Abstract
RanBP type proteins have been reported to increase the catalytic efficiency of the RanGAP-mediated GTPase reaction on Ran. Since the structure of the Ran-RanBP1-RanGAP complex showed RanBP1 to be located away from the active site, we reinvestigated the reaction using fluorescence spectroscopy under pre-steady-state conditions. We can show that RanBP1 indeed does not influence the rate-limiting step of the reaction, which is the cleavage of GTP and/or the release of product Pi. It does, however, influence the dynamics of the Ran-RanGAP interaction, its most dramatic effect being the 20-fold stimulation of the already very fast association reaction such that it is under diffusion control (4.5 × 108 M−1 s−1). Having established a valuable kinetic system for the interaction analysis, we also found, in contrast to previous findings, that the highly conserved acidic C-terminal end of RanGAP is not required for the switch-off reaction. Rather, genetic experiments in Saccharomyces cerevisiae demonstrate a profound effect of the acidic tail on microtubule organization during mitosis. We propose that the acidic tail of RanGAP is required for a process during mitosis.
The GTP-binding protein Ran, which belongs to the superfamily of Ras-like guanine-nucleotide binding proteins, is both a key regulator of nuclear transport (22, 44) and a marker of chromosome position in spindle formation and nuclear envelope assembly (29) of eukaryotic cells. RCC1, the nucleotide exchange factor for Ran, localizes to histones H2A and H2B (52) and consequently creates a high concentration of Ran-GTP in the vicinity of the chromatin (11, 54). RanGAP, the GTPase-activating protein specific for Ran, increases the hydrolysis rate 105-fold (35). During interphase, it is exclusively cytosolic (32) and, together with RCC1, creates a gradient of Ran-GTP across the nuclear membrane (34), which is the major driving force and determiner of directionality for nuclear transport (22, 51). During mitosis, Ran-GTP induces spindle formation (75) and nuclear envelope assembly (78, 79). For the latter, Ran-GTP hydrolysis accelerated by RanGAP is required (28), although it is not obvious how RanGAP is prevented from abolishing the Ran-GTP gradient. It is also not clear how the function of Ran in spindle and nuclear envelope formation can be applied to lower eukaryotes such as Saccharomyces cerevisiae and Schizosaccharomyces pombe, which undergo a closed mitosis, whereas vertebrates go through the process of disassembly and reassembly of the nuclear membrane (open mitosis).
The presence or absence of Ran-GTP in nucleosol or cytosol, respectively, is the determining factor for Ran-dependent cargo transport (22). Import receptors of the importin-β family bind to cargo in the absence of Ran-GTP and release it in the nucleus, where they bind with high affinity to Ran-GTP. In contrast, export cargo binding to export receptors (exportins) requires the presence of Ran-GTP. The dissociation of Ran-GTP complexes with importins and exportins on the cytoplasmic side of the nuclear pore complexes (NPCs) requires the action of Ran-binding proteins (RanBPs) (7, 20, 42) and is made irreversible by RanGAP-catalyzed hydrolysis. RanBP1 is a 23-kDa protein and contains a single conserved Ran-binding domain (RanBD) (14). Because of its small size, RanBP1 can diffuse into the nucleus. However, it is also actively transported into the cytoplasm due to an amino-terminal nuclear export sequence (59). RanBP2 is a 3,224-residue large protein attached to the cytosolic side of the NPC and comprises an amino-terminal leucine-rich region, four RanBDs, eight zinc-finger motifs, and a carboxy terminus with homology to cyclophilin (77). Whereas RanBP1 is an essential component of the nuclear transport system in eukaryotes, RanBP2 could not be identified in yeast organisms and is dispensable for transport in metazoans (74). RanBDs fulfill the definition for guanine-nucleotide binding effector proteins because they bind with a higher affinity to the GTP-bound (Kd ≈ 1 nM) than the GDP-bound (Kd ≈ 10 μM) form of Ran (40, 72). However, their effect on Ran is decisively different from that of importins and exportins, the true biological effectors. Whereas the latter inhibit both the intrinsic and RanGAP-stimulated hydrolysis of Ran-GTP (19, 23), RanBDs were shown to costimulate RanGAP-catalyzed hydrolysis (10).
RanGAP was first reported as a Saccharomyces cerevisiae mutant (rna1-1) defective in mRNA processing or transport (27, 31). Bischoff et al. purified RanGAP from HeLa cells (8) and found it to be homologous to RNA1 (6, 9). All known RanGAP proteins show a modular structure. They share a 330-to-350-residue large leucine-rich repeat (LRR) domain, followed by an acidic region of 35 to 50 residues. In higher eukaryotes, an additional domain localizes RanGAP to the NPC. In animals, this is achieved via modification with the ubiquitin-like protein SUMO-1 at the carboxy terminus of RanGAP (43, 45). The modified RanGAP then binds to RanBP2 (46). In plant cells, for which no homologue of RanBP2 is known, RanGAPs have an amino-terminal domain with a conserved WPP motif, which also leads to NPC localization (48, 60). RanGAP-stimulated Ran-GTP hydrolysis occurs either in the cytoplasm or on the cytoplasmic fibrils of the NPC but is not directly coupled to the translocation process itself. This explains why up to 1,000 translocations per NPC per s can be observed (57).
Spindle formation in mitotic cells is believed to be induced by a high local concentration of Ran-GTP, which in turn is created by the attachment of RCC1 (and possibly Ran itself) on chromatin. The mechanism of nuclear envelope formation may be due to a similar preferred RCC1 localization. In HeLa cells, RanGAP is localized to kinetochores and mitotic spindles and the localization is at least partially dependent on RanBP2 (33). Much less is known about the targeting of RanGAP in yeast, but it is mandatory for both yeast and mammalian cells that the localization should be different from that of RCC1 in order to establish and maintain the Ran-GTP gradient around chromatin.
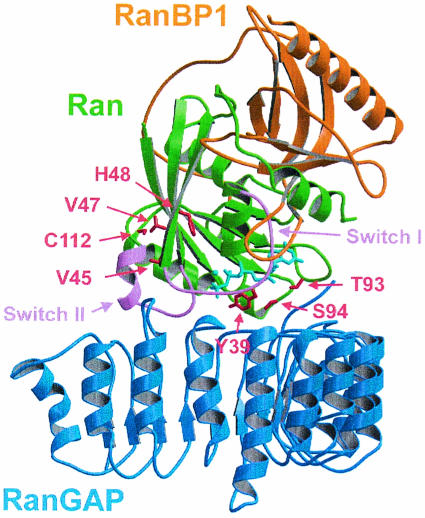
RanGAP-mediated GTP hydrolysis has many features that are distinct from the canonical RasGAP and RhoGAP reactions. While the maximal 105-fold stimulation of GTP hydrolysis is similar (2, 4, 16, 25, 30, 35), RanGAP does not employ an arginine finger to stabilize the transition state of the reaction (66). Instead, the crystal structure of Ran in complex with RanBP1 and RanGAP (Fig. 1) shows that Ran itself encodes the basic machinery for GTP hydrolysis and that the positioning of the catalytic glutamine residue appears to be essential for the reaction. Furthermore, while RanBDs were shown to costimulate RanGAP-catalyzed hydrolysis (10, 72), the structure of the ternary complex and multiple turnover kinetics suggested that the catalytic machinery was entirely located on Ran. Previously, it was suggested that the conserved C-terminal highly acidic region of RanGAP is required for Ran binding and for stimulation of GTP hydrolysis (26). Although this acidic region was not visible in the structure, it did not appear to participate in catalysis. We thus wanted to reinvestigate the contribution of this region, using the structure of the LRR domain as a guideline for a deletion construct missing the C terminus of RNA1. Previously, the interaction between RanGAP and Ran was investigated by multiple turnover kinetics using radioactive nucleotides to determine kcat and Km values (6, 35). In order to investigate individual steps of the RanGAP catalyzed reaction, we decided to employ pre-steady-state kinetics by using stopped-flow and fluorescence spectroscopy methods. Although the fluorescent 2′(3′)-O-(N-methylanthraniloyl) (mant) analogues of GDP and GTP have been used successfully for the Ras-RasGAP system (2-4, 15) and for Ran-nucleotide and Ran-effector interactions (36, 40, 72), they did not show fluorescence changes in the Ran-RanGAP reaction. In analogy to a previously developed assay to characterize interactions of Ras and Rap with their respective GAPs (38), a highly sensitive, fluorescence-based test system was established with 5-((((2-iodoacetyl) amino) ethyl) amino) naphthalene-1-sulfonic acid (1,5-IAEDANS)-labeled Ran. The modified proteins could be used to characterize the dynamics of the Ran-RanGAP interaction, to analyze equilibrium binding of Ran to RanGAP, and to follow the time course of Ran-GTP hydrolysis. We found that RanBP has an effect on the dynamics of the interaction, whereas the acidic region of RanGAP does not affect catalysis. Instead, deletion of the acidic region on RNA1 in Saccharomyces cerevisiae via homologous recombination shows a pleiotropic phenotype, with major defects in the spindle apparatus.
FIG. 1.
Structure of the Ran-GppNHp-RanBP1-RanGAP complex (66) indicating the positions of residues of Ran (green) selected for site-directed mutagenesis, the switch regions (pink), nucleotide (cyan), and binding partners RanBP1 (orange) and RanGAP (blue). The indicated amino acids were mutated to cysteine, C112 was mutated to Ser, and Y39, which is required for catalysis, was not mutated. The figure was prepared with MOLSCRIPT (39) and RASTER3D (49).
MATERIALS AND METHODS
Protein expression and modification.
RanGAP from Schizosaccharomyces pombe, human Ran, and human RanBP1 were prepared as described previously (30, 40, 66). Site-directed mutagenesis was performed according to the Quikchange protocol (Stratagene, Amsterdam, The Netherlands). Surface-exposed cysteine residues of Ran were labeled in thiol-free buffer (20 mM Tris-HCl [pH 7.5], 5% glycerol [vol/vol], 5 mM MgCl2, 2 mM ascorbate) with a 10-fold excess of 1,5-IAEDANS (Molecular Probes, Leiden, The Netherlands). The reaction was carried out at 4°C for 12 h and subsequently stopped by buffer exchange via ultrafiltration. Labeling specificity was confirmed by electrospray ionization-mass spectrometry. Protein concentrations were determined according to the method of Gill and von Hippel (21). Nucleotide exchange of Ran was performed by incubating Ran with catalytic amounts of Prp20, the RanGEF from Schizosaccharomyces pombe, which was purified in a single step as a glutathione S-transferase fusion protein on glutathione-Sepharose.
Fluorescence measurements and data analysis.
All fluorescence measurements were performed at 20°C in a buffer containing 100 mM Tris-HCl, pH 7.5, 5% (vol/vol) glycerol, 5 mM MgCl2, and 5 mM dithioerythritol. Equilibrium measurements were carried out in a FluoroMax II spectrofluorimeter (SPEX Instruments, Edison, N.J.) at an excitation wavelength of 350 nm and a band pass of 1.5 nm. For emission spectra, fluorescence emission was monitored with a band pass of 3 nm between 380 and 600 nm. For titrations, emission was followed at a fixed wavelength of 450 nm with a band pass of 3 nm. Stopped-flow experiments were performed on an SX18 MV Applied Photophysics apparatus (Leatherhead, Surrey, United Kingdom). The fluorescent probes were excited at 350 nm with a band pass of 6.4 nm, and emission at wavelengths of >408 or 470 nm was monitored with cutoff filters. Binding curves, titration experiments, and dissociation experiments were analyzed with Perl scripts and GRAFIT 5.0 (Erithacus Software, Surrey, United Kingdom).
Genetic techniques.
For functional analysis, the strain Saccharomyces cerevisiae FY1679 (76) was used. PCR-based gene insertion was performed as described previously (41) using plasmid pFA6a-kanMX4 (73). Primers were designed to allow for amplification of the Geneticin cassette of the plasmid and for subsequent homologous recombination. Transformation was carried out by the LiAc/PEG method (1). Specific insertion was confirmed by PCR.
Fluorescence microscopy.
Overnight cultures of Saccharomyces cerevisiae FY1679 were diluted to an A600 of 0.2 and grown in yeast-peptone-dextrose medium to early log phase (A600 = 0.6). Ten milliliters of the culture was then fixed by the addition of 4% (vol/vol) formaldehyde (5 min at room temperature [RT]), subjected to centrifugation (800 × g, 3 min, RT), and resuspended in phosphate-buffered saline with 4% (vol/vol) formaldehyde. After 1 h of incubation at RT, cells were washed three times in phosphate-buffered saline, and the resultant pellet was resuspended in buffer KP (50 mM potassium phosphate [pH 7], 1.2 M sorbitol, 1 mM MgCl2). Cell walls were digested with 40 μl of Zymolyase 100T and 5 μl of glucuronidase (15 min, 30°C) and washed two times in buffer KP. Twenty microliters of the resultant suspension of fixed cells was treated with monoclonal anti-α-tubulin antibodies or with DAPI (4′,6′-diamidino-2-phenylindole; Sigma-Aldrich) as described previously (17). Microscopy was carried out on an Axiophot microscope equipped with a ×100 Neofluar lens (Carl Zeiss Inc., Jena, Germany). Images were recorded with a mounted charge-coupled device camera, and Adobe Photoshop was used to optimize contrast and brightness.
RESULTS
Protein modification and characterization.
Based on the crystal structures of Ran-GDP (62), Ran-GppNHp-RanBD1 (71), and Ran-GppNHp-RanBP1-RanGAP (66), six positions of Ran were chosen for cysteine mutagenesis and fluorophore coupling (Fig. 1) and were analyzed for accessibility with NACCESS (University College, London, United Kingdom) (Table 1). Of the three cysteines in wild-type Ran, only cysteine 112 was exposed and was therefore mutated to serine for all following experiments. The modification of the conserved tyrosine at position 32 was successful for the Ras-RasGAP characterization (38), but the homologous position 39 of Ran was excluded due to the presumed catalytic role of this residue in Ran (66). Positions 45, 47, 48, 93, and 94 were mutated to cysteine. Coupling with 1,5-IAEDANS (Molecular Probes) was successful at positions 47 and 94. Ran C112S/V47C-EDANS (referred to as Ran47 for the rest of this report) was most sensitive to the nucleotide state and RanBP1 binding, whereas Ran C112S/S94C-EDANS (referred to as Ran94 for the rest of this report) showed the greatest sensitivity to RanGAP binding (Fig. 2 and 3). The nucleotide binding properties of Ran47 and Ran94 were not affected, as the mant nucleotide dissociation was very similar to that of wild-type Ran (data not shown).
TABLE 1.
Solvent accessibility of positions selected for mutagenesis
| Position | Solvent accessibilitya of:
|
|||||
|---|---|---|---|---|---|---|
| Ran-GDP
|
Ran-GppNHp-RanBD1
|
Ran-GppNHp- BP1-GAP
|
||||
| Absolute (Å2) | Relative (%) | Absolute (Å2) | Relative (%) | Absolute (Å2) | Relative (%) | |
| Tyr 39 | 112.14 | 63.2 | 75.81 | 42.7 | 2.23 | 1.3 |
| Val 45 | 0.00 | 0.0 | 16.86 | 14.8 | 19.72 | 17.3 |
| Val 47 | 15.02 | 13.1 | 43.41 | 38.0 | 48.55 | 42.5 |
| His 48 | 0.00 | 0.0 | 21.32 | 14.5 | 46.14 | 31.4 |
| Cys 85 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 |
| Thr 93 | 19.66 | 19.3 | 33.85 | 33.3 | 21.62 | 21.3 |
| Ser 94 | 32.6 | 41.7 | 26.61 | 34.1 | 3.69 | 4.7 |
| Cys 112 | 15.02 | 15.5 | 14.99 | 15.5 | 7.48 | 7.7 |
| Cys 120 | 0.00 | 0.0 | 0.00 | 0.0 | 0.00 | 0.0 |
Relative and absolute solvent accessibilities were calculated based on the crystal structures of Ran-GDP (62), Ran-GppNHp-RanBD1 (71), and the ternary complex Ran-GppNHp-RanBP1-RanGAP (66) using the program NACCESS. Shown is the accessible surface of a given side chain in absolute and relative measures, the latter based on comparison with an Ala-X-Ala tripeptide.
FIG. 2.
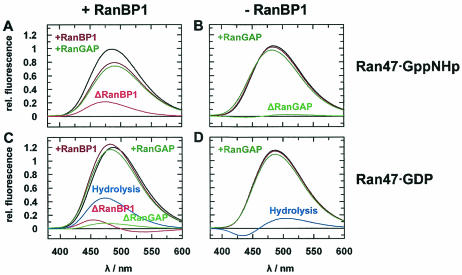
Fluorescence emission spectra of Ran47. Spectra were recorded at 20°C in 0.1 M Tris-HCl (pH 7.5), 2 mM DTE, 2 mM MgCl2, and 5% glycerol and were normalized to the emission maximum of 0.5 μM Ran47-GppNHp. The excitation wavelength was 350 nm, with 5-nm slits. The emission wavelength was 380 to 600 nm, integrated in steps of 1 nm with a 1-s integration time and 5-nm slits. Starting with Ran (black curve), interaction partners were added (green curve, RanGAP; brown curve, RanBP1) and difference spectra were calculated (red curve, after the addition of RanBP1; light green, after the addition of RanGAP; blue curve, difference spectra calculated from the curves for Ran-GppNHp and Ran-GDP). The following were added to 0.5 μM Ran-GppNHp (A and B) or 0.5 μM Ran-GDP (C and D): 1 μM (A) or 10 μM (C) RanBP1 and 10 μM RanGAP (A to D) (final concentrations).
FIG. 3.
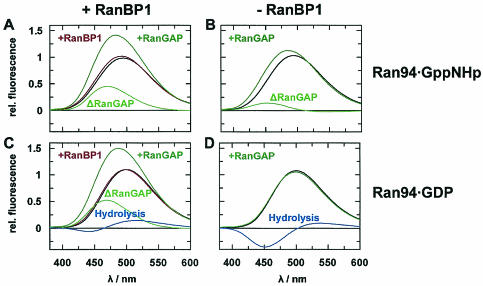
Fluorescence emission spectra of Ran94. Spectra were normalized to the emission maximum of 0.5 μM Ran94-GppNHp. For conditions and protein concentrations, see the legend for Fig. 2.
Interaction of Ran with RanGAP.
Ran94 was used to study RanBP1 and Ran nucleotide-dependent binding of RanGAP (Fig. 4A and B). For Ran bound to the nonhydrolyzable analog GppNHp, RanBP1 binding increases Ran's affinity for RanGAP 3.5-fold, from 7 to 2 μM (Kd). In the GDP form, Ran binds weakly to RanGAP (Kd, approximately 100 μM), and its affinity to RanGAP is dramatically increased upon RanBP1 binding (Kd = 2 μM). Although the affinity of RanGAP for the Ran-GTP-RanBP1 complex (at least with GTP instead of GppNHp) is higher (see below), Nevertheless RanBP1 induces a more Ran-GTP-like conformation on Ran-GDP. This is similar to the interaction of Ran-GDP with transport receptors which is also induced by RanBP-type proteins (13, 72).
FIG. 4.
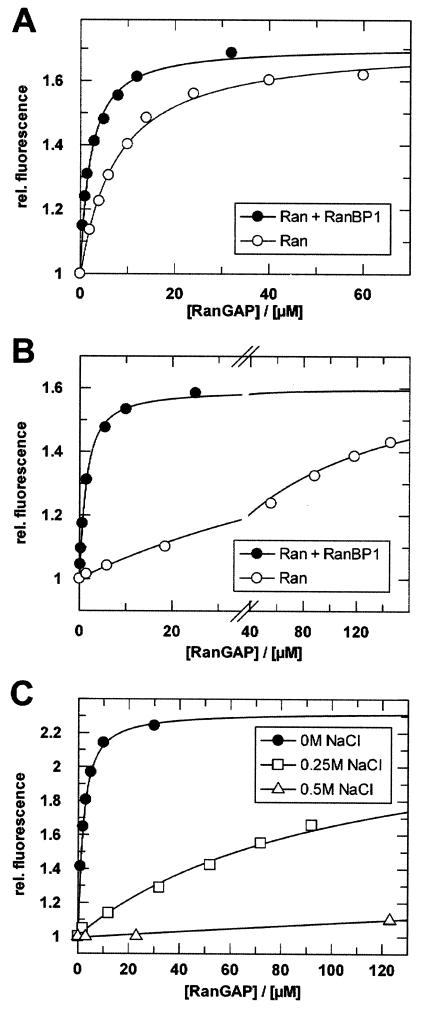
Titrations of Ran with RanGAP. (A) Ran94-GppNHp (0.5 μM) was titrated in the absence (○) or presence (•) of 1 μM RanBP1 with Schizosaccharomyces pombe RanGAP (Kd, 7 or 2 μM, respectively). (B) Ran94-GDP (0.75 μM) was titrated in the absence (○) or presence (•) of 40 μM RanBP1 with Schizosaccharomyces pombe RanGAP (Kd, ∼100 or 2 μM, respectively). (C) Salt dependence of the Ran-RanGAP interaction. Ran94-GppNHp-RanBP1 (0.75 μM) was titrated at 8°C with Schizosaccharomyces pombe RanGAP in buffer (as described above) containing 0 M NaCl (•) (Kd, 1 μM), 250 mM NaCl (□) (Kd, 220 μM), or 500 mM NaCl (▵) (Kd, ≥1 mM). Fluorescence spectra were recorded, and calculation of Kd values was done as described in Materials and Methods. Note that the affinities (Kd) of RanBP1 for Ran-GppNHp and Ran-GDP are 1 nM and 10 μM, respectively (40).
In the crystal structure of the Ran-RanBP1-RanGAP complex, it was found that the protein interface between Ran and RanGAP is polar and dominated by charge-charge interactions (66). Supporting this hypothesis, we find that the interaction is strongly salt dependent and can be easily shielded by NaCl. An increase in the salt concentration in the standard buffer (containing 100 mM Tris, equivalent in ion strength to approximately 80 mM NaCl) to 250 mM NaCl strongly decreases the affinity (Kd ≈ 220 μM) (Fig. 4C).
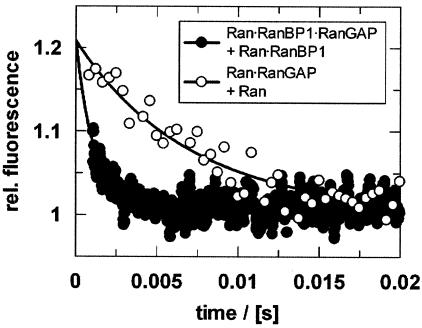
To further investigate the dynamic nature of the interaction, the dissociation rate for the Ran-RanGAP complex was determined by competition experiments. This approach was feasible for both the binary Ran-GppNHp-RanGAP and the ternary Ran-GppNHp-RanBP1-RanGAP complexes, because both RanBP1 and nucleotide dissociate slowly from Ran (36, 72), while RanGAP dissociation is fast. A fluorescent complex of Ran94-GppNHp-RanGAP was preformed, and a 50-fold excess of unlabeled Ran-GppNHp was added in the stopped-flow apparatus. The decrease in fluorescence was fitted with a single exponential and reflected the koff value (150 s−1). If RanBP1 is present in the experiment, the dissociation rate constant is about sixfold higher (880 s−1) (Fig. 5).
FIG. 5.
Dissociation of RanGAP from the binary Ran-GppNHp-RanGAP (○) or the ternary Ran-GppNHp-RanBP1-RanGAP (•) complex after adding excess Ran or Ran-RanBP1 complex, respectively. To observe the Ran-RanGAP dissociation, 4 μM Ran94-GppNHp-RanGAP was mixed at a 1:1 ratio (vol/vol) with 80 μM Ran-GppNHp (○) (koff = 150 s−1). To determine the dissociation constant in the presence of RanBP1, 1.5 μM Ran94-GppNHp-RanBP1-RanGAP was mixed at a 1:1 ratio (vol/vol) with 80 μM Ran-GppNHp-RanBP1 (•) (koff = 880 s−1). Measurements were performed by stopped-flow experiments and analyzed as described in Materials and Methods.
Calculating kon from the equilibrium affinity and dissociation rate constants yields association constants of 2.1 × 107 M−1 s−1 in the absence of RanBP1 and a 20-fold increase to 4.5 × 108 M−1 s−1 in the presence of RanBP1. These rates are clearly above typical values for protein-protein interactions (105 to 106 M−1 s−1) and are indicative of an electrostatically enhanced association (67). Direct measurements of the association rates were not feasible due to the high koff value and the micromolar Kd.
Ras-GDP can be induced to bind to RasGAP by adding aluminum fluoride, and this is dependent on the presence of an arginine finger in RasGAP (3, 50). Similar complexes have also been observed for other Ras-like GTP-binding proteins, such as Rho (3) and Rap1, which does not use an arginine for GTPase stimulation (38). In contrast, and for reasons that are unclear to us, Ran94-GDP shows no observable increase in binding affinity for RanGAP in the presence of fluoroaluminate (AlFx). In the presence of RanBP, which already induces tight binding between Ran47-GDP and RanGAP, there is no observable fluorescence change upon binding of AlFx, although we have shown that AlFx binds under these conditions (66).
RanGAP-catalyzed hydrolysis of Ran-GTP.
The RanGAP-catalyzed hydrolysis of Ran-GTP was monitored by single-turnover stopped-flow reactions in order to learn more about individual and possibly rate-limiting steps in the overall reaction and whether or not RanBP-type proteins have a modulatory effect on such steps. For the interaction of Ras with its cognate RasGAP proteins, different kinetic mechanisms have been observed. For p120GAP, very fast association of substrates and release of products were found, and the chemical reaction is believed to be rate limiting (2, 38, 65). A completely different situation is observed during neurofibromin (NF1)-catalyzed hydrolysis of Ras-GTP, during which binding is also fast, but the overall reaction is limited by either the phosphate or Ras-GDP release step (5, 38). Here, measurements were done with Ran47 because this mutant showed the largest difference in fluorescence between the GDP- and GTP-bound states.
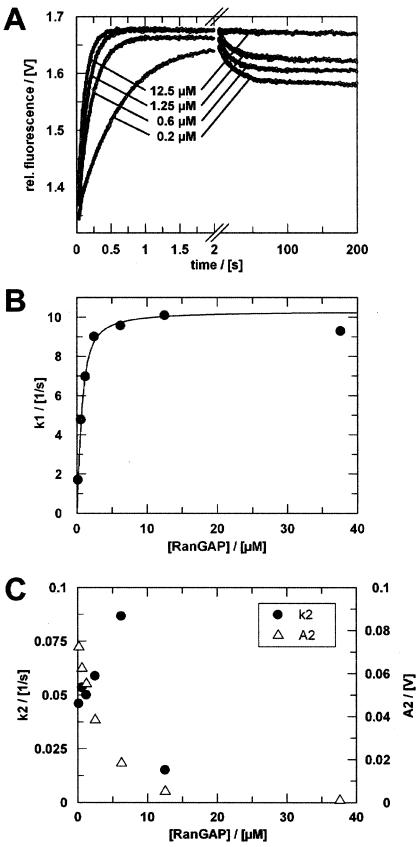
Mixing 1.5 μM Ran47-GTP-RanBP1 with an excess of 12.5 μM RanGAP showed a monophasic fluorescence increase (Fig. 6A). At lower concentrations of RanGAP, a second, slower phase was observed. For all concentrations, a single- or double-exponential function (Fig. 6B and C) can describe the resultant fluorescence transients. The fast, initial phase is characteristic of saturation kinetics and reaches its maximum rate at about 2.5 μM RanGAP. The corresponding rate of 10 s−1 is similar to the kcat that was observed with simple phosphate detection multiple-turnover assays in the absence of RanBP1 (30, 35) and suggests that this phase reflects the hydrolysis reaction and/or the phosphate release. This also supports the notion that the reporter group and the V47C mutation do not affect the rate-limiting step of the GTPase reaction by Ran.
FIG. 6.
Single-turnover measurements of RanGAP-catalyzed hydrolysis of Ran-GTP in the presence of RanBP1. (A) Ran47-GTP-RanBP1 (1.5 μM) was mixed with different concentrations of RanGAP (only four curves are shown) in the stopped-flow apparatus. Fluorescence transients were fitted as double exponentials. (B) Rate constants (•) of the first phase plotted against RanGAP concentration. Assuming a simple saturation model (16) and given that koff ≫ kcat, observed rate constants can be fitted to a quadratic equation (Kdapp = 0.3 μM; kcat = 10.3 s−1). (C) Rate constants (•) and amplitudes (▵) of the second phase (k2 ≈ 0.05 s−1) plotted against RanGAP concentration. Measurements were performed and analyzed as described in Materials and Methods.
Assuming a simple saturation model and given that koff ≫ kcat, the observed rate constants can be fitted to a quadratic equation to yield the Kd (16). Fitting the initial phase according to this formula yields a Kd of ≈0.3 μM. This value corresponds well to the previously determined Michaelis constant of 0.4 μM in the absence of RanBP1 (30, 35) and is lower than the Kd between Ran-GppNHp-RanBP1 and RanGAP determined by fluorescence equilibrium measurements (Fig. 4), which is approximately sixfold higher. Presumably, the difference arises because GppNHp is not a perfect analog of GTP and leads to a higher dissociation rate.
The second phase of the fluorescence transient disappears when the RanGAP concentration is increased (Fig. 6C). Its amplitude curve has the shape of a titration curve and probably reflects the binding equilibrium of Ran-GDP-RanBP1, which is pushed towards a ternary complex at high concentrations of RanGAP and is in the same range as the 2-μM dissociation constant determined independently. While the rate constant for the second step, described as k2, can be determined with reasonable accuracy (for [RanGAP] of <5 μM), it does not change with the RanGAP concentration, but is constant at 0.05 s−1.
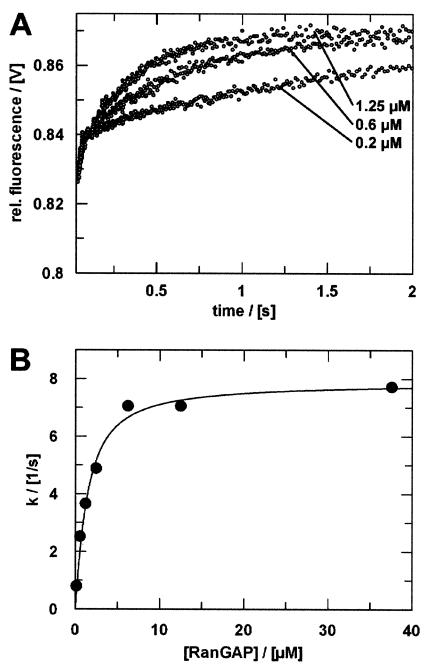
The transient obtained in the reaction between Ran-GTP and RanGAP (no RanBP) can be described by a single exponential (Fig. 7). The time scale corresponds to the initial, fast phase observed in the presence of RanBP1. As before, a plot of the rate constants versus [RanGAP] shows saturation kinetics. The maximum rate of 7.9 s−1 is slightly lower than that in the presence of RanBP1. The half-maximal saturation of 1 μM [RanGAP] is again in good agreement with the Kd and is higher than the Km determined previously (30, 35). As with Ran-GppNHp, the affinity of Ran-GTP is about 3.3-fold higher in the presence of RanBP1.
FIG. 7.
RanGAP-catalyzed hydrolysis of Ran-GTP. (A) Ran47-GTP (1.5 μM) was mixed at a 1:1 ratio (vol/vol) with RanGAP in the stopped-flow apparatus (only three curves are shown). The fluorescence transients were fitted as single exponentials. (B) Plot of rate constants (•) versus [RanGAP]. Assuming a simple saturation model (16) and given that koff ≫ kcat, the observed rate constants can be fitted to a quadratic equation (Kd = 1.0 μM; kcat = 7.9 s−1). Measurements were performed and analyzed as described in Materials and Methods.
Interaction of Ran with RanBP1.
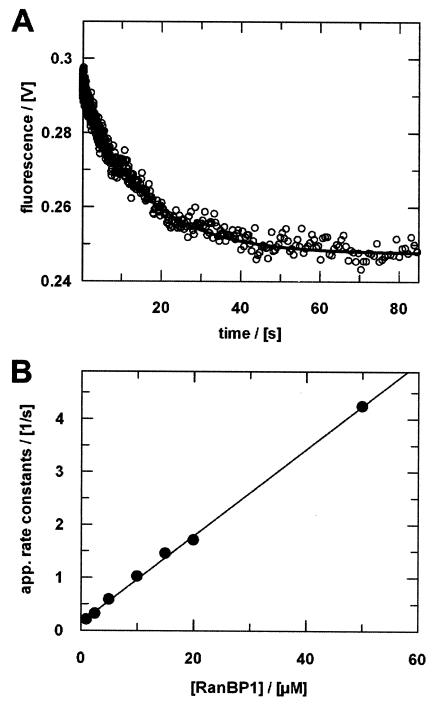
Interactions between Ran-GppNHp and the RanBDs of RanBPs have been characterized previously by use of fluorescent mant nucleotides (40, 72). However, the dissociation rate of Ran-GDP from the complex with RanBP1 could only be approximated from a plot of apparent association rates. We thus characterized the dynamics of the Ran-GDP-RanBP1 interaction both directly and indirectly via kobs using Ran94-GDP (Fig. 8) and Ran47-GDP (not shown). The dissociation rates of 0.05 s−1 (for Ran47-GDP) or 0.06 s−1 (for Ran94-GDP) thus obtained are significantly smaller than the values (0.2 to 1 s−1) determined previously (40). However, the value for Ran47 nicely matches the observed slow rate in the hydrolysis experiments with RanBP1 (Fig. 6) and supports the notion that the second step is the dissociation of RanBP1 from Ran-GDP. Further support of this conclusion comes from the observations that the second phase is only observed in the presence of RanBP1 and that its rate is independent of the RanGAP concentration. The association rate for the Ran47-RanBP1 interaction determined here (8 × 104 M−1 s−1) is in agreement with previous data obtained with mant nucleotide (40, 72).
FIG. 8.
Ran-RanBP1 interaction. (A) Dissociation of the Ran-GDP-RanBP1 complex after adding excess Ran-GDP. Ran94-GDP-RanBP1 (6 μM) was mixed at a 1:1 ratio (vol/vol) with 250 μM RanBP1 in the stopped-flow apparatus. Fitting the curve to a single exponential yields a dissociation rate (koff) of 0.06 s−1. (B) Association of Ran-GDP and RanBP1. Ran94-GDP (1 μM final concentration) was mixed with increasing concentrations of RanBP1 in the stopped-flow apparatus. Kinetics were analyzed assuming pseudo-first-order conditions. The apparent rate constants were plotted against [RanBP1] to yield the results kon = 0.08 μM−1 s−1 (slope) and koff = 0.1 s−1 (intercept). Measurements were performed and analyzed as described in Materials and Methods.
The acidic region does not contribute to catalysis.
Having established a reliable kinetic system for the investigation of the RanGTPase reaction, we set out to investigate the role of the RanGAP C terminus. In all known RanGAP proteins, the LRR region is followed by a region of approximately 40 amino acids (Fig. 9), whose extreme acidity but not its sequence per se is conserved. In mammalian RanGAPs, it consists almost entirely of glutamate and aspartate. Haberland et al. (26), using C-terminal deletion mutants of Schizosaccharomyces pombe and Saccharomyces cerevisiae RanGAP and phosphate release assays, suggested that the C terminus is required for GAP activity and Ran binding. It should be noted that the structure of RanGAP was not available at that time (30) and that those deletion constructs were based on incorrectly assigned LRRs which in turn were based on the structure of RNase A inhibitor (37). The crystal structure showed RanGAP to contain 11 LRRs. Thus, the partial deletion of the last LRR in RanGAPΔC330 and RanGAPΔ341 may have destabilized the proteins.
FIG. 9.
Acidic carboxy-terminal region of RanGAP proteins. Aspartic acid and glutamic acid residues are highlighted (black). The first two letters of the sequence names indicate the organism as follows: Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Mf, Macaca fascicularis; Mm, Mus musculus; Xl, Xenopus laevis; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Ms, Medicago sativa; Os, Oryza sativa.
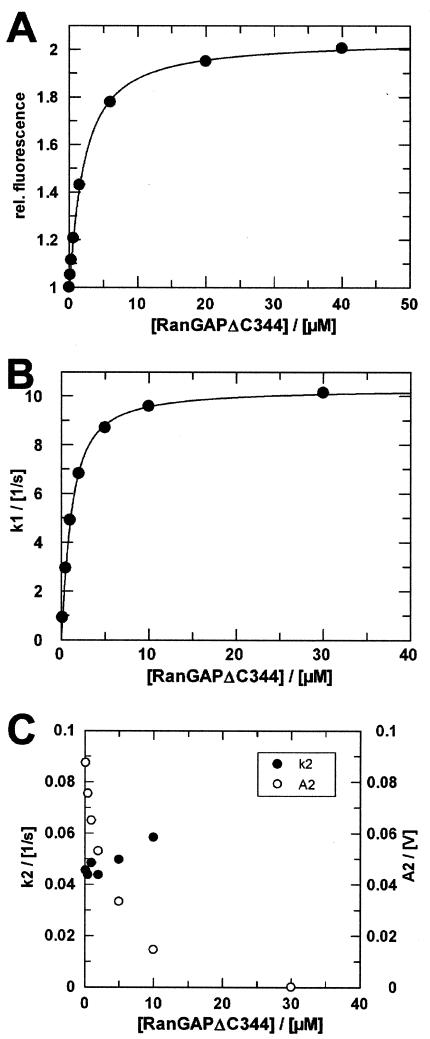
We constructed a deletion mutant of Schizosaccharomyces pombe RanGAP comprising residues 1 to 344 (RanGAPΔC344 from here on). Titration of Ran-GppNHp-RanBP1 with RanGAPΔC344 shows that the deletion mutant has the same affinity as the wild-type RanGAP protein (Fig. 10A) and does not change the dynamics of the interaction (data not shown). Using stopped-flow kinetic experiments as described above (see Fig. 6), we investigated the RanGAP-mediated Ran-GTPase reaction with the deletion construct. Here again, the fluorescent transient and the RanGAP concentration dependence of the first and the second rate are indistinguishable from those for wild-type RanGAP (Fig. 10B and C), indicating that at least the rate-limiting step(s) of the GAP-stimulated hydrolysis reaction is not influenced by the acidic C-terminal tail.
FIG. 10.
Titrations of Ran with RanGAPΔC344. (A) Ran94-GppNHp-RanBP1 (0.5 μM) was titrated with Schizosaccharomyces pombe RanGAPΔC344 (Kd = 1.7 μM). Buffer conditions and instrument settings were as described for Fig. 4B and C. Single-turnover measurements of RanGAPΔC344 catalyzed hydrolysis of Ran-GTP in the presence of RanBP1. Ran47-GTP-RanBP1 (1.5 μM) was mixed with different concentrations of RanGAPΔC344 in the stopped-flow apparatus. Fluorescence transients were fitted as double exponentials. (B) Rate constants (•) of the first phase plotted against RanGAP concentration. Assuming a simple saturation model and assuming that koff ≫ kcat, observed rate constants can be fitted to a quadratic equation (Kdapp = 0.5 μM; kcat = 10.5 s−1). (C) Rate constants (•) and amplitudes (○) of the second phase plotted against RanGAP concentration (k2 ≈ 0.05 s−1). Measurements were performed and analyzed as described in Materials and Methods.
Deletion of RanGAP's acidic tail leads to defects in spindle formation.
In order to obtain further information about the function of the acidic region, we deleted it from Saccharomyces cerevisiae RanGAP (residues 363 to 407) by homologous recombination and studied the resultant phenotype in vivo. A similar experiment was done previously (70). However, in this work amino acids 359 to 397 of Saccharomyces cerevisiae RanGAP were deleted, with residues 359 to 362 being part of the LRR domain, and again, deletion of these residues presumably renders RanGAP unstable. In addition, RanGAP's function for nuclear transport was not known at this time. It was believed to participate in RNA processing and transport (31) and, consequently, Traglia et al. looked for phenotypes of RNA metabolism (70).
Heterozygous diploid RanGAPΔC362 transformants do not show any evident phenotype. The effect of the deletion was then characterized by random spore analysis. Two equally strong populations of haploid cells were identified, and their genotypes were checked by PCR. The fast-growing population represented the wild type, whereas the slow-growing population represented haploid cells with the desired deletion in RanGAP's carboxy-terminal region.
Under the light microscope, mutant cells showed a pleiotropic phenotype (Fig. 11). Many of the cells were larger than wild-type cells and had unusual shapes. A large number of cells was found in various stages of unsuccessful attempts to complete mitosis. Chromatin staining with DAPI yielded further information about the mutant phenotypes (Fig. 12A) and helped to quantify the results (Table 2). Compared to wild-type cells in mitosis, a significant number of the cells (∼5%) was enlarged, and some of the cells, predominantly the larger ones, were anucleate (∼5%). In rare cases, cells with two nuclei were observed (∼0.2%). Approximately 17% of the cells were found in mitosis, which is significantly reduced compared to wild-type cells during exponential growth (∼30%). It was not clear whether all of the mitotic cells would have completed mitosis. Rather, it appeared that many cells were arrested in mitosis. For example, cells 3 and 4 (Fig. 12A) show an uneven separation of chromatin. The mother and bud marked “4” were able to separate but are still connected by chromatin. If in these cases septum formation was successful, the resultant cells were probably not provided with a complete set of chromosomes or a nucleus.
FIG. 11.
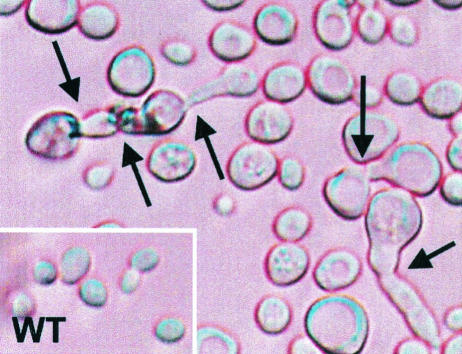
Haploid cells of Saccharomyces cerevisiae FY1679 rangapΔC362 under a light microscope (100-fold primary magnification). Arrows indicate mutant cells that were unable to complete mitosis. WT, wild type.
FIG. 12.
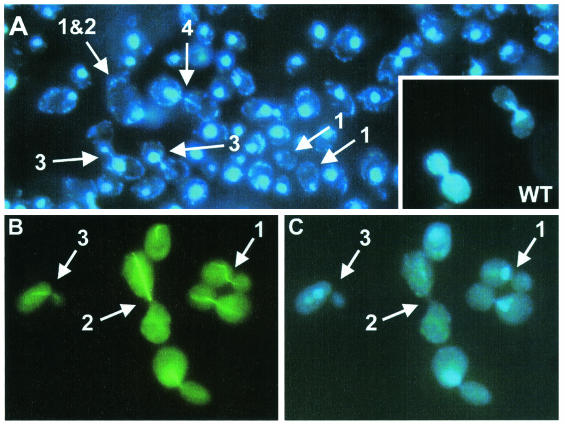
Chromatin structure. (A) Haploid Saccharomyces cerevisiae FY1679 rangapΔC362 cells stained with DAPI. Numbered arrows denote different phenotypes as follows: 1, cell without nucleus; 2, very large cell; 3, mitotic cell which apparently has arrested in mitosis; 4, mitotic cell which failed in separating the chromatin evenly between the mother and bud cells. For comparison, dividing wild-type cells (WT) are shown in the lower right corner. Pictures were taken with an Axiophot fluorescence microscope (Zeiss) at 40-fold primary magnification. (B and C) α-Tubulin and chromatin structure. In haploid Saccharomyces cerevisiae FY1679 rangapΔC362 cells, α-tubulin was stained with antibodies (fluorescein isothiocyanate-labeled secondary antibody) (B) and chromatin was stained with DAPI (C). Numbered arrows denote different phenotypes as follows: 1, wild type-like cell division; 2 and 3, malformed microtubule and nucleus structures. Pictures were taken with an Axiophot fluorescence microscope (Zeiss) at 100-fold primary magnification.
TABLE 2.
Phenotypes of haploid Saccharomyces cerevisiae FY1679 rangapΔC362 cells
| Phenotype | Absolute no. of cells | % of cells |
|---|---|---|
| Cells in mitosis | 247 | 17 |
| Large cellsa | 76 | 5 |
| Cells without nucleusb | 78 | 5 |
| Cells with two nucleib | 3 | 0.2 |
| Wild-type-like cells | 1,106 | 77 |
| Total | 1,429 | 100 |
Cells were compared to wild-type cells in mitosis.
Many of these cells were dividing or very large and were also counted in the corresponding categories. Thus, counts in all categories do not add up to the total number of cells.
As the observed phenotypes pointed toward defects in mitosis and in the cytoskeleton dynamics, tubulin and actin were stained and cells were analyzed. Whereas actin staining appeared normal (not shown), tubulin staining showed various malformations of the microtubules (Fig. 12B) and irregular chromatin distribution (Fig. 12C). The cells labeled “1” show one example of wild type-like microtubules. However, these cells are also affected by the mutation, because the separation of the nucleus between mother and bud is not properly performed. While the large bud size clearly indicates that the cell is in the middle of anaphase, the location of the nucleus at the entrance of the bud neck points toward an earlier state in the cell cycle. A different situation can be observed for cells 2 and 3, which, according to the sizes of the cells, are in anaphase. Their microtubules, however, span only one cell volume, most likely the one of the mother cell.
DISCUSSION
Effect of RanBP1 on RanGAP-mediated GTPase activity.
The fluorescence-based test system employed for this study has allowed us to reevaluate the role of RanBP1. Previously, it was observed that under nonsaturating conditions RanBP1 increases catalytic efficiency (kcat/Km) by 1 order of magnitude, although the nature of the effect was not elucidated. These findings, however, were obtained with a filter binding assay of low time resolution, and the authors describe problems with Ran stability (10). Here, by using equilibrium and single-turnover studies, we compare the kinetic parameters of the RanGAP-stimulated hydrolysis reaction with and without RanBP1 and show that the catalytic efficiency is increased by a factor of 4 and that this is almost entirely due to a change in Kd. The most dramatic consequence of RanBP is the 20-fold stimulation of the association rate with RanGAP.
Typical association rates of protein-protein complexes are on the order of 105 to 106 M−1 s−1 (63), and in very rare cases rates of up to 109 M−1 s−1 have been observed (64). For very fast association rates, strong electrostatic complementarity and steering are required. Apparently, this is true for the interaction surface of Ran and RanGAP, which is dominated by large, complementary electrostatic potentials (66). Based on the fluorescence-based test system presented here, we confirmed that Ran and RanGAP indeed interact on a very fast time scale. Ran-GppNHp-RanBP1 and RanGAP bind with an association constant (kon) of 4.5 × 108 M−1 s−1 and dissociate quickly at 900 s−1 (koff). Thus, the interaction is highly dynamic and has a micromolar affinity (Kd) of ≈2 μM. The increase in affinity due to RanBP1 is 3- to 4-fold and is not simply a decrease in the dissociation and/or an increase in the association rate, as expected, but rather an increase in both the dissociation (6-fold) and association (20-fold) rates, with a net positive effect on affinity. Although the kinetics of the interaction of Ran-GTP with RanGAP have not been determined directly, the similar increase in affinity from 0.3 to 1 μM suggests a corresponding increase in association and dissociation rate constants.
Thus, the major effect of RanBP-type proteins is to increase the dynamics of the interaction between Ran and RanGAP, while the chemical reaction itself is barely affected and increases from 7.9 to 10 s−1, which might not be significant. Although the structural basis for an increase in rates can only be speculated about, we assume that it is due to the localization of the C-terminal end of Ran. This C-terminal extension is tightly bound to the body of the G domain in Ran-GDP (62) and is detached from it and wrapped around RanBP in Ran-GppNHp-RanBD or in the ternary complex shown in Fig. 1. Although the structure of Ran-GTP alone is not available, we can assume from a number of biochemical studies (summarized in reference 71) that the C-terminal end is not stably bound to the G domain and may thus prevent binding of RanGAP (or transport factors). Removal of the C-terminal extension may thus expose more of the polar surface and/or present an additional interaction surface to RanGAP and thereby increase the rate of association and dissociation reactions.
A fast acceleration caused by favorable electrostatic interactions is typically found in cases where speed is crucial for biological function (63). One example is protease inhibitors, for which an accelerated association leads to increased efficiency. Among these inhibitors, very high association rates of up to 5 × 109 M−1 s−1 were observed (64). Highly dynamic interactions have also been found in signal transduction for the binding of Ras to effector molecules. In the case of the interaction between Ras-GppNHp and RafRBD, association and dissociation rate constants of 5 × 107 M−1 s−1 and 7.7 s−1, respectively, have been observed (69), suggesting that a single Ras-GTP molecule is able to activate multiple effector molecules and that even in the presence of an effector(s), Ras can be efficiently downregulated by GAPs.
In the context of nuclear transport, an optimization for speed appears to be favorable for biological function. When Ran-GTP-receptor complexes exit the nucleus, they should be unloaded quickly. This is supported by the fast association between Ran-GTP and RanGAP, the even faster association in the presence of RanBP1 and RanBP2, and the catalytic activity of RanGAP. According to the accepted model, Ran-GTP hydrolysis and translocation through the NPC are not directly coupled (58). While the localization of RanGAP at the NPCs of higher eukaryotes suggests that Ran-GTP hydrolysis occurs in the vicinity of the cytoplasmic filaments, it is clear from measurements (57) and simulations (24, 68) of nuclear transport kinetics that the observed fast translocation rates are only possible if several RanGAP molecules per NPC are acting on Ran-GTP molecules exiting the nucleus and if catalysis by RanGAP is not rate limiting.
Kinetic scheme for Ran inactivation.
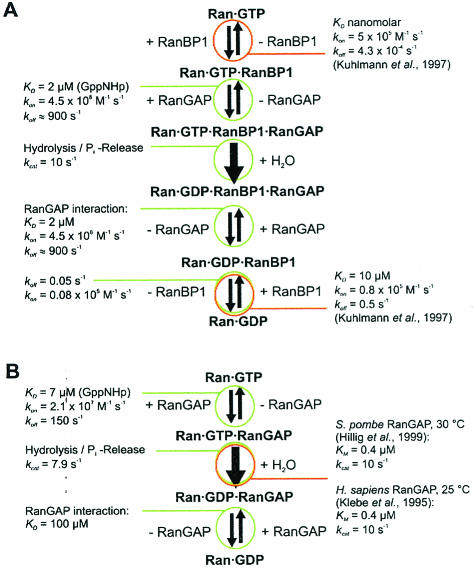
Binding and association constants determined in this study can be used to derive a kinetic model for the interactions between Ran, RanBP1, and RanGAP (Fig. 13). Based on this model, simulations can be performed that show the Ran-GTP gradient and the flux of Ran and receptor molecules between nucleosol and cytosol (24, 68).
FIG. 13.
Kinetic models for RanGAP-catalyzed Ran-GTP hydrolysis and corresponding rate and equilibrium constants. (A) With RanBP1. (B) Without RanBP1. From the data presented here, it cannot be determined whether the rate limiting step kcat denotes the actual cleavage reaction or the Pi release step and whether or not Pi is released before or after RanGAP dissociation.
The model for the inactivation of Ran-GTP by combined action of RanBP1 and RanGAP is drawn based on the assumption that Ran-GTP binding to RanBP1 precedes binding to RanGAP (Fig. 13A). This model is close to the situation in vivo, because RanBP1 (or RanBP2) supports the dissociation of Ran-GTP-receptor complexes (7, 20, 72). Once the receptor complex is dissociated, the very slow dissociation rate of the Ran-GTP-RanBP1 complex (koff ≈ 4 × 10−4 s−1) (40) and the very fast association rate shown above induce ternary Ran-GTP-RanBP1-RanGAP complex formation. After the quasi-irreversible hydrolysis step, phosphate (Pi), RanGAP, and RanBP1 can dissociate from the Ran-GDP product. Rate constants determined in this work suggest that RanGAP dissociates first, and then RanBP1 dissociates. Previous work on different RasGAPs and RapGAP has shown that the chemical step and/or the Pi release step can be fully or partially rate limiting (5, 38, 53). Based on the data presented here, it cannot be decided at which point Pi leaves the complex and whether hydrolysis or Pi release is rate limiting. Time-resolved Fourier-transform infrared spectroscopy experiments are being undertaken to resolve this issue.
RanGAP has a function during mitosis.
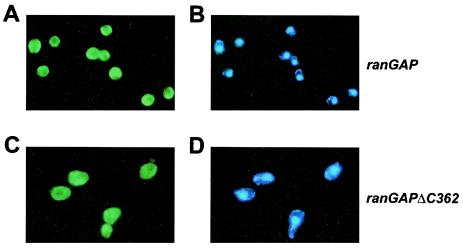
Contrary to previous findings of Haberland et al. (26), we find that RanGAP's acidic carboxy-terminal region does not play a measurable role in the Ran-RanGAP reaction, since the deletion mutant RanGAPΔC344 is fully functional with respect to Ran-GTP binding and stimulation of Ran-GTP hydrolysis. Presumably, the acidic motif does not play any role in interphase cells, where the main function of Ran is in the transport of cargo in and out of the nucleus. We can also show that the deletion of the acidic region in RanGAP does not affect its cytoplasmic localization in yeast cells (Fig. 14). However, the phenotype of deletion of RanGAP's acidic region suggests that, apart from regulating nuclear transport, RanGAP may play an important role in mitosis and mitotic spindle formation, at least in Saccharomyces cerevisiae.
FIG. 14.
Localization of RanGAP (A and C) or chromatin (B and D) in either Saccharomyces cerevisiae wild-type (A and B) or rangapΔC362 (C and D) cells. RanGAP staining was performed using primary anti-RanGAP antibodies and secondary fluorescein isothiocyanate-labeled antibodies. Chromatin was stained with DAPI.
Mitotic spindle defects were not yet observed in RanGAP mutants from Schizosaccharomyces pombe and Saccharomyces cerevisiae. In contrast, a Schizosaccharomyces pombe Ran point mutant, spi1-25, as well as eight different nuclear transport-competent Schizosaccharomyces pombe RanGEF mutants (pim1) influence microtubule integrity independently of the nuclear transport function (18, 61). Aberrant spindle structures and improperly separated chromosomes in these mutants resemble the phenotype which we observed for RanGAPΔC362. Overproduction of Rna1 (Saccharomyces cerevisiae RanGAP) from the strong GAL1 promoter causes chromosome instability (55), which is consistent with uneven chromatin separation of the RanGAPΔC mutant.
Previously, it was shown in Schizosaccharomyces pombe that disturbing the appropriate balance of Ran-GDP and Ran-GTP by inactivation or overexpression of RanGEF, RanGAP, and RanBP1 has severe effects on the passage of cells through mitosis (47, 61). It has been argued that it is difficult to decide whether these effects are directly related to a role of Ran in mitosis or caused indirectly due to a defect in nuclear transport. Here we show that deletion of the C-terminal end of Rna1 from Saccharomyces cerevisiae seems to uncouple the transport effect from the mitotic effects of RanGAP.
In metaphase of HeLa cells, RanGAP localizes at the kinetochores and in the vicinity of the spindle poles (33). Chromosome segregation appears to be guided by a gradient of Ran-GTP that possibly spans between RanGEF and RanGAP. In dividing tobacco cells, RanGAP colocalizes with tubulin and consequently must be close to the spindle apparatus during all steps of mitosis (56). It is also known that hydrolysis of Ran-GTP is required for nuclear envelope formation (12, 28). This implies that RanGAP is involved in the process, since the intrinsic hydrolysis of Ran is very slow. Because of the differences between open mitosis in higher eukaryotes and closed mitosis in lower eukaryotes, it is not clear how these findings can be applied to yeast organisms, which do not have an open mitosis. It has, however, been shown that Ran acts on mitotic microtubules in Schizosaccharomyces pombe and that the imbalance of Ran-nucleotide has severe effects on passage through mitosis (18, 47, 61).
It is generally agreed that a high local concentration of Ran-GTP is required for the mitotic effects of Ran and that attachment of the RanGEF RCC1 to chromatin supplies the positional information for localized Ran activation in higher eukaryotic cells. However, an equal distribution of RanGAP in the mitotic fluid would oppose the formation of the Ran-GTP gradient. This implies that an active process of keeping RanGAP away from RanGEF should be very helpful for the establishment and maintenance of such a gradient. We imagine that the conserved acidic region of RanGAP could be involved in an as yet uncharacterized localization mechanism which involves the acidic region. While our findings on RanGAP having a role beyond regulating nuclear transport strictly apply only to yeast organisms, it will be necessary to investigate whether the conserved acidic region of RanGAP is important for the regulation of the mitotic effects of Ran in higher organisms also.
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft (AZ Ku 1339/1-1) and the Louis-Jeantet foundation for support.
REFERENCES
- 1.Agatep, R., R. D. Kirkpatrick, D. L. Parchaliuk, R. A. Woods, and R. D. Gietz. 1998. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Tech. Tips Online 1:P01525. [Online.] http://research.bmn.com/tto. [Google Scholar]
- 2.Ahmadian, M. R., U. Hoffmann, R. S. Goody, and A. Wittinghofer. 1997. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry 36:4535-4541. [DOI] [PubMed] [Google Scholar]
- 3.Ahmadian, M. R., R. Mittal, A. Hall, and A. Wittinghofer. 1997. Aluminum fluoride associates with the small guanine nucleotide binding proteins. FEBS Lett. 408:315-318. [DOI] [PubMed] [Google Scholar]
- 4.Ahmadian, M. R., P. Stege, K. Scheffzek, and A. Wittinghofer. 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4:686-689. [DOI] [PubMed] [Google Scholar]
- 5.Allin, C., M. R. Ahmadian, A. Wittinghofer, and K. Gerwert. 2001. Monitoring the GAP catalyzed H-Ras GTPase reaction at atomic resolution in real time. Proc. Natl. Acad. Sci. USA 98:7754-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, J., F. Melchior, V. Gerke, F. R. Bischoff, H. Ponstingl, and A. Wittinghofer. 1995. Rna1 encodes a GTPase-activating protein-specific for Gsp1P, the Ran/Tc4 homolog of Saccharomyces cerevisiae. J. Biol. Chem. 270:11860-11865. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff, F. R., and D. Görlich. 1997. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 419:249-254. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff, F. R., C. Klebe, J. Kretschmer, A. Wittinghofer, and H. Ponstingl. 1994. Rangap1 induced GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91:2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff, F. R., H. Krebber, T. Kempf, I. Hermes, and H. Ponstingl. 1995. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc. Natl. Acad. Sci. USA 92:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bischoff, F. R., H. Krebber, E. Smirnova, W. Dong, and H. Ponstingl. 1995. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 14:705-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischoff, F. R., and H. Ponstingl. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354:80-82. [DOI] [PubMed] [Google Scholar]
- 12.Boman, A. L., M. R. Delannoy, and K. L. Wilson. 1992. GTP hydrolysis is required for vesicle fusion during nuclear envelope assembly in vitro. J. Cell Biol. 116:281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi, N. C., E. J. Adam, G. D. Visser, and S. A. Adam. 1996. RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J. Cell Biol. 135:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutavas, E., M. Ren, J. D. Oppenheim, P. D'Eustachio, and M. G. Rush. 1993. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature 366:585-587. [DOI] [PubMed] [Google Scholar]
- 15.Eccleston, J. F., K. J. Moore, G. G. Brownbridge, M. R. Webb, and P. N. Lowe. 1991. Fluorescence approaches to the study of the p21ras GTPase mechanism. Biochem. Soc. Trans. 19:432-437. [DOI] [PubMed] [Google Scholar]
- 16.Eccleston, J. F., K. J. Moore, L. Morgan, R. H. Skinner, and P. N. Lowe. 1993. Kinetics of interaction between normal and proline 12 Ras and the GTPase-activating proteins, p120-GAP and neurofibromin. The significance of the intrinsic GTPase rate in determining the transforming ability of ras. J. Biol. Chem. 268:27012-27019. [PubMed] [Google Scholar]
- 17.Farkasovsky, M., and H. Küntzel. 1995. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J. Cell Biol. 131:1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleig, U., S. S. Salus, I. Karig, and S. Sazer. 2000. The fission yeast Ran GTPase is required for microtubule integrity. J. Cell Biol. 151:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floer, M., and G. Blobel. 1996. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem. 271:5313-5316. [DOI] [PubMed] [Google Scholar]
- 20.Floer, M., G. Blobel, and M. Rexach. 1997. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J. Biol. Chem. 272:19538-19546. [DOI] [PubMed] [Google Scholar]
- 21.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 22.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 23.Görlich, D., N. Pante, U. Kutay, U. Aebi, and F. R. Bischoff. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584-5594. [PMC free article] [PubMed] [Google Scholar]
- 24.Görlich, D., M. J. Seewald, and K. Ribbeck. 2003. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22:1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham, D. L., J. F. Eccleston, and P. N. Lowe. 1999. The conserved arginine in Rho-GTPase-activating protein is essential for efficient catalysis but not for complex formation with Rho GDP and aluminum fluoride. Biochemistry 38:985-991. [DOI] [PubMed] [Google Scholar]
- 26.Haberland, J., J. Becker, and V. Gerke. 1997. The acidic C-terminal domain of rna1p is required for the binding of Ran GTP and for RanGAP activity. J. Biol. Chem. 272:24717-24726. [DOI] [PubMed] [Google Scholar]
- 27.Hartwell, L. H. 1967. Macromolecule synthesis in temperature-sensitive mutants of yeast. J. Bacteriol. 93:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hetzer, M., D. Bilbao-Cortes, T. C. Walther, O. J. Gruss, and I. W. Mattaj. 2000. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell 5:1013-1024. [DOI] [PubMed] [Google Scholar]
- 29.Hetzer, M., O. J. Gruss, and I. W. Mattaj. 2002. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 4:E177-E184. [DOI] [PubMed] [Google Scholar]
- 30.Hillig, R. C., L. Renault, I. R. Vetter, T. Drell, A. Wittinghofer, and J. Becker. 1999. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol. Cell 3:781-791. [DOI] [PubMed] [Google Scholar]
- 31.Hopper, A. K., and F. Banks. 1978. A yeast mutant which accumulates precursor tRNAs. Cell 14:211-219. [DOI] [PubMed] [Google Scholar]
- 32.Hopper, A. K., H. M. Traglia, and R. W. Dunst. 1990. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J. Cell Biol. 111:309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joseph, J., S. H. Tan, T. S. Karpova, J. G. McNally, and M. Dasso. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalab, P., K. Weis, and R. Heald. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295:2452-2456. [DOI] [PubMed] [Google Scholar]
- 35.Klebe, C., F. R. Bischoff, H. Ponstingl, and A. Wittinghofer. 1995. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins Rcc1 and RanGAP1. Biochemistry 34:639-647. [DOI] [PubMed] [Google Scholar]
- 36.Klebe, C., H. Prinz, A. Wittinghofer, and R. S. Goody. 1995. The kinetic mechanism of Ran-nucleotide exchange catalyzed by Rcc1. Biochemistry 34:12543-12552. [DOI] [PubMed] [Google Scholar]
- 37.Kobe, B., and J. Deisenhofer. 1993. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 366:751-756. [DOI] [PubMed] [Google Scholar]
- 38.Kraemer, A., T. Brinkmann, I. Plettner, R. Goody, and A. Wittinghofer. 2002. Fluorescently labelled guanine nucleotide binding proteins to analyse elementary steps of GAP-catalysed reactions. J. Mol. Biol. 324:763-774. [DOI] [PubMed] [Google Scholar]
- 39.Kraulis, P. J. 1991. Molscript-A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 40.Kuhlmann, J., I. Macara, and A. Wittinghofer. 1997. Dynamic and equilibrium studies on the interaction of Ran with its effector, RanBP1. Biochemistry 36:12027-12035. [DOI] [PubMed] [Google Scholar]
- 41.Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 42.Lounsbury, K. M., and I. G. Macara. 1997. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J. Biol. Chem. 272:551-555. [DOI] [PubMed] [Google Scholar]
- 43.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 44.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 45.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matunis, M. J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matynia, A., K. Dimitrov, U. Mueller, X. He, and S. Sazer. 1996. Perturbations in the spi1p GTPase cycle of Schizosaccharomyces pombe through its GTPase-activating protein and guanine nucleotide exchange factor components result in similar phenotypic consequences. Mol. Cell. Biol. 16:6352-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meier, I. 2000. A novel link between Ran signal transduction and nuclear envelope proteins in plants. Plant Physiol. 124:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Macromol. Crystallogr. B 277:505-524. [DOI] [PubMed] [Google Scholar]
- 50.Mittal, R., M. R. Ahmadian, R. S. Goody, and A. Wittinghofer. 1996. Formation of a transition-state analog of the Ras GTPase reaction by Ras center dot GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science 273:115-117. [DOI] [PubMed] [Google Scholar]
- 51.Nachury, M. V., and K. Weis. 1999. The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA 96:9622-9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nemergut, M. E., C. A. Mizzen, T. Stukenberg, C. D. Allis, and I. G. Macara. 2001. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292:1540-1543. [DOI] [PubMed] [Google Scholar]
- 53.Nixon, A. E., M. Brune, P. N. Lowe, and M. R. Webb. 1995. Kinetics of inorganic phosphate release during interaction of p21Ras with the GTPase-activating proteins, p120-GAP and neurofibromin. Biochemistry 34:15592-15598. [DOI] [PubMed] [Google Scholar]
- 54.Ohtsubo, M., H. Okazaki, and T. Nishimoto. 1989. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 109:1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ouspenski, I. I., S. J. Elledge, and B. R. Brinkley. 1999. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 27:3001-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pay, A., K. Resch, H. Frohnmeyer, E. Fejes, F. Nagy, and P. Nick. 2002. Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 30:699-709. [DOI] [PubMed] [Google Scholar]
- 57.Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ribbeck, K., and D. Görlich. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21:2664-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richards, S. A., K. M. Lounsbury, K. L. Carey, and I. G. Macara. 1996. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J. Cell Biol. 134:1157-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rose, A., and I. Meier. 2001. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98:15377-15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salus, S. S., J. Demeter, and S. Sazer. 2002. The Ran GTPase system in fission yeast affects microtubules and cytokinesis in cells that are competent for nucleocytoplasmic protein transport. Mol. Cell. Biol. 22:8491-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheffzek, K., C. Klebe, K. Fritzwolf, W. Kabsch, and A. Wittinghofer. 1995. Crystal-structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature 374:378-381. [DOI] [PubMed] [Google Scholar]
- 63.Schreiber, G. 2002. Kinetic studies of protein-protein interactions. Curr. Opin. Struct. Biol. 12:41-47. [DOI] [PubMed] [Google Scholar]
- 64.Schreiber, G., and A. R. Fersht. 1996. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 3:427-431. [DOI] [PubMed] [Google Scholar]
- 65.Schweins, T., K. Scheffzek, R. Assheuer, and A. Wittinghofer. 1997. The role of the metal ion in the p2l(ras) catalysed GTP-hydrolysis: Mn2+ versus Mg2+. J. Mol. Biol. 266:847-856. [DOI] [PubMed] [Google Scholar]
- 66.Seewald, M. J., C. Körner, A. Wittinghofer, and I. R. Vetter. 2002. RanGAP mediates GTP hydrolysis without an arginine finger. Nature 415:662-666. [DOI] [PubMed] [Google Scholar]
- 67.Sheinerman, F. B., R. Norel, and B. Honig. 2000. Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 10:153-159. [DOI] [PubMed] [Google Scholar]
- 68.Smith, A. E., B. M. Slepchenko, J. C. Schaff, L. M. Loew, and I. G. Macara. 2002. Systems analysis of Ran transport. Science 295:488-491. [DOI] [PubMed] [Google Scholar]
- 69.Sydor, J. R., M. Engelhard, A. Wittinghofer, R. S. Goody, and C. Herrmann. 1998. Transient kinetic studies on the interaction of Ras and the Ras-binding domain of c-Raf-1 reveal rapid equilibration of the complex. Biochemistry 37:14292-14299. [DOI] [PubMed] [Google Scholar]
- 70.Traglia, H. M., N. S. Atkinson, and A. K. Hopper. 1989. Structural and functional analyses of Saccharomyces cerevisiae wild-type and mutant RNA1 genes. Mol. Cell. Biol. 9:2989-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vetter, I. R., C. Nowak, T. Nishimoto, J. Kuhlmann, and A. Wittinghofer. 1999. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398:39-46. [DOI] [PubMed] [Google Scholar]
- 72.Villa-Braslavsky, C. I., C. Nowak, D. Görlich, A. Wittinghofer, and J. Kuhlmann. 2000. Different structural and kinetic requirements for the interaction of Ran with the Ran-binding domains from RanBP2 and importin-beta. Biochemistry 39:11629-11639. [DOI] [PubMed] [Google Scholar]
- 73.Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065-1075. [DOI] [PubMed] [Google Scholar]
- 74.Walther, T. C., H. S. Pickersgill, V. C. Cordes, M. W. Goldberg, T. D. Allen, I. W. Mattaj, and M. Fornerod. 2002. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158:63-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilde, A., and Y. Zheng. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284:1359-1362. [DOI] [PubMed] [Google Scholar]
- 76.Winston, F., C. Dollard, and S. L. Ricuperohovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 77.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, C., and P. R. Clarke. 2000. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288:1429-1432. [DOI] [PubMed] [Google Scholar]
- 79.Zhang, C., and P. R. Clarke. 2001. Roles of Ran-GTP and Ran-GDP in precursor vesicle recruitment and fusion during nuclear envelope assembly in a human cell-free system. Curr. Biol. 11:208-212. [DOI] [PubMed] [Google Scholar]