Abstract
A mutation in the POU4F3 gene (BRN-3.1, BRN3C) is responsible for DFNA15 (MIM 602459), autosomal-dominant nonsyndromic hearing loss. POU4F3 is a member of the POU family of transcription factors and is essential for inner-ear hair cell maintenance. To test the potential effects of the human POU4F3 mutation, we performed a series of experiments in cell culture to mimic the human mutation. Mutant POU4F3 loses most of its transcriptional activity and most of its ability to bind to DNA and does not function in a dominant-negative manner. Moreover, whereas wild-type POU4F3 is found exclusively in the nucleus, our studies demonstrate that the mutant protein is localized both to the nucleus and the cytoplasm. Two nuclear localization signals were identified; both are essential for proper nuclear entry of POU4F3 protein. We found that the mutant protein half-life is longer than that of the wild type. We propose that the combination of defects caused by the mutation on the function of the POU4F3 transcription factor eventually leads to hair cell morbidity in affected family H members.
The POU domain transcription factors play an important role in tissue-specific gene regulation. They were originally defined by sequence homology between four transcription factors: mammalian Pit-1, Oct-1, and Oct-2 and nematode Caenorhabditis elegans Unc-86 (4, 7, 15, 16). The region of homology, referred to as the POU domain, is a bipartite DNA-binding domain that contains a POU-specific (POUS) domain and the POU-homeodomain (POUHD) joined by a variable linker. High-affinity DNA binding of POU transcription factors requires both domains (1, 32). Based on sequence homology in the POU domain, the POU transcription factors were divided into six subclasses from I to VI (37). The class IV POU subfamily includes POU4F1 (Brn3a/Brn-3.0), POU4F2 (Brn3b/Brn-3.2), and POU4F3 (Brn3c/Brn-3.1) in mammals (11, 14, 21, 25, 35, 44), C. elegans Unc-86, and Drosophila factors I-POU and tI-POU (37).
The three mammalian members of the POU IV subfamily are expressed in distinct but overlapping populations of neuronal cells during development and in the adult organism (11, 25, 35). Gene-targeted mutagenesis in the mouse of each of these class IV transcription factors demonstrates that the cell type in which it is first expressed is the system that is most affected (6, 10, 23, 39, 40). In the inner ear, Pou4f3 is uniquely and strongly expressed in cochlear and vestibular hair cells. Targeted deletion of Pou4f3 results in profound deafness and impaired balance due to complete loss of auditory and vestibular hair cells. This is followed by a partial secondary loss of spiral and vestibular ganglion neurons (6, 39, 42). Pou4f3 is required for the final differentiation and survival of hair cells (41).
A mutation in the POU4F3 gene is associated with hearing loss in a large Israeli Jewish family, family H (36). Affected members of family H suffer from progressive autosomal-dominant sensorineural hearing loss (9). In hearing impaired members of family H, an 8-bp deletion was identified in exon 2 of the human POU4F3 gene. This deletion leads to a frameshift, predicted to cause a stop codon to be formed prematurely in the first helix of the POU-homeodomain.
Hearing loss is the most common form of sensory impairment in humans, affecting normal communication in 10% of people aged 65 years or older. Deafness can result from genetic or environmental causes or a combination of both (38). The loss of sensory hair cells of the inner ear is a common phenomena in progressive hearing loss, but the reasons underlying hair cell loss remain an enigma. Transcription factors play a particularly vital role in the cascade of events leading to proper spatial and temporal expression patterns of their downstream targets. A multitude of transcription factors are involved in tight regulation of developmental processes in the mammalian inner ear, including the determination of hair cell fate (reviewed in references 8 and 29). Nevertheless, the mechanisms leading to hearing loss and hair cell dysfunction due to the POU4F3 mutation are unknown.
The aim of the present study was to characterize the POU4F3 mutation associated with human hereditary hearing loss. We demonstrate that this deletion affects DNA binding, transcription activation, protein stability, and nuclear localization of the transcription factor. Furthermore, we have identified the nuclear localization signals (NLS) of POU4F3 and demonstrated their relevance to the POU4F3 deafness mutation.
MATERIALS AND METHODS
Plasmid constructs.
A full-length human POU4F3 cDNA was amplified from human cochlea RNA (kindly provided by Cynthia Morton, Brigham and Women's Hospital and Harvard Medical School, Boston, Mass. [28]) and cloned into the pBluescript vector (Stratagene). This clone served as a template for in vitro mutagenesis to create the family H del8 cDNA. The NLS mutant cDNAs were also subcloned into pBluescript.
The wild-type and different mutant cDNAs were subcloned into the pHM6 expression vector (Roche Applied Science) by using oligonucleotides that introduced KpnI and EcoRI restriction sites into the 5′ and 3′ of the cDNA by using pBluescript constructs as templates.
The correct sequence of all clones was confirmed on an ABI Prism 3100 genetic analyzer. All primer sequences are available upon request from authors.
In vitro mutagenesis.
Primers were designed to introduce the deleted 8-bp family H mutation into the wild-type sequence by PCR (Expand High-Fidelity PCR System; Roche Applied Science). Two separate PCRs were performed by using pBluescript-POU4F3 cDNA as a template. The two products were annealed together by using a third PCR.
To examine the POU4F3 NLS, pHM6-POU4F3 served as a template, and point mutations were introduced by using oligonucleotides containing the desired mutations as primers for the QuikChange site-directed mutagenesis kit (Stratagene).
Cell culture.
PC12 cells were grown in Dulbecco modified Eagle medium (DMEM; Biological Industries) supplemented with 8% fetal calf serum (FCS), 8% donor horse serum, 2 mM l-glutamine, and 100 U of penicillin-streptomycin-nystatin/ml. HEK293 and COS-7 cells were grown in DMEM supplemented with 10% FCS, 2 mM l-glutamine, and 100 U of penicillin-streptomycin-nystatin/ml. Cells from the established cochlear cell line UB/OC-2 (kindly provided by Matthew Holley [27]) were grown in minimal essential medium with Earle's salts and Glutamax (Gibco-BRL/Life Technologies) supplemented with 10% FCS. Cells were grown at 33°C with 50 U of gamma interferon (Gibco-BRL/Life Technologies)/ml.
Transient and stable transfection of cells.
HEK293 and COS-7 cells were transiently transfected with the Fugene transfection reagent (Roche Applied Science). Transfection was performed according to manufacturer's instructions by using a 1:3 ratio. Stable transfectants were selected in a medium containing G418 (800 μg/ml), and clones were grown routinely in DMEM containing 10% FCS and G418 (800 μg/ml).
PC12 and UB/OC-2 cells were transiently transfected with the Lipofectamine reagent (Gibco-BRL/Life Technologies) according to the manufacturer's instructions.
Luciferase assay.
PC12 cells and UB/OC-2 cells were transfected with the Snap25 promoter region cloned into the pGL2-Basic vector (Promega) with either pHM6 vector, pHM6-wt POU4F3 cDNA, or pHM6-del8 POU4F3 cDNA. Luciferase activities were measured 24 h after transfection by using the Luciferase Assay System (Promega) according to the manufacturer's instructions. All luciferase activities were normalized with the β-galactosidase activities derived from the control plasmid pHM6-lacZ (Roche Applied Science). Experiments were performed in triplicate and repeated three times.
Western blot analysis.
Transfected HEK293 and COS-7 cells were grown in six-well plates. Cells were scraped into 250 μl of radioimmunoprecipitation assay (RIPA) lysis buffer containing 150 mM NaCl, 10 mM EDTA, 1% Triton X-100, 1% deoxycholic acid, 0.1% sodium dodecyl sulfate (SDS), 15 mM HEPES, 1 mM dithiothreitol (DTT) and protease inhibitor cocktail set III (Calbiochem). Equal amounts of protein were electrophoresed on 12.5% polyacrylamide gels (SDS-PAGE). Gels were transblotted to BioTraceNT nitrocellulose membrane (Pall Corp.). Membranes were blocked overnight at 4°C in blocking solution (3% bovine serum albumin in TTBS), incubated for 60 min with primary monoclonal anti-HA.11 antibody (1:1,000; Covance), and incubated for 45 min with a secondary antibody [ImmunoPure goat anti-mouse immunoglobulin G(H+L) peroxidase-conjugated antibody (Pierce) at 1:40,000], and detection was done by enhanced chemiluminescence. The SuperSignal chemiluminescene substrate kit (Pierce) was used according to the manufacturer's instructions.
Immunocytochemistry.
Coverslip-grown HEK293 or COS-7 transfected cells were fixed for 10 min in 4% paraformaldehyde and permeabilized for 10 min in 0.5% Triton at room temperature. After the cells were stained for 10 min with DAPI (4′,6′-diamidino-2-phenylindole; Sigma) and blocked for 20 min in 10% normal goat serum, they were incubated for 60 min with primary monoclonal anti-HA.11 antibody (Covance) at 1:1,000. After incubation for 45 min with ImmunoPure goat anti-mouse IgG(H+L) fluorescein-conjugated antibodies (Pierce) at 1:100, chamber slides were mounted in Gel/Mount (Biomeda). Imaging of confluent cells was done by using a laser scanning confocal microscope (CLSM 410; Zeiss).
To compare the relative amounts of immunostaining with the antihemagglutinin (anti-HA) antibody in the nucleus, we analyzed confocal images by using the histogram function of the Photoshop package (version 6.0). Nuclear localizaton of the different proteins was calculated as the ratio between the fluorescence intensity in the area colocalized with the DAPI staining and the fluorescence intensity in the total cellular area.
Electrophoretic mobility shift assay (EMSA).
Pou4f2 and the wild-type and mutant Pou4f3 proteins were produced by the coupled TNT transcription/translation system (Promega) with expression plasmids described elsewhere (22). For the probe, DNA oligonucleotides containing a consensus Pou4f binding site were end radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase. Binding reactions were carried out at room temperature for 20 to 30 min in a final volume of 20 μl containing 10 mM HEPES (pH 7.5), 50 mM KCl, 1 mM EDTA, 0.1% Triton X-100, 5% glycerol, 0.1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 1 mg of poly(dI-dC), 5 × 105 cpm of labeled probe, and 3 μl of each desired protein lysate. Competition was performed by adding a 500-fold excess amount of cold oligonucleotides to the reactions. Free and bound probes were resolved on 5% nondenaturing polyacrylamide gels. The normal specific oligonucleotide used for probe and competition contained the consensus Pou4f binding site 5′-CACGCATAATTAATCGC-3′ (13). The nonspecific mutant oligonucleotide M4 (5′-CACAGCTCAGCAACGCGC-3′) was described elsewhere (43).
Metabolic labeling.
For metabolic labeling, POU4F3-transfected COS-7 cells were first starved for methionine for 1 h at 37°C in DMEM lacking methionine and cysteine (Sigma) supplemented with 0.5% dialyzed FCS and 1% glutamine. Cells were pulse-labeled with [35S]methionine (EXPRE35S35S Protein labeling mix; New England Nuclear). After a 30-min pulse, the cells were washed with phosphate-buffered saline and chased with complete culture medium. After the indicated times, cells were washed in cold phosphate-buffered saline and lysed in 1 ml of RIPA lysis buffer. Lysates were clarified by centrifugation at 20,000 × g for 30 min.
Immunoprecipitations.
Lysates from the metabolic labeling experiments were precleared by rotating for 2 h at 4°C with normal mouse serum (Jackson Immunoresearch Laboratories) captured on protein AG-agarose beads (Jackson Immunoresearch Laboratories). Precleared samples were measured by using a β-counter, and 107 cpm of each sample was immunoprecipitated with primary monoclonal anti-HA.11 antibody (Covance) captured on protein AG-agarose beads by rotating them for 2 h at 4°C. The beads were washed three times in wash buffer containing 150 mM NaCl, 10 mM EDTA, 0.1% Triton, 0.1% Doc, 0.01% SDS, 15 mM HEPES, 1 mM DTT, and protease inhibitor cocktail (Calbiochem). Bound proteins were eluted in 40 μl of sample buffer at 100°C and subjected to SDS-12.5% polyacrylamide gel electrophoresis (PAGE). Gels were dried under a vacuum. After the signal was enhanced with NAMP100 Amplify (Amersham), the gels were exposed to Kodak Biomax MR film.
Bioinformatics analysis.
Two putative NLS sequences were revealed by using PSORT software (24). For evolutionary analysis, a set of 67 homologues was obtained by using basic local alignment search tool (BLAST) through the SwissProt database (www.expasy.org/sprot/). A second set of 99 homologues included, in addition to POU domain proteins, homeodomain proteins lacking the POUS domain. The two sets were aligned by using CLUSTAL W with default parameters (34). Phylogenetic analysis was performed by using the neighbor-joining tree, which is the output of the Rate4Site program (26).
A three-dimensional (3D) structure model for POU4F3 was built by using the Nest facility of the Jackal protein structure modeling package (http://trantor.bioc.columbia.edu/∼xiang/jackal/#nest). The 3D structure of the Oct-1 POU domain (PDB code 1OCT, chain C [19]) was used as a template to build a model of the corresponding domain in POU4F3. The reliability of the model can be assessed through the 47% sequence identity between POU4F3 and the template or through the 53% sequence identity when the variable linker region that consists of 18 amino acids is disregarded. This linker region, connecting the POUS domain to the POUHD, lacks any sequence similarity between POU4F3 and the templates and is highly variable throughout the POU family. The model applies only to amino acids 180 to 334 of the protein and contains both the POUS domain and the POUHD.
RESULTS
We previously reported a novel mutation in the POU4F3 transcription factor, leading to progressive deafness in an extended Israeli family (36). The expression of this protein is mostly restricted to hair cells of the inner ear (6, 39). Due to the impossibility of obtaining material for in vivo experiments, we performed a series of experiments in cell culture to mimic the human mutation in order to test the potential effects of the human POU4F3 mutation on sensory hair cells of the inner ear. Wild-type and mutant forms of the POU4F3 cDNA were introduced into COS-7, HEK293, PC12, and UB/OC2 cells (UB/OC-2 is a cell line derived from mouse embryonic day 13 inner ear sensory epithelium and is commonly used as an in vitro system for studies of the auditory sensory epithelium [27]). We evaluated the DNA binding, transcriptional activation, subcellular localization, and protein stability of mutant POU4F3 and identified the NLS of the wild-type protein.
Family H mutation alters DNA binding and transcriptional activity of POU4F3.
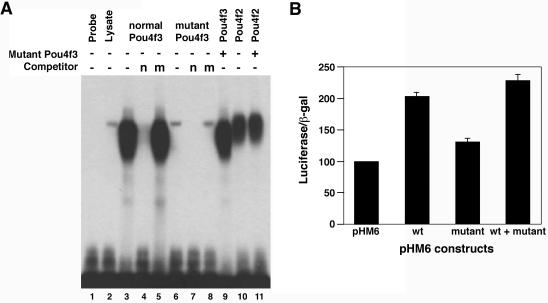
In family H, the 8-bp deletion removes the carboxy-terminal portion of the POU4F3 transcription factor (Fig. 1). The truncation occurs in the first alpha helix of the POUHD, with the POUS domain predicted to remain intact. To identify the mechanism by which this mutation alters normal hearing, we assessed the ability of the wild-type and mutated proteins to bind to target DNA-binding sequence by using EMSA. In vitro studies have revealed a consensus DNA binding site that is recognized by each of the POU IV family members (ATAATTAAT) (13). This site differs from binding sites previously described for other POU transcription factors. We used labeled DNA oligonucleotides containing the POU4F consensus binding site, along with in vitro-translated wild-type and mutant POU4F3 proteins. As can be seen in Fig. 2A, the wild-type protein can bind the DNA and form a specific DNA-protein complex. Competition experiments, with unlabeled consensus DNA oligonucleotides, almost completely reduced the protein-DNA complex. On the other hand, there is hardly any binding of mutant POU4F3 to the POU4F consensus DNA-binding site. These experiments also revealed that mutant POU4F3 does not appear to form complexes with normal POU4F3 or POU4F2, nor does it interfere with binding of the normal proteins to the consensus site.
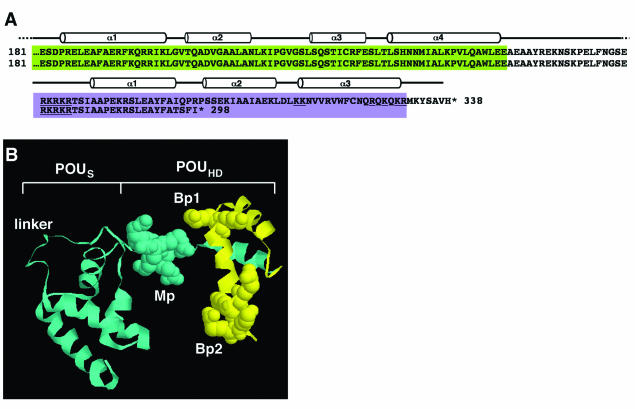
FIG. 1.
Sequence and structure of POU domain in wild-type and family H mutant POU4F3. (A) Sequence comparison of the POU domain in wild-type (top) and mutant POU4F3 (bottom). The POUS domain is colored green, and the POUHD is colored purple. The helical domains are drawn above the sequence. Mutant POU4F3 contains four novel amino acids (amino acids 294 to 298), followed by a premature stop codon. (B) A ribbon and spacefill representation of a 3D model of the POU4F3 POUS domain and POUHD, separated by a linker. The model was obtained by using the experimentally determined structure of the Oct-1 POU domain as a template (19) as described in Materials and Methods. The monopartite (Mp) and bipartitite (Bp1 and Bp2) NLS are represented by the space-fill regions. The portion of the homeodomain shown in yellow is presumably truncated due to the family H POU4F3 mutation.
FIG. 2.
DNA binding and transcriptional activities of wild-type and mutant POU4F3 proteins. (A) EMSA was performed with a radiolabeled POU4F binding site as a probe. Reactions contained in vitro-translated normal POU4F3 (lanes 3 to 5) or mutant POU4F3 (lanes 6 to 8). Competition was performed by using a 500-fold excess amount of cold consensus POU4F binding site (n; lanes 4 and 7) or nonspecific competitor (m; lanes 5 and 8). To assess whether mutant POU4F3 interferes with DNA binding, reactions containing in vitro translated POU4F3 (lane 3 and 9) and POU4F2 (lanes 10 and 11) were incubated in the presence (lanes 9 and 11) or absence (lanes 3 and 10) of in vitro-translated mutant POU4F3. (B) The transcriptional activity is shown as relative luciferase activity after cotransfection of different expression constructs with the pGL2-SNAP25 reporter into PC12 cells. Luciferase activity was measured 48 h after transfection. The transfection efficiency was controlled by measuring β-galactosidase activity after transfection with the pHM6-lacZ plasmid. The basic transcriptional activity for the promoter with pHM6 empty vector was set arbitrarily to 100. The data shown represent three separate experiments performed in triplicate in each experiment (mean ± the standard deviation [SD]).
The next step was to assess whether the family H mutation affects the transcriptional activity of POU4F3. To date, no bona fide downstream targets of POU4F3 are known. The POU IV family members are the closest mammalian homologs to the nematode Unc-86. This, together with their restricted expression in the developing and adult nervous system, has led to the idea that they may play a role in regulating gene expression in neuronal cells. SNAP-25 is a presynaptic nerve terminal protein, and its promoter has been shown to be activated by POU4F3 only in neuron-derived cell lines such as PC12 and ND7. This activation depends on a neuron-specific activation domain in the amino terminus of POU4F3 (30). PC12 cells transfected with the pHM6 vector, pHM6-wt (wild-type POU4F3 cDNA), or pHM6-mut (del8 POU4F3 cDNA) constructs were examined for their ability to activate the SNAP-25 promoter connected to a luciferase reporter gene. The luciferase values were normalized for β-galactosidase levels. We found that the mutant product had only ca. 30% activity compared to the wild-type product [(1.3 ± 0.07)-fold versus (2.02 ± 0.06)-fold] (Fig. 2B). Cotransfection of both normal and mutant constructs gave additive activity levels [(2.28 ± 0.09)-fold]. Similar results were obtained with transiently transfected UB/OC-2 cells.
The human POU4F3 mutation affects protein stability.
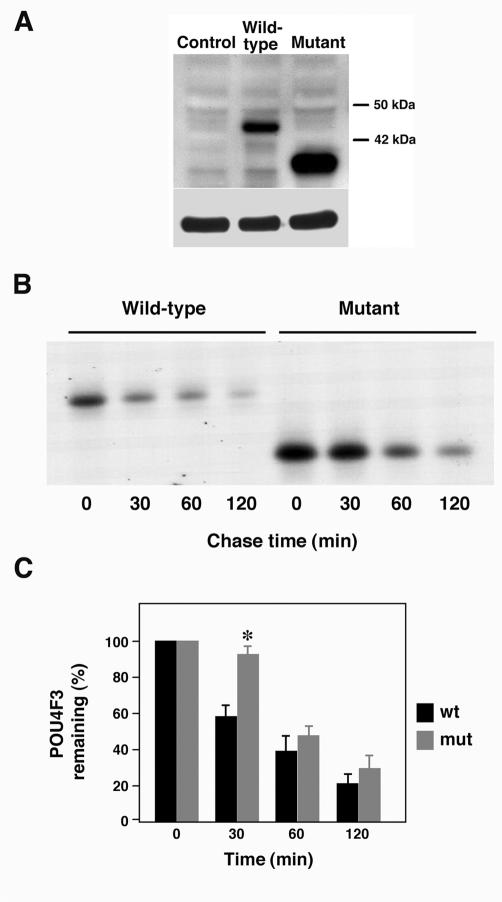
The POU4F3 family H mutation occurs in the final coding exon and is predicted to produce a mutant protein of 298 amino acids in length rather than the normal 338 amino acids. Immunoblot analysis demonstrated that the POU4F3 family H transcript is translated in cell culture, leading to a truncated protein relative to a wild-type POU4F3 protein (Fig. 3A). Not only does the mutant protein appear to be stably expressed, but its expression appears to be stronger than that of the normal product. Similar results were obtained when we used protein extracted from transiently transfected HEK293 and stably transfected COS-7 and HEK293 cell lines (data not shown).
FIG. 3.
The family H mutation in POU4F3 affects protein stability. (A) Western blot analysis was performed to determine whether mutant POU4F3 was successfully expressed in transfected COS-7 cells. Control, lysate from cells transfected with empty pHM6 vector; Wild-type, lysate from cells transfected with pHM6-wt POU4F3 cDNA; Mutant, lysate from cells transfected with pHM6-Mut POU4F3 cDNA. Western blotting was done with a monoclonal anti-HA antibody (top) and a monoclonal antitubulin antibody to confirm equivalent protein loading (bottom). (B and C) The turnover of POU4F3 proteins was determined in a pulse-chase experiment. COS-7 cells were transfected with pHM6-wt or pHM6-Mut, labeled with [35S]methionine for 30 min, and immunoprecipitated with monoclonal anti-HA antibody after 0, 30, 60, and 120 min of chase. The immunoprecipitated proteins were run on an SDS-PAGE gel and analyzed by autoradiography. (C) A representative autoradiograph is shown in panel B. The relative amounts of POU4F3 (mean ± the SD) were plotted against the chase period. Wild-type POU4F3 is indicated in black, and mutant POU4F3 is indicated in gray. ✽, P < 0.001 as determined by using the Student two-tailed t test with unequal variances.
Taking into account the disparity in protein accumulation in transfected cells, we speculated that this may be due to differences in protein stability. To compare the stability of the normal and mutant proteins, we conducted pulse-chase experiments in transiently transfected COS-7 cells. HA-tagged proteins were immunoprecipitated after 0, 30, 60, and 120 min (Fig. 3B). When we quantified the amounts of radioactivity present in immunoprecipitated anti-HA tagged proteins, it was clear that, after 30 min, almost half of the wild-type protein was degraded, indicating that POU4F3 is a very short-lived protein (Fig. 3C). In contrast, the half-life of the mutant protein was ca. 60 min.
Changes in subcellular localization of POU4F3 mutant protein.
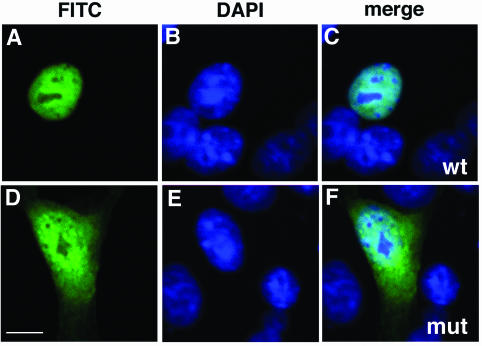
POU4F3 is a transcription factor and is localized to the nucleus (43). A defect in subcellular localization may prevent POU4F3 from functioning as a transcription regulator. Since transcriptional activation is affected by the mutation, we examined its localization. In order to test this hypothesis, we transfected COS-7 cells with pHM6, pHM6-wt, or pHM6-mut plasmids and examined the cells by confocal microscopy. As expected, the wild-type form of POU4F3 is localized exclusively in the nuclei of transfected cells (Fig. 4A to C). The mutant POU4F3 caused a severe defect in protein trafficking and was found in both the cytoplasm and nucleus (Fig. 4D to F). The same results were obtained when we repeated the experiments in HEK293 cells (data not shown), as well as in both transient and stable transfections.
FIG. 4.
The family H mutation changes the subcellular localization of POU4F3. In transfected COS-7 cells, the localization of wild-type (wt) and family H mutant (mut) pHM6 constructs were analyzed. Nuclei were stained with DAPI (blue), and HA-fused proteins (green) were visualized by confocal microscopy. For each construct, we show fluorescein isothiocyanate staining alone (A and D), DAPI staining alone (B and E), and double staining (merge) (C and F). The wild-type protein (A to C) was localized exclusively to the nucleus, whereas the mutant protein was localized both to the cytoplasm and nucleus (D to F). Scale bar, 10 μm.
Identification of NLS for POU4F3.
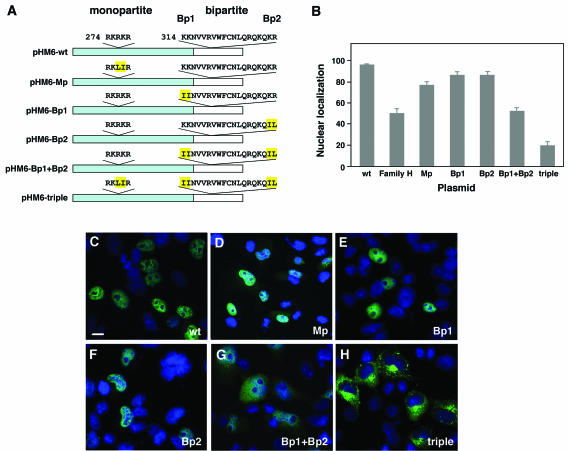
Based on the observation that the family H mutant protein lost its nuclear specificity, we began a search for the POU4F3 NLS. Although it appeared likely that the signal would be in the portion of the protein that is missing in the family H truncated protein, the entire sequence was analyzed by using bioinformatics tools. Two putative NLS sequences were revealed by using the PSORT software (24). The first is a monopartite NLS (amino acids 274 to 278; RKRKR), and the second is a bipartite NLS (amino acids 314 to 331; KKNVVRVWFCNLQRQKQKR). Both suggested signals are located in the POU homeodomain, although only the bipartite NLS is in the portion predicted to be missing in the family H mutant protein (Fig. 1). To determine which of the proposed NLS are functional, several amino acid residues were changed to either leucine or isoleucine, both of which are neutral amino acids, so that conformational changes are minimized. Each of the constructs contains two amino acid substitutions affecting part of the putative NLS (Fig. 5A). COS-7 cells were transfected with the wild-type POU4F3 (pHM6-wt), pHM6-monopartite (pHM6-Mp), pHM6-bipartite1 (pHM6-Bp1), and pHM6-bipartite2 (pHM6-Bp2) constructs, proteins were extracted, and Western blot analysis was performed. Both wild-type and mutant NLS proteins are expressed and appear similar in size and quantity (data not shown).
FIG. 5.
POU4F3 contains two functional NLS. (A) Schematic representation of the constructs containing wild-type POU4F3 and NLS mutations. The portion missing in the family H mutant protein is shown in white. The monopartite (Mp) and bipartite (Bp1 and Bp2) NLS sequences are indicated. Amino acids that were mutated are highlighted in yellow. (B) The graph displays the relative fluorescence intensities in the nucleus versus the whole cell after immunostaining with anti-HA antibody (mean ± the SD). (C to H) In transfected COS-7 cells, the localization of different mutant NLS constructs was analyzed. Nuclei were stained with DAPI (blue), and HA-fused proteins (green) were visualized by confocal microscopy. Immunolocalization of pHM6-Mp was both nuclear and cytoplasmic (D), and that of pHM6-Bp1 (E) and pHM6-Bp2 (F) was nuclear, similar to the cellular localization of pHM6-wild-type POU4F3 (wt) (C). Mutating both parts of the bipartite NLS (pHM6-Bp1+Bp2) led to partial cytoplasmic localization (G). Mutating both the monopartite and bipartite NLS (pHM6-Mp+Bp1+Bp2 or triple) resulted in complete cytoplasmic accumulation of the protein (H), indicating that both NLS are required for nuclear localization of POU4F3. Scale bar, 10 μm.
The relative intensity of immunostaining with the anti-HA antibody in the nucleus versus the whole cell was measured, providing a quantitative assessment of POU4F3 expression in the nuclei of the transfected cells. Cells transfected with wild-type POU4F3 demonstrated that 97.38% ± 0.7% was localized to the nucleus (Fig. 5B and C). Immunolocalization of the Mp mutant protein demonstrated that although the majority of the protein was in nucleus, a portion was in the cytoplasm as well (77.63% ± 3.28%) (Fig. 5B and D). The vast majority of Bp1 (87.78% ± 2.97%) and Bp2 (87.80% ± 2.66%) mutant proteins was localized to the nucleus (Fig. 5B, E, and F). A plasmid containing both the Bp1 and Bp2 mutations was constructed, disrupting both parts of the bipartite NLS (pHM6-Bp1+Bp2). Figure 5G shows that the protein localizes both to the nucleus and the cytoplasm (53.21% ± 2.46%). This was similar to the levels of nuclear expression for family H mutant POU4F3 (50.31% ± 4.59%) (Fig. 5B). Since both putative NLS seemed to have an effect on the cellular localization of the protein to some extent, another construct was built containing sequence alterations of both the monopartite and bipartite NLS sequences (the triple construct, pHM6-Mp+Bp1+Bp2). This protein was found to be localized mostly to the cytoplasm (20.06% ± 4.25%), indicating loss of function of the NLS (Fig. 5B and H).
Evolutionary analysis of the NLS was performed by using a set of homologue proteins obtained through the SwissProt database. The set of homologues was divided into clades based on its phylogenetic tree. The data indicate that the monopartite NLS is highly conserved throughout the POU domain family; a homologous segment of five to six basic residues appears in all but two POU proteins (data not shown). This segment is unique to POU proteins and does not appear in homeodomain proteins outside the POU family. The bipartite NLS, on the other hand, appears exclusively in the closest POU4F3 clade, which comprises the POU domain class IV subfamily (data not shown). These results suggest two NLS are unique to the POU family: a monopartite that appears throughout the POU proteins and a bipartite NLS solely exhibited in the class IV subfamily.
DISCUSSION
Inherited hearing loss is genetically heterogeneous and is caused by mutations in genes encoding proteins responsible for a variety of processes, among them maintenance of hair cell structure, neuronal innervation, and trafficking. The first human nonsyndromic deafness-causing gene was identified in 1995 and, since then, more than 30 additional genes have been discovered (http://www.uia.ac.be/dnalab/hhh/). Our major challenge today is to understand the biological consequences of the mutations and how they affect the function of the inner ear.
We checked the effects of the POU4F3 mutation, which causes deafness in a large Israeli family, on DNA-binding ability, transcriptional activation, protein stability, and subcellular localization. In addition, we defined the NLS sequences required for nuclear entry of POU4F3.
The mechanism by which the POU4F3 mutation leads to hearing loss is unknown. One hypothesis was that haploinsufficiency of POU4F3 may downregulate genes responsible for hair cell survival, causing a progressive loss of hair cells that is clinically significant only after an extended period of time. However, brainstem-evoked responses were evaluated in Pou4f3 knockout mice up to 24 months of age, revealing that hearing of heterozygote animals is indistinguishable from that of wild-type animals, suggesting that haploinsufficiency of POU4F3 is most probably not the explanation for the deafness phenotype (18). Alternatively, the truncated protein may have novel DNA-binding properties or may act as a dominant-negative mutation by pairing with the wild-type form of POU4F3 or other transcription factors. DNA-binding experiments indicate that mutant POU4F3 loses its ability to bind to the predicted POU4F recognition sequence but does not interfere with binding of normal POU4F3 or POU4F2 to DNA. Reporter gene experiments have shown that the truncated protein loses most of its transcriptional activity but, again, does not affect the activity of the wild-type protein. Based on these two experiments, we have ruled out the possibility of dominant-negative activity of the mutant protein. Little is known about the protein-protein interactions involving POU4F family members. Previous studies have shown that POU4F1 and POU4F2 can heterodimerize with each other (33). Our studies have shown that mutant POU4F3 does not interfere with POU4F2 binding to DNA in vitro but, since it is reasonable to assume that POU4F3 does interact with other proteins, we cannot rule out the possibility that mutant POU4F3 interferes with these interactions.
Immunolocalization experiments have demonstrated that the POU4F3 normal product localizes to the nucleus, whereas the mutant protein is localized both to the cytoplasm and to the nucleus. Cytoplasmic localization of transcription factors obviously affects their ability to activate downstream targets. This correlates with our findings that mutant POU4F3 loses most of its ability to activate the SNAP-25 promoter.
Nuclear proteins usually have a common sequence, defined as an NLS, that is important for active protein transport into the nucleus. Two classical types of NLS are usually found: monopartite and bipartite. The first is a simple cluster of four to six basic amino acids. The second consists of two adjacent basic amino acids, a spacer of 9 to 12 different amino acids and an additional 3 basic amino acids (5, 17). We have shown that POU4F3 contains both the monopartite (amino acids 274 to 278 [RKRKR]) and the bipartite NLS (amino acids 314 to 331 [KKNVVRVWFCNLQRQKQKR]). The presence of the two signals and the potential difference in their efficiencies raise the possibility of a regulatory mechanism for the POU4F3 import process. The family H mutant protein has a cellular distribution similar to that of the Bp1+2 mutant protein. This observation correlates with the fact that the family H-POU4F3 protein is predicted to lose the bipartite NLS.
Evolutionary analysis revealed that the monopartite residues are part of all POU proteins. This signal is not found in other homeodomain proteins lacking the POU-specific domain. The bipartite signal appears only in class IV POU domain proteins. Thus, it is reasonable that this NLS is distinct to this subfamily. We suggest that a monopartite NLS is functional in the entire POU family and that the bipartite NLS is unique to class IV. This hypothesis is strengthened by the identification of a lone NLS in another POU protein, Oct-6, which belongs to the class III POU subfamily (31). Indeed, this NLS is identical to the POU4F3 monopartite NLS. At present, the NLS of this protein is the only other explored NLS from the POU family.
Our studies have shown that wild-type POU4F3 is a very short-lived protein in cultured cells. Pulse-chase analysis has revealed that the family H mutant POU4F3 is more stable than the wild-type protein. Many short-lived proteins, among them some transcription factors, are degraded by the proteasome-ubiquitin system (reviewed in reference 3). Until recently, the accepted paradigm was that proteasomal degradation takes place uniquely in the cytoplasm, but recent studies have provided evidence for nuclear proteasomal degradation. The degradation can take place solely in the nucleus or in the cytosol, but the ubiquitin-protein ligase (E3) is localized to the nucleus (reviewed in reference 12). Increased turnover of the substrate correlated with its nuclear localization, whereas slow proteolysis is correlated with cytosolic localization (20). This dependence on localization signals is most probably caused by the restriction of components of the ubiquitin system to certain compartments. For example, prevention of Far1 protein translocation into the nucleus stabilizes the protein (2). Since we observed differences in cellular localization of wild-type and mutant POU4F3 proteins, this may explain the extended half-life of mutant POU4F3.
In summary, the present study represents a detailed analysis of the deafness-causing POU4F3 deletion mutation and provides evidence that this mutation causes a spectrum of defects affecting the function of the transcription factor. We speculate that this eventually leads to the death of hair cells in the inner ear, causing progressive hearing loss.
Acknowledgments
We thank Yosef Shaul for insight and contributions at the beginning of this research project, Meital Cohen for help with the 3D model, Cynthia Morton and Matthew Holley for reagents, Leonid Mittelman and Yair Andegeko for confocal microscopy and analysis, and Tama Sobe for invaluable help throughout.
This work was supported by grants from the Israel Science Foundation-The Dorot Science Fellowships Foundation (grant no. 740/01) (K.B.A.), by NIH/Fogarty International Center grant R03 TW01108 (K.B.A.), by the European Commission (QLG2-CT-1999-00988) (K.B.A.), and by the National Institutes of Health (DC04594 and EY12020) (M.X.).
REFERENCES
- 1.Aurora, R., and W. Herr. 1992. Segments of the POU domain influence one another's DNA-binding specificity. Mol. Cell. Biol. 12:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondel, M., J. M. Galan, Y. Chi, C. Lafourcade, C. Longaretti, R. J. Deshaies, and M. Peter. 2000. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 19:6085-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22:442-451. [DOI] [PubMed] [Google Scholar]
- 4.Clerc, R. G., L. M. Corcoran, J. H. LeBowitz, D. Baltimore, and P. A. Sharp. 1988. The B-cell-specific Oct-2 protein contains POU box- and homeo box-type domains. Genes Dev. 2:1570-1581. [DOI] [PubMed] [Google Scholar]
- 5.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences: a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 6.Erkman, L., R. J. McEvilly, L. Luo, A. K. Ryan, F. Hooshmand, S. M. O'Connell, E. M. Keithley, D. H. Rapaport, A. F. Ryan, and M. G. Rosenfeld. 1996. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature 381:603-606. [DOI] [PubMed] [Google Scholar]
- 7.Finney, M., G. Ruvkun, and H. R. Horvitz. 1988. The Caenorhabditis elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell 55:757-769. [DOI] [PubMed] [Google Scholar]
- 8.Fritzsch, B., and K. W. Beisel. 2001. Evolution and development of the vertebrate ear. Brain Res. Bull. 55:711-721. [DOI] [PubMed] [Google Scholar]
- 9.Frydman, M., S. Vreugde, B. I. Nageris, S. Weiss, O. Vahava, and K. B. Avraham. 2000. Clinical characterization of genetic hearing loss caused by a mutation in the POU4F3 transcription factor. Arch. Otolaryngol. Head Neck Surg. 126:633-637. [DOI] [PubMed] [Google Scholar]
- 10.Gan, L., M. Xiang, L. Zhou, D. S. Wagner, W. H. Klein, and J. Nathans. 1996. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc. Natl. Acad. Sci. USA 93:3920-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerrero, M. R., R. J. McEvilly, E. Turner, C. R. Lin, S. O'Connell, K. J. Jenne, M. V. Hobbs, and M. G. Rosenfeld. 1993. Brn-3.0: a POU-domain protein expressed in the sensory, immune, and endocrine systems that functions on elements distinct from known octamer motifs. Proc. Natl. Acad. Sci. USA 90:10841-10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 13.Gruber, C. A., J. M. Rhee, A. Gleiberman, and E. E. Turner. 1997. POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol. Cell. Biol. 17:2391-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, X., M. N. Treacy, D. M. Simmons, H. A. Ingraham, L. W. Swanson, and M. G. Rosenfeld. 1989. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340:35-41. [DOI] [PubMed] [Google Scholar]
- 15.Herr, W., R. A. Sturm, R. G. Clerc, L. M. Corcoran, D. Baltimore, P. A. Sharp, H. A. Ingraham, M. G. Rosenfeld, M. Finney, G. Ruvkun, et al. 1988. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2 and Caenorhabditis elegans unc-86 gene products. Genes Dev. 2:1513-1516. [DOI] [PubMed] [Google Scholar]
- 16.Ingraham, H. A., R. P. Chen, H. J. Mangalam, H. P. Elsholtz, S. E. Flynn, C. R. Lin, D. M. Simmons, L. Swanson, and M. G. Rosenfeld. 1988. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell 55:519-529. [DOI] [PubMed] [Google Scholar]
- 17.Jans, D. A., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 18.Keithley, E. M., L. Erkman, T. Bennett, L. Lou, and A. F. Ryan. 1999. Effects of a hair cell transcription factor, Brn-3.1, gene deletion on homozygous and heterozygous mouse cochleas in adulthood and aging. Hearing Res. 134:71-76. [DOI] [PubMed] [Google Scholar]
- 19.Klemm, J. D., M. A. Rould, R. Aurora, W. Herr, and C. O. Pabo. 1994. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77:21-32. [DOI] [PubMed] [Google Scholar]
- 20.Lenk, U., and T. Sommer. 2000. Ubiquitin-mediated proteolysis of a short-lived regulatory protein depends on its cellular localization. J. Biol. Chem. 275:39403-39410. [DOI] [PubMed] [Google Scholar]
- 21.Lillycrop, K. A., V. S. Budrahan, N. D. Lakin, G. Terrenghi, J. N. Wood, J. M. Polak, and D. S. Latchman. 1992. A novel POU family transcription factor is closely related to Brn-3 but has a distinct expression pattern in neuronal cells. Nucleic Acids Res. 20:5093-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, W., S. L. Khare, X. Liang, M. A. Peters, X. Liu, C. L. Cepko, and M. Xiang. 2000. All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development 127:3237-3247. [DOI] [PubMed] [Google Scholar]
- 23.McEvilly, R. J., L. Erkman, L. Luo, P. E. Sawchenko, A. F. Ryan, and M. G. Rosenfeld. 1996. Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature 384:574-577. [DOI] [PubMed] [Google Scholar]
- 24.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ninkina, N. N., G. E. Stevens, J. N. Wood, and W. D. Richardson. 1993. A novel Brn3-like POU transcription factor expressed in subsets of rat sensory and spinal cord neurons. Nucleic Acids Res. 21:3175-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pupko, T., R. E. Bell, I. Mayrose, F. Glaser, and N. Ben-Tal. 2002. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics 18(Suppl. 1):S71-S17. [DOI] [PubMed] [Google Scholar]
- 27.Rivolta, M. N., N. Grix, P. Lawlor, J. F. Ashmore, D. J. Jagger, and M. C. Holley. 1998. Auditory hair cell precursors immortalized from the mammalian inner ear. Proc. R. Soc. Lond. B Biol. Sci. 265:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson, N. G., U. Khetarpal, G. A. Gutierrez-Espeleta, F. R. Bieber, and C. C. Morton. 1994. Isolation of novel and known genes from a human fetal cochlear cDNA library using subtractive hybridization and differential screening. Genomics 23:42-50. [DOI] [PubMed] [Google Scholar]
- 29.Ryan, A. F. 1997. Transcription factors and the control of inner ear development. Semin. Cell Dev. Biol. 8:249-256. [DOI] [PubMed] [Google Scholar]
- 30.Smith, M. D., P. J. Morris, and D. S. Latchman. 1998. The Brn-3c transcription factor contains a neuronal-specific activation domain. Neuroreport 9:851-856. [DOI] [PubMed] [Google Scholar]
- 31.Sock, E., J. Enderich, M. G. Rosenfeld, and M. Wegner. 1996. Identification of the nuclear localization signal of the POU domain protein Tst-1/Oct6. J. Biol. Chem. 271:17512-17518. [DOI] [PubMed] [Google Scholar]
- 32.Sturm, R. A., and W. Herr. 1988. The POU domain is a bipartite DNA-binding structure. Nature 336:601-604. [DOI] [PubMed] [Google Scholar]
- 33.Theil, T., B. Rodel, F. Spiegelhalter, and T. Moroy. 1995. Short isoform of POU factor Brn-3b can form a heterodimer with Brn-3a that is inactive for octamer motif binding. J. Biol. Chem. 270:30958-30964. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner, E. E., K. J. Jenne, and M. G. Rosenfeld. 1994. Brn-3.2: a Brn-3-related transcription factor with distinctive central nervous system expression and regulation by retinoic acid. Neuron 12:205-218. [DOI] [PubMed] [Google Scholar]
- 36.Vahava, O., R. Morell, E. D. Lynch, S. Weiss, M. E. Kagan, N. Ahituv, J. E. Morrow, M. K. Lee, A. B. Skvorak, C. C. Morton, A. Blumenfeld, M. Frydman, T. B. Friedman, M. C. King, and K. B. Avraham. 1998. Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science 279:1950-1954. [DOI] [PubMed] [Google Scholar]
- 37.Wegner, M., D. W. Drolet, and M. G. Rosenfeld. 1993. POU-domain proteins: structure and function of developmental regulators. Curr. Opin. Cell Biol. 5:488-498. [DOI] [PubMed] [Google Scholar]
- 38.Willems, P. J. 2000. Genetic causes of hearing loss. N. Engl. J. Med. 342:1101-1109. [DOI] [PubMed] [Google Scholar]
- 39.Xiang, M., L. Gan, D. Li, Z. Y. Chen, L. Zhou, B. W. O'Malley, Jr., W. Klein, and J. Nathans. 1997. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc. Natl. Acad. Sci. USA 94:9445-9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang, M., L. Gan, L. Zhou, W. H. Klein, and J. Nathans. 1996. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc. Natl. Acad. Sci. USA 93:11950-11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang, M., W. Q. Gao, T. Hasson, and J. J. Shin. 1998. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development 125:3935-3946. [DOI] [PubMed] [Google Scholar]
- 42.Xiang, M., A. Maklad, U. Pirvola, and B. Fritzsch. 2003. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC Neurosci. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang, M., L. Zhou, J. P. Macke, T. Yoshioka, S. H. Hendry, R. L. Eddy, T. B. Shows, and J. Nathans. 1995. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J. Neurosci. 15:4762-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang, M., L. Zhou, Y. W. Peng, R. L. Eddy, T. B. Shows, and J. Nathans. 1993. Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron 11:689-701. [DOI] [PubMed] [Google Scholar]