Doetsch, M. Dizdaroglu, and S. Boiteux. 1998. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 26:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shapiro, R., and R. S. Klein. 1966. The deamination of cytidine and cytosine by acidic buffer solutions: mutagenic implications. Biochemistry 5:2358-2362. [DOI] [PubMed] [Google Scholar]
- 50.Sherman, F., and J. Hicks. 1991. Micromanipulation and dissection of asci. Academic, San Diego, Calif. [DOI] [PubMed]
- 51.Swanson, Rebecca L., Natalie J. Morey, Paul W. Doetsch, and Sue Jinks-Robertson. 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2929-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas, D., A. D. Scot, R. Barbey, M. Padula, and S. Boiteux. 1997. Inactivation of OGG1 increases the incidence of G. C→T. A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet. 254:171-178. [DOI] [PubMed] [Google Scholar]
- 53.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 54.Tomilin, N. V., and O. N. Aprelikova. 1989. Uracil-DNA glycosylases and DNA uracil repair. Int. Rev. Cytol. 114:125-179. [DOI] [PubMed] [Google Scholar]
- 55.Torres-Ramos, Carlos A., Robert E. Johnson, Louise Prakash, and Satya Prakash. 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unk, I., L. Haracska, R. E. Johnson, S. Prakash, and L. Prakash. 2000. Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem. 275:22427-22434. [DOI] [PubMed] [Google Scholar]
- 57.Unk, Ildiko, Lajos Haracska, Satya Prakash, and Louise Prakash. 2001. 3′-phosphodiesterase and 3′→5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell. Biol. 21:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Kemp, P. A., D. Thomas, R. Barbey, R. de Oliveira, and S. Boiteux. 1996. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7, 8-dihydro-8-oxoguanine and 2, 6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 93:5197-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao, W., B. L. Chow, M. Hanna, and P. W. Doetsch. 2001. Deletion of the MAG1 DNA glycosylase gene suppresses alkylation-induced killing and mutagenesis in yeast cells lacking AP endonucleases. Mutat. Res. 487:137-147. [DOI] [PubMed] [Google Scholar]
- 60.Yu, Sung-Lim, Sung-Kun Lee, Robert E. Johnson, Louise Prakash, and Satya Prakash. 2003. The stalling of transcription at abasic sites is highly mutagenic. Mol. Cell. Biol. 23:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]