Abstract
Gene activation requires alteration of chromatin structure to facilitate active transcription complex formation at a gene promoter. Nucleosome remodeling complexes and histone modifying complexes each play unique and interdependent roles in bringing about these changes. The role of distant enhancers in these structural alterations is not well understood. We studied nucleosome remodeling and covalent histone modification mediated by the β-globin locus control region HS2 enhancer at nucleosome-level resolution throughout a 5.5-kb globin gene model locus in vivo in K562 cells. We compared the transcriptionally active locus to one in which HS2 was inactivated by mutations in the core NF-E2 sites. In contrast to inactive templates, nucleosomes were mobilized in discrete areas of the active locus, including the HS2 core and the proximal promoter. Large differences in restriction enzyme accessibility between the active and inactive templates were limited to the regions of nucleosome mobilization, which subsumed the DNase I hypersensitive sites. In contrast to this discrete pattern, histone H3 and H4 acetylation and H3 K4 methylation were elevated across the entire active locus, accompanied by depletion of linker histone H1. The coding region of the gene differed from the regulatory regions, demonstrating both nucleosome mobilization and histone hyperacetylation, but lacked differences in restriction enzyme accessibility between transcriptionally active and inactive genes. Thus, although the histone modification pattern we observe is consistent with the spreading of histone modifying activity from the distant enhancer, the pattern of nucleosome mobilization is more compatible with direct contact between an enhancer and promoter.
During gene activation, ATP-dependent nucleosome remodeling complexes and complexes that covalently modify histones, such as acetyl transferases (HATs) and methyl transferases, alter the repressive nucleosomal structure of chromatin and provide a permissive environment for transcription. Remodeling by ATP-dependent complexes such as yeast SWI/SNF and ISWI results in a positional change (mobilization or sliding) and/or a conformational (structural) change in nucleosomes in vitro (54). Histone acetylation modifies lysine residues in the N-terminal tails of the core histones H3 and H4, decreasing the stability of histone-DNA interactions and loosening the compaction of nucleosomal arrays (7). The structural effects of histone methylation are less clear and can be associated with activation (H3 K4 methylation) or repression (H3 K9 methylation) of chromatin (8). Together, these alterations may create an unstable and accessible nucleosome structure that favors transcriptional initiation and, possibly, elongation. In addition to a structural role, covalent histone modifications participate in gene activation as part of a “histone code,” marking genes for subsequent steps in the activation pathway (51).
SWI/SNF-type alterations of chromatin structure are generally considered to be localized at regulatory regions of active genes, such as promoters, to which the complex is targeted by activators (42). Examples include the yeast PHO8 gene (44) and the human c-myc (36) and beta interferon (34) genes. However, evidence also exists for more widespread alterations of chromatin structure requiring SWI/SNF at the yeast FLO1 and HIS3 genes (11, 26).
Hyperacetylation of histones is also associated with actively transcribed genes and may be targeted or widespread. Yeast Gcn5p mediates promoter-specific H3 acetylation (29), and at the induced human beta interferon gene, H3 and H4 acetylation by p300/CBP was limited to 600 bp at the transcription start site (41). In contrast, the human c-myc gene linked to a locus control region (LCR) enhancer was widely hyperacetylated both upstream and downstream of the transcription start site (36). In vitro studies indicate that the property of mediating localized versus widespread hyperacetylation may be particular to different HAT complexes, such as the yeast SAGA and Nu4A complexes, respectively (55). Both nucleosome remodeling complexes and HAT complexes may also modify transcribed regions of genes in advance of, or concomitant with, RNA polymerase II passage (38, 40).
In complex gene loci, such as the globin and growth hormone loci, which are regulated by LCR enhancers, both locus-wide histone hyperacetylation and localized patterns of modification limited to regulatory regions such as enhancers and promoters are observed (5, 12). The human β-globin LCR comprises four core regions (HS1 to -4) that are hypersensitive to DNase I in the chromatin of erythroid cells and are located from 6 to 60 kb upstream of the genes they regulate (50). Although deletion of the human LCR in a chromosomal setting has shown the LCR to be dispensable for locus-wide histone modification, it is required for promoter hyperacetylation and remodeling at the distant globin genes when they are transcriptionally activated (46).
Although a large body of work details recruitment of chromatin remodeling complexes to promoters via sequence-specific DNA binding transcription activators acting through cis elements (18), it is less clear how a distant LCR enhancer participates in such recruitment. If distant LCR enhancers recruit HAT and nucleosome remodeling complexes, like activator elements near gene promoters, then how do such complexes access the promoters they activate? One possibility is that the complexes migrate by tracking (53) across the intervening chromatin, as has been proposed to underlie widespread acetylation in the globin locus (12). Alternatively, an LCR enhancer and promoter may exist in proximity in vivo by looping (31), as suggested by recent work in which distant enhancers and promoters are occupied simultaneously by the HAT CBP (19, 35, 48, 49) and the SWI/SNF components BRG1 (19) and BAF 155 (17). The work of Hatzis and Talianidis (19) elegantly illustrates that these models may not be mutually exclusive, as CBP and BRG1 maintain enhancer association as they progress through intervening sequences to the promoter. These associations also corresponded with unidirectional spreading of histone acetylation. However, for these experiments, the effects of SWI/SNF nucleosome remodeling were assayed at the enhancer and promoter regions only.
To investigate how distant LCR enhancers work, we have studied enhancer-associated changes in chromatin structure of model globin gene loci maintained on chromatinized episomes in human erythroid K562 cells. The β-globin LCR HS2 enhancer was fused to a complete ɛ-globin gene, creating a model locus of over 5 kb in which the HS2 enhancer core is situated 2.5 kb from the ɛ-globin promoter. Since individual nucleosomes are the targets of ATP-dependent nucleosome remodeling complexes and histone modifying complexes, we conducted our analysis with nucleosome-level resolution. We found that enhancer-dependent nucleosome remodeling is highly localized to the regulatory regions of an active chromatin template, has abrupt transitions, and does not affect intervening sequences. In contrast, enhancer-mediated histone modifications across the active locus span the enhancer, gene, and intervening sequences. Surprisingly, distinct from earlier studies, this high-resolution analysis revealed no peak of histone modification at the proximal ɛ-globin promoter or at the HS2 core. Rather, the HS2 core is substantially depleted of histones and indeed appears to be nucleosome-free. In contrast, the gene promoter is not depleted of histones, but contains histones that are modified to a lesser extent than are those in the coding sequences. We discuss these results with respect to models of enhancer action.
MATERIALS AND METHODS
Minichromosome construction, cell culture conditions, and transfection.
Minichromosomes containing HS2 and the ɛ-globin gene were constructed in an Epstein-Barr virus vector containing the Epstein-Barr virus orip, EBNA-1, and the hygromycin gene for minichromosome maintenance. HS2 was contained within a 1.46-kb KpnI-BglII fragment, and the ɛ-globin gene was a 3.7-kb EcoRI fragment. The tandem NF-E2 binding motifs in HS2 were mutated by site-directed mutagenesis (15). Minichromosomes were transfected into K562 cells, and multiple individual clones were selected and maintained as described previously (15). Structural studies were replicated on two clones of each construct.
RNase protection assay.
RNA was prepared from 5 × 106 K562 cells carrying minichromosomes by use of the PUREscript kit (Gentra). The episomal copy of the ɛ-globin gene was marked by a distinguishing mutation in the 5′-untranslated region (15). RNase digestion and gel analyses were performed as suggested by the manufacturer of the reagents (RPA II kit; Ambion). RNase protection assay results were normalized to the actin control signal and corrected for copy number. Clones used in these studies carried between 10 and 30 copies of the minichromosome. Endogenous ɛ-globin mRNA signals varied among K562 cell clones but were unrelated to minichromosome transcription.
Preparation of nuclei and nuclease digestion.
Nuclei were prepared from 5 × 107 K562 cells and digested with 3 to 12 μg of DNase I in a 1-ml volume for 10 min at room temperature or with 50 to 200 U of micrococcal nuclease (MNase) in a 1-ml volume for 5 min at 37°C (15). For restriction enzyme cleavage analyses, nuclei from 1.5 × 107 to 2.0 × 107 cells were incubated with 100 U of restriction enzymes in 400 μl (250-U/ml concentration of each enzyme used) at 37°C for 30 min, conditions previously determined to yield maximum cleavage (15, 16). DNA was purified, electrophoresed on 1% agarose gels (1.2% gels in the case of MNase-digested chromatin), and transferred to nylon membranes. Membranes were hybridized with 32P-labeled probes as indicated in the text and figure legends.
ChIP.
Chromatin immunoprecipitation (ChIP) for analysis of histone modification was carried out as described previously (33). Briefly, nuclei were prepared from 5 × 107 K562 cells, and equal aliquots were digested with different MNase concentrations (0.0025, 0.01, and 0.04 U/μl) for 10 min at 37°C and then combined. Soluble chromatin was fractionated on a sucrose gradient (5 to 30%), and mono- and dinucleosomes were pooled. Fifteen to 20 μg of chromatin was precleared with 100 μl of protein A-agarose for 1 h at 4°C by gentle rocking in IP buffer (50 mM NaCl, 10 mM Tris [pH 7.4], 1 mM EDTA, 10 mM sodium butyrate, and protease inhibitors). The purified chromatin was then reacted with antibodies for 2 h, 100 μl of protein A-agarose was added, and incubation was continued overnight at 4°C. The protein A-agarose was washed five times and bound material was eluted twice. Unbound chromatin in the no-antibody sample was used as input. DNA from both unbound and eluted chromatin was purified by phenol extraction and ethanol precipitation. For analysis of histone H1, 2 × 107 cells were first cross-linked with 0.4% formaldehyde for 10 min at room temperature and then sonicated to an average chromatin fragment size of 200 to 400 bp (23). Soluble chromatin was reacted with anti-H1 antibody for 3 h, and then protein A-agarose was added. After a 2-h incubation, the protein A-agarose was washed five times and eluted as described above. After overnight incubation at 65°C to reverse cross-links, the DNA was purified as described above. The antibodies used were anti-di-acetylated (K9, K14) histone H3, anti-tetra-acetylated (K4, K7, K11, K15) histone H4, anti-di-methylated H3 (K4), anti-di-methylated H3 (K9), and anti-H1 (Upstate Biotechnology, Lake Placid, N.Y.).
Quantitative real-time PCR analysis, primers, and Taqman probes.
Immunoprecipitated DNA was analyzed by quantitative real-time PCR (ABI Prism 7700) using Taqman probes and primers (Table 1) designed by Primer Express 1.0 (PE Applied Biosystems). DNA was quantified by use of picogreen, and 2 ng of DNA was amplified with 200 nmol of Taqman probes and 900 nmol of primers in a 50-μl reaction volume. PCR amplification was performed for 1 cycle at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Data were collected at the threshold, where amplification was linear. The fold difference for each primer pair was determined by comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in input DNA (33). Primers selected from human β-globin sequences generated 60- to 110-bp amplicons at intervals of 100 bp or less. Intervals occasionally ranged up to 300 to 400 bp when sequences unfavorable to primer design were encountered. The sequences of primers and Taqman probes are given in Table 1.
TABLE 1.
Taqman primers and probes
| Target area (starting nt/ending nt) | Primer or probe (sequence) |
|---|---|
| HS2 5′ (−3447/−3367) | Forward (5′AGTCACTCTGTCTCACTGTGTCTTAGC) |
| Reverse (5′TTGTTGGAGGATACCCATTCTCTAT) | |
| Probe (5′6FAM-AGTTCCTTACAGCTTGCCCTGATGGGA-TAMRA) | |
| HS2 HindIII (−2735/−2664) | Forward (5′CTTAGTTCCTGTTACATTTCTGTGTGTCT) |
| Reverse (5′GGCCAGAACTGCTCATGCTT) | |
| Probe (5′6FAM-TAGTGACCTCCCATAGTC-TAMRA) | |
| HS2 NF-E2 (−2594/−2511) | Forward (5′GGCTCAAGCACAGCAATGC) |
| Reverse (5′CATCACTCTAGGCTGAGAACATCTG) | |
| Probe (5′6FAM-AGTCATGCTGAGGCTTAGGGTGTGTGC-TAMRA) | |
| HS2 XbaI (−2459/−2383) | Forward (5′AGGAGAAGCTGACCACCTGACT) |
| Reverse (5′CACATTCTGTCTCAGGCATCCA) | |
| Probe (5′6FAM-AAACTCCACCTCAAACGGCATCATAAAGAAA-TAMRA) | |
| HS2 AT-rich (−2299/−2209) | Forward (5′TGTTGTTATCAATTGCCATAGAATGA) |
| Reverse (5′CTGATTCTGCCGCTTCTAGGTATAG) | |
| Probe (5′6FAM-TTATCTTGCAGGTGGCC-TAMRA) | |
| HS2 3′ (−2124/−2051) | Forward (5′AAACAGGAAAAGAAAACAAACCTTGT) |
| Reverse (5′CACAGGCCTTTTGCCACCTA) | |
| Probe (5′6FAM-TGCCATTTTAAGGCACCCCTGGACAG-TAMRA) | |
| N6− (−1708/−1634) | Forward (5′GAATCCCCAAAGGCACTTGA) |
| Reverse (5′GGCCCTAGAGGTTCTTCGTCTT) | |
| Probe (5′6FAM-TGGGAGACAGGAGCCATACTGCTAAGTGA-TAMRA) | |
| N5− (−1204/−1107) | Forward (5′ATGGTGATGTAAGGGACTTTTATAGAATT) |
| Reverse (5′CTATCAATGTGACAGAAGTAATGATTGC) | |
| Probe (5′6FAM-CAATGCTGGAATTTGTGGAACTCTGCTTCTA-TAMRA) | |
| N4− (−725/−651) | Forward (5′GCCCCAGGCAATACTCACAA) |
| Reverse (5′CCAGGGAATCCTCTCTCTTTTCA) | |
| Probe (5′6FAM-AGCCAGTGTCCCCTGTGGTCATAGAGAA-TAMRA) | |
| N2− (−324/−240) | Forward (5′GATTTGCTCCTTTATATGAGGCTTTC) |
| Reverse (5′TTTTACCCTCTTCATCATCTTCCAA) | |
| Probe (5′6FAM-TGGAAAAGGAGAATGGGAGAGATGGATATCA-TAMRA) | |
| N1− (−161/−52) | Forward (5′CACAAACTTAGTGTCCATCCATCAC) |
| Reverse (5′CCCTGTTCTCCATGGTACTTAAAAG) | |
| Probe (5′6FAM-CTGACCCTCTCCGGACCTGACTCCAC-TAMRA) | |
| N1+ (18/86) | Forward (5′CAGCTGCAATCACTAGCAAGCT) |
| Reverse (5′GCAGCCTTCTCCTCAGCAGTA) | |
| Probe (5′6FAM-CAGGCCTGGCATCATGGTGCA-TAMRA) | |
| N2+ (300/371) | Forward (5′TTTTTTGACAGCTTTGGAAACCT) |
| Reverse (5′GCCATGGGCCTTGACCTT) | |
| Probe (5′6FAM-TCGTCTCCCTCTGCCATCCTGGG-TAMRA) | |
| N3+ (422/494) | Forward (5′CAAGCCCGCCTTTGCTAAG) |
| Reverse (5′CACCTTGAAGTTCTCAGGATCCA) | |
| Probe (5′6FAM-TGAGTGAGCTGCACTGTGACAAGCTGC-TAMRA) | |
| N4+ (940/1017) | Forward (5′ATTTCCCATCAGTATTGTGACCAA) |
| Reverse (5′AGAGTTCTCAGCGGGAGTTTAAAA) | |
| Probe (5′6FAM-TGAAGGCTTGTTTCCGAATTTGTTGGG-TAMRA) | |
| N5+ (137/1446) | Forward (5′CTACTCACTTTGGCAAGGAGTTCA) |
| Reverse (5′CGACAGCAGACACCAGCTTCT) | |
| Probe (5′6FAM-CCCTGAAGTGCAGGCTGCCTGG-TAMRA) |
RESULTS
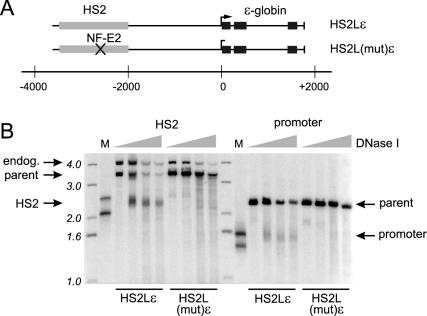
We previously observed nucleosome mobilization and histone hyperacetylation at the promoter of the transcriptionally active ɛ-globin gene linked with the 374-bp core HS2 enhancer on stably maintained minichromosomes in K562 cells (16, 17). To investigate the chromatin anatomy of a more complete enhancer-gene locus, we created minichromosomes linking ɛ-globin to a longer 1.46-kb KpnI-to-BglII LCR HS2 enhancer fragment (Fig. 1A, HS2Lɛ). The promoter and the core of the long enhancer are separated by 2.5 kb in this locus. In a second minichromosome, designated HS2L(mut)ɛ, the tandemly repeated NF-E2 sites in HS2 were destroyed by clustered point mutations. The mutation resulted in the loss of the DNase I hypersensitive region at HS2 (Fig. 1B) and at the ɛ-globin promoter, both of which had been preserved in HS2Lɛ. In HS2Lɛ, the hypersensitive regions spanned −2,420 to −2,820 (relative to the ɛ-globin transcription start site in the minichromosome) in HS2 and −180 to +20 in the ɛ-globin gene. No other hypersensitive sites were detected in sequences flanking the HS2 core, in the extended 5′ flanking region of the ɛ-globin gene, or in transcribed sequences.
FIG. 1.
Formation of HS2 and the ɛ-globin promoter DNase I hypersensitive site are enhancer dependent. (A) A 1.46-kb HS2 fragment containing the 400-bp HS2 core with 5′ and 3′ flanking sequences was linked to a complete genomic ɛ-globin gene (see Materials and Methods) in HS2Lɛ. HS2L(mut)ɛ was identical, except the tandem NF-E2 sites in the HS2 core were mutated by clustered point mutations. The core of HS2 lies about 2.5 kb distant from the ɛ-globin promoter in these constructs. Gray bars, HS2 sequences; black bars, exons of the gene. (B) Nuclei of cells carrying the wild-type or mutant constructs were prepared and digested with various amounts of DNase I as described in Materials and Methods. The purified DNA was completely digested with either EcoRV (HS2 analysis) or BglII (promoter analysis) to produce a parent fragment, run on 1% agarose gels, and analyzed by Southern blotting with appropriate radiolabeled probes. Representative results are presented. Increasing DNase I concentrations are indicated above the lanes. The signal from the endogenous K562 chromosome is indicated (endog.). M lanes, restriction enzyme fragments used to map the location of the DNase I-cleaved bands. The migration positions of radiolabeled 1-kb ladder fragments are indicated.
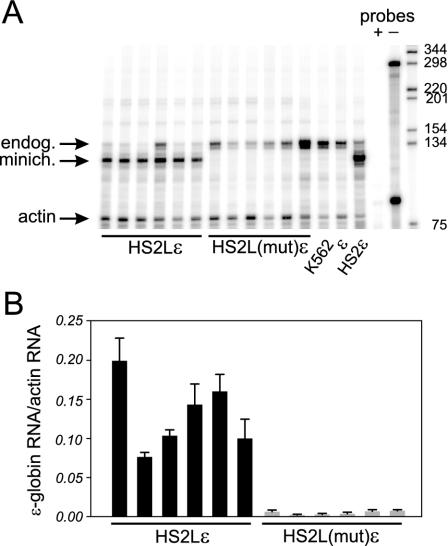
The NF-E2 mutation in the context of the 374-bp core had inactivated the enhancer function of HS2 (15). To investigate the effect on transcription activation of the NF-E2 mutation in the context of an extended HS2, we performed RNase protection assays on RNA isolated from clones with the wild-type (HS2Lɛ) or mutated [HS2L(mut)ɛ] enhancer. The assay distinguishes episomal ɛ-globin transcripts from endogenous transcripts because the marked episomal copy results in shorter protected probe fragments. A representative experiment is depicted in Fig. 2A, and the results of multiple determinations for six different clones of HS2Lɛ and HS2L(mut)ɛ are summarized in Fig. 2B. Abundant transcription of ɛ-globin RNA is seen when the gene is linked to the longer 1.46-kb HS2 sequence (HS2Lɛ). Destruction of the tandem NF-E2 binding sites in HS2 abolished transcription of the ɛ-globin gene [HS2L(mut)ɛ]. These results are consistent with our previous findings using a short 374-bp HS2 core (15).
FIG. 2.
Transcription of the ɛ-globin gene depends on the HS2 NF-E2 sites. (A) Representative RNase protection analysis of RNA from multiple clones carrying the wild-type (HS2Lɛ) or mutant [HS2L(mut)ɛ] minichromosome. Controls include K562 RNA and RNA from a clone carrying an enhancerless ɛ-globin gene (ɛ) or the gene linked to a 400-bp HS2 core fragment (HS2ɛ). The signal from endogenous K562 RNA (endog.) is indicated. RNase protection probes, with (+) or without (−) RNase treatment, were included. Actin served as the loading control. Sizes of marker fragments are indicated in nucleotides. (B) Bar graph representing the results of three independent experiments (plus standard errors of the means). The ɛ-globin RNA signal is normalized to the actin RNA loading control and for minichromosome copy number.
Discrete nucleosome mobilization in HS2 and the ɛ-globin gene.
The presence of DNase I hypersensitive sites implies remodeling of nucleosomes at HS2 and at the promoter of the ɛ-globin gene. ATP-dependent remodeling complexes can mobilize nucleosomes and/or bring about nucleosomal conformational changes in vitro (54). To directly study nucleosome mobilization in vivo across the model locus, we examined MNase-derived nucleosome ladder patterns by Southern blotting with small DNA probes. In this assay, a distinct ladder of mono- and oligonucleosome fragments indicates the presence of stable nucleosomes that protect probe sequences from MNase digestion, while a disrupted or smeared ladder indicates nucleosome mobilization and/or loss, resulting in vulnerability of underlying sequences to nuclease digestion. For simplicity, we will refer to this change as mobilization.
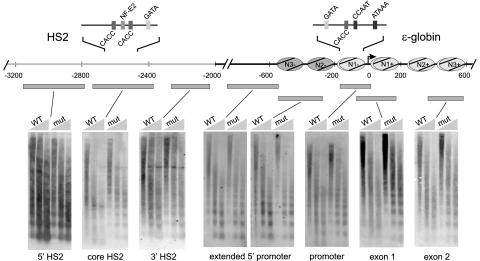
Nucleosome ladders obtained from MNase treatment of nuclei of HS2Lɛ or HS2L(mut)ɛ cells were successively hybridized with three small probes corresponding to the HS2 core and its 5′ and 3′ flanking sequences or with five probes spanning 1.4 kb of sequence surrounding the ɛ-globin gene. As illustrated in Fig. 3, HS2L(mut)ɛ nucleosome ladders were distinct at all locations in the template, indicating the presence of stable nucleosomes throughout. However, an HS2Lɛ nucleosome ladder probed within the HS2 core was smeared and reduced in intensity, indicating nucleosome mobilization. In contrast, the 5′ and 3′ HS2-flanking regions yielded distinct nucleosomal ladders, indicating stable nucleosomes. The resolution of this HS2 analysis suggests the mobilization of two nucleosomes, at the most. We conclude that nucleosome mobilization at the HS2 enhancer is confined to two nucleosomes at the HS2 core and has discrete limits on both flanks.
FIG. 3.
Discrete nucleosome mobilization in the HS2 core and gene promoter. The positions of nucleosomes as determined by indirect end labeling over ɛ-globin sequences are shown as shaded ovals (15). Filled bars represent the restriction enzyme fragments, with respect to the locus illustrated above, which were hybridized to Southern blots of MNase ladders. Nuclei of K562 cells carrying the wild-type or mutant constructs were prepared, digested with various amounts of MNase, and processed as described in Materials and Methods. The purified DNA was run on 1.2% agarose gels and analyzed by Southern blotting and sequential hybridization with each of the indicated radiolabeled probes. Representative results are shown. Increasing MNase concentrations are indicated above the lanes. Light gray, mobilized nucleosomes; dark gray, stable nucleosomes.
Stable nucleosomes were apparent across the sequences separating HS2 and the gene promoter in HS2Lɛ. This pattern abruptly changed at the proximal promoter, at which smearing of the ladder pattern which extended through the coding region of the actively transcribed gene was observed. In earlier work we mapped the positions of nucleosomes surrounding the ɛ-globin promoter (labeled in Fig. 3). We conclude that mobilization at the ɛ-globin gene commences sharply at N1− and continues at least through the second exon to N3+.
Restriction enzyme accessibility is variable across the locus.
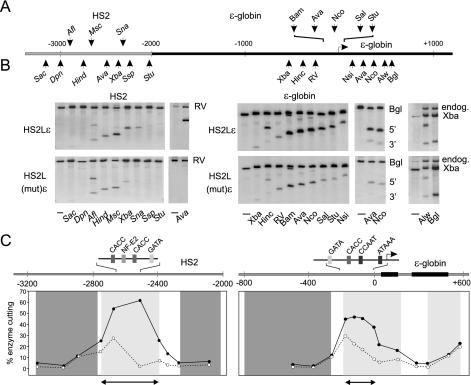
To assess conformational changes in nucleosomes as a consequence of remodeling, we measured restriction enzyme accessibility in chromatin in the active and inactive templates. Figure 4A illustrates the locations of the 10 sites surrounding HS2 and the 13 sites across the extended promoter and transcribed regions of the ɛ-globin gene that were studied, and representative results are presented in Fig. 4B. The percent cut for each enzyme was calculated, and the data are presented in Fig. 4C as line graphs superimposed on the MNase ladder results from Fig. 3.
FIG. 4.
Restriction enzyme accessibility is limited to the HS2 core and ɛ-globin proximal promoter. (A) The locations of the restriction enzyme sites studied within HS2 and the ɛ-globin gene are indicated. The gray bar represents HS2 sequences, and the black bar represents ɛ-globin sequences. (B) Nuclei of 1.5 × 107 to 2.0 × 107 K562 cells carrying HS2Lɛ or the mutant HS2L(mut)ɛ were prepared and digested with different restriction enzymes. After purification, the DNA was cleaved to completion with either EcoRV, BglII, or XbaI to yield a parent fragment. Southern blots of the cleaved DNA separated on 1% agarose gels were hybridized with internal radiolabeled probes. The parent bands are indicated by the restriction enzyme used to generate them. Where visible, the endogenous chromosomal band is indicated (endog.). Representative results are presented. (C) Restriction enzyme accessibility and nucleosome mobilization across the locus are compared. Light gray panels, extent of mobilized nucleosomes; dark gray panels, regions of stable nucleosomes within actively transcribed HS2Lɛ (see Fig. 3). The average percent (for three independent determinations) restriction enzyme accessibilities [cut/(cut + uncut)] for HS2Lɛ (solid line) and HS2(mut)ɛ (dotted line) are plotted (see panel B). The heavy horizontal arrow represents the extent of the HS2 and promoter DNase I hypersensitive sites (Fig. 1B). Black rectangles, exons 1 and 2 of ɛ-globin.
Large increases in restriction enzyme accessibility at the active locus compared to the inactive locus were confined to the core of HS2 and the proximal promoter of the ɛ-globin gene. These regions corresponded to regions of nucleosome mobilization (Fig. 4C, light gray panels), DNase I hypersensitive sites (data from Fig. 1) (Fig. 4C, horizontal arrows), and transcription activator motifs. In particular, AvaII showed extremely high cutting levels in the HS2 core of active templates which were greater than at the proximal-promoter AvaII site. Thus, HS2 has an altered chromatin structure which is highly susceptible to restriction enzyme cleavage. In contrast, both the active and inactive loci showed similar levels of restriction enzyme accessibility and stable nucleosomes (Fig. 4C, dark gray panels) in sequences flanking the HS2 core and in sequences between the enhancer and the promoter.
The ɛ-globin transcribed region showed unique structural characteristics. Nucleosomes are mobilized in the actively transcribed gene coding sequences, but restriction enzyme accessibility is similar between the transcribed and nontranscribed genes (including at AvaII and NcoI sites, which are cut strongly in the active promoter). Thus, nucleosome remodeling at the enhancer and gene promoter appears to be fundamentally different from the type of nucleosome remodeling observed in the coding region.
Histone modification and H1 depletion characterize the entire active locus.
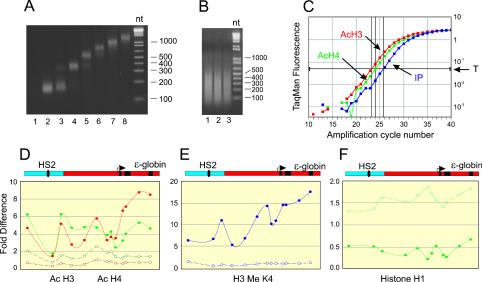
HS2 transactivation of the ɛ-globin gene is associated with hyperacetylation of H3 and H4 at HS2 and at the gene promoter in active templates relative to inactive templates (16, 17). To examine whether covalent histone modification is localized at these regulatory regions or whether it extends broadly in the intervening sequences between promoter and enhancer and in the transcribed region, we determined the histone modification levels by ChIP at nucleosome-level resolution across the 5.5-kb loci. To compensate for potential differences in sensitivity to MNase across the locus, nuclei were digested with graded concentrations of MNase, combined, and fractionated through a sucrose gradient. The mono- and dinucleosome-containing fractions (Fig. 5A, lanes 2 to 4) were pooled for ChIP assays. Samples were reacted with antibodies to di-acetylated H3, tetra-acetylated H4, and H3 dimethylated at either K4 or K9. The specifically immunoprecipitated DNA was amplified by quantitative real-time PCR using Taqman probes at 13 points distributed across the locus. To the extent that was possible, amplicons were selected to fall within known nucleosome positions. The relative abundance at each point was calculated by comparing the threshold cycle number in immunoprecipitated DNA with the number for input DNA. An example of an amplification plot using primer N4− (Table 1) appears in Fig. 5C.
FIG. 5.
Histone modification is widespread across the locus. (A) Nuclei from K562 cells carrying HS2Lɛ or HS2L(mut)ɛ were digested with three different concentrations of MNase (see Materials and Methods). Soluble chromatin from combined digests was fractionated on a sucrose gradient. Fractions were analyzed on a 2% agarose gel. The mono- and dinucleosome-containing fractions (2-4) were pooled. (B) For H1 analysis, cells were cross-linked with formaldehyde and sonicated to an average size of 200 to 400 bp as described in Materials and Methods. An ethidium bromide-stained image of aliquots of DNA from three different preparations (lanes 1 to 3) is shown. (C) Specifically immunoprecipitated DNA and input DNA were quantitatively analyzed by real-time PCR using primers and Taqman probes (Table 1) that spanned the locus. The fold difference for each primer pair was determined by comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in input DNA (IP), as described previously (33). An example of an amplification plot obtained with N4− primers is shown. T, threshold signal used to calculate fold increase. (D) ChIP was performed with antibodies to di-acetylated H3 (9, 14) (red) and hyperacetylated H4 (green). At least three individual chromatin preparations for each clone were analyzed, with highly reproducible results. Average results are shown for HS2Lɛ (solid lines) or HS2L(mut)ɛ (dashed lines) in this and the following panels. (E) ChIP was performed as for panel D with antibodies to H3 dimethylated at K4. (F) ChIP was performed with antibodies to H1. Diagrams of HS2 (blue) and ɛ-globin (red) sequences in the almost 6-kb locus appear above panels D to F. Black oval, core HS2 factor binding sites; black rectangles, exons 1 to 3 of ɛ-globin.
At all the points analyzed, transcriptionally active HS2Lɛ showed elevated H3 and H4 acetylation levels compared to those seen in HS2L(mut)ɛ (Fig. 5D). Enrichment of at least twofold, and up to ninefold, was observed. Our analysis, at approximately nucleosome-level resolution, revealed varying histone acetylation across the extended promoter of the active ɛ-globin gene. Nucleosome-to-nucleosome variation was also seen for the endogenous ɛ-globin gene of K562 cells, but its significance is not clear (A. Kim and A. Dean, unpublished observations). The highest levels of modification for acetylated H3 were observed across the coding sequences of the ɛ-globin gene. As noted above, the transcribed region contained mobilized but not remodeled nucleosomes, as they did not demonstrate increased restriction enzyme accessibility (Fig. 4C).
These results reveal that the pattern of histone acetylation established by the enhancer is not localized to regulatory regions but is established across the locus, including the sequences surrounding the enhancer core, the intervening sequences between the promoter and enhancer, and the transcribed region. The H3 and H4 acetylation profiles corresponded with H3 K4 methylation, a modification which is also associated with active genes (Fig. 5E). Again, the highest levels of methylation of H3 K4 appeared in the coding region. In contrast, H3 K9 methylation, generally associated with repressive chromatin, occurred at a very low level in the active template and was inversely related to H3 K4 methylation (data not shown). Finally, immunoprecipitation of formaldehyde-cross-linked chromatin, sonicated to 200- to 500-bp fragments (Fig. 5B), with antibodies to linker histone H1 (Fig. 5F) indicated depletion of H1 across the active locus.
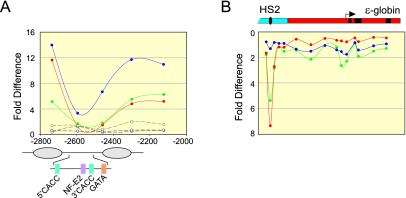
Unexpectedly, we found that neither HS2 nor the gene promoter appeared as peaks of histone modification. In fact, HS2 was significantly under-acetylated in this analysis. We repeated the analysis with more closely spaced primers within HS2 and found that sequences 5′ and 3′ of the core were highly acetylated at H3 and H4 and methylated at H3 K4 (Fig. 6A); H3 and H4 acetylation, however, was absent at the HS2 core NF-E2 sites and at a position 60 bp 3′, while K4 methylation was uniquely low at the NF-E2 sites.
FIG. 6.
A nucleosome is evicted at HS2. Nuclei from K562 cell clones carrying HS2Lɛ or HS2L(mut)ɛ were prepared by digestion with three different concentrations of MNase and analyzed as described in the legend for Fig. 5. (A) Anti-acH3 (red), -acH4 (green), and -H3-meK4 (blue) antibodies were used for immunoprecipitation, followed by analysis by real-time PCR as described in the legend to Fig. 5C, using three additional primer pairs (Table 1). Solid lines, HS2Lɛ; dashed lines, HS2L(mut)ɛ. (B) Total DNA from K562 cells carrying HS2Lɛ, HS2L(mut)ɛ, or no minichromosome was lightly digested with EcoRI, and samples were amplified by real-time PCR with the primers and probes used for analysis in Fig. 5D to F and 6A. The fold difference for each primer pair was determined by comparing the amount of target sequence in total DNA to the amount of target sequence in input DNA. HS2 and ɛ-globin sequences are diagrammed above. Green line, HS2Lɛ; blue line, HS2L(mut)ɛ; red line, K562 with no minichromosome.
We postulated that depletion of HS2 sequences from the input material due to high sensitivity to digestion by MNase could explain this result, even though the input represented a pool of mono- and dinucleosomes obtained from graded MNase digestions. To test this possibility, we compared the genomic DNA signal to the input DNA signal across the locus (Fig. 6B). For this analysis, values that are significantly >1 indicate depleted sequences and appear as a depression in the inverted plot. Primers encompassing the NF-E2 sites revealed that sequences corresponding to this region are indeed depleted from the active locus, consistent with the idea that most HS2 sequences are nucleosome-free and therefore highly susceptible to MNase digestion. Significantly, this pattern is not unique to the minichromosome, since identical results were obtained at the endogenous locus in K562 cells (Fig. 6B). The extent of the depleted region corresponds to one nucleosome encompassing the tandem NF-E2 sites and nearby activator motifs in the HS2 core. Proximal promoter sequences do not appear to be depleted from input DNA, either on minichromosomes or in the endogenous locus. These results are consistent with the absence of a nucleosome at HS2 but the persistence of a remodeled nucleosome at the active ɛ-globin gene promoter.
DISCUSSION
To gain insight into how an enhancer changes chromatin structure when it activates a distant gene, we linked the HS2 enhancer of the β-globin LCR to a complete ɛ-globin gene on chromatinized episomes in human cells. Our study of the chromatin anatomy of this model locus at near-nucleosome-level resolution revealed sharp transitions between mobilized and stable nucleosomes at the core of the HS2 enhancer and at the ɛ-globin gene promoter. In the 2.5 kb between the HS2 core and the active ɛ-globin gene promoter, nucleosomes were not mobilized, nor was restriction enzyme accessibility different from that of the inactive template. In contrast, histone modification was widespread across the active locus. Surprisingly, histone modification was most pronounced within coding sequences which appeared to have unique chromatin structural properties. Thus, the enhancer acts to recruit both nucleosome remodeling and histone modifying complexes, but each complex manifests a distinct spatial pattern of modification along the chromatin.
Nucleosome remodeling.
The HS2 enhancer and the ɛ-globin promoter regulatory regions where nucleosomes were mobilized correspond precisely with regions of increased restriction enzyme accessibility, indicative of ATP-dependent SWI/SNF-type remodeling (42, 54). Although the identity of the complex involved in remodeling HS2 and the promoter is unclear, we have shown that HS2 recruits BAF 155, a component of human SWI/SNF, to HS2 and to the ɛ-globin promoter (17). An erythroid SWI/SNF-type remodeling complex, ERC-1, described in vitro, also contains BAF 155 (3). Consistent with recruitment of SWI/SNF components to this and other enhancers (17, 49), these experiments indicated that SWI/SNF-type remodeling is clearly a component of enhancer activity. Interestingly, the transitions between remodeled and stable nucleosomes in our model locus were quite sharp, occurring at both borders of core HS2 and at the 5′ border of the promoter nucleosome N1−. Possibly, a stable barrier to nucleosome mobility exists in these locations. A stable nucleosome with an adjacent occupied GATA-1 motif exists at the 3′ border of HS2 in K562 cells (25). The GATA-1 site at −210 in the ɛ-globin promoter could serve a similar function here.
How SWI/SNF or another remodeler which is recruited to an enhancer acts at a distant promoter is not clear. We observed no chromatin changes attributable to nucleosome remodeling between the enhancer and promoter. If a remodeling complex is recruited to HS2 and traverses the intervening chromatin to the promoter, it leaves no trace of its passage. Possibly, the complex is transferred directly to the promoter, as has been proposed for transfer of RNA polymerase II from the globin LCR to the β-globin promoter (24). Alternatively, recruitment to the promoter might be a consequence of enhancer-promoter contact, consistent with the looping model of enhancer-promoter communication and as demonstrated for HS2 and the mouse β-globin gene (6, 52). Since only the BAF 155 and BRG1 subunits of SWI/SNF have been detected at enhancers, we cannot rule out the possibility that an incomplete or catalytically inactive complex tracks across intervening chromatin to the promoter. BRG1, the catalytic ATPase of human SWI/SNF, was detected at intermediate positions between the enhancer and promoter of the HNF4-α gene (19), although structural effects of SWI/SNF were not assayed in those studies. Since SWI/SNF and HAT complexes can act synergistically (14, 38), it has been speculated that one function of BRG1 in transit across intervening chromatin is to facilitate the histone acetylation process (see below) (19).
Histone modification.
We found that an active HS2 enhancer was associated with covalent histone modification across the model gene locus, suggesting, as have other studies, that HAT activity is a component of enhancer function (28, 36). In some cases, recruitment to an enhancer has been correlated with physical passage of the HAT over the intervening DNA associated with histone modification (19, 35, 48, 49). Spreading of histone acetylation leading to hyperacetylated domains of chromatin has been proposed (10, 12). Tissue-specific domains of histone acetylation have been found in both globin (13, 20, 33, 46, 47) and human growth hormone (9) loci, all of which are regulated by a distant LCR. In the chicken globin locus, H3 and H4 acetylation directly correlated with H3 K4 methylation and was inversely correlated with H3 K9 methylation (32). This pattern was replicated in our model locus in an enhancer-dependent fashion. Deletion studies, performed by others, of the mouse and human globin loci have shown that the LCR hypersensitive sites are dispensable for locus-wide histone acetylation (4, 46, 47). Since HAT recruitment is a function of HS2 in the model locus we studied, we suggest that this capacity is likely retained in the native locus as well and that HAT activity is redundant outside of the LCR rather than absent from the LCR sequences deleted in earlier studies.
In addition to locus-wide acetylation, the mouse and human globin loci exhibit peaks of acetylated histones at the LCR and at transcribed genes, although such peaks were not obvious in the chicken globin locus (5). These local modifications are dependent on the LCR per se in the human locus (46). Surprisingly, in our studies, HS2 and the gene promoter were not seen as peaks in histone modification. The promoter had a relatively low acetylation level compared to adjacent regions, and at the HS2 core, acetylation was similar between the active HS2 and the inactive mutant. Despite technical adjustments, comparison with total DNA indicated significant depletion of HS2 core sequences from the input DNA. The very high sensitivity of the HS2 core to MNase suggests that there is no nucleosome at HS2. This conclusion is consistent with the relatively weak signal obtained with the core HS2 probe in the MNase ladder experiments with HS2Lɛ (Fig. 3). Although DNase I hypersensitive sites have been assumed to be nucleosome-free, this is far from clear. We have earlier shown that the ɛ-globin gene promoter, in spite of its DNase I hypersensitive structure, retains a nucleosome (16), which is consistent with the current data. These experiments make the important distinction between eviction of a nucleosome at HS2 and the presence of a remodeled nucleosome at the active ɛ-globin gene promoter. Although Fig. 6B confirms that these structural aspects of the minichromosomal globin locus are shared with the endogenous globin locus in K562 cells, it will be important to carry out a complete analysis of the endogenous human globin locus.
It is likely that the small size of the chromatin we used (200 bp) and the small size of the PCR amplicons (<100 bp) allowed us to describe these previously unobserved structural aspects of HS2 and the gene promoter. When our fold-enrichment data were normalized for comparison to the results of other investigators (5), at points other than core HS2 and at N1−, there was a high degree of agreement. Quantitative modification at the LCR in our model locus was similar to that in the endogenous murine locus, and modification of the active ɛ-globin gene promoter was similar to that of the active β-globin gene in other studies. One site of difference between investigators, however, was located between LCR HS1 and the ɛ-globin gene. The modification level was very low in a previous study (13), while it was high in our case. This is likely attributable to the fact that the embryonic gene is active in K562 cells, studied here, but is inactive in MEL cells, studied by others, in which it is part of an under-acetylated condensed chromatin domain.
The role of linker histone H1.
We found that linker histone H1 was depleted from the entire model locus in an HS2-dependent fashion, as described in earlier work with the active ɛ-globin promoter (16). Others have described a similar magnitude of depletion of H1 (twofold) in the active versus the inactive chicken β-globin locus (43). H1 acts as a general repressor of transcription (30), perhaps by stabilizing higher-order chromatin structure. It also acts as a specific inhibitor of histone acetylation (21) and SWI/SNF activity (22) in vitro. Interestingly, we found that H1 and acetylated H3 and H4 (and K4 methylated H3) were inversely correlated in the model locus. The internuclear organization of H1 is dynamic (37), and its binding to any particular site on chromatin is transient. Therefore, it is possible that in the absence of H1, acetylation of histones and SWI/SNF remodeling can progress and in turn inhibit redeposition of H1. Phosphorylation of H1 may produce local regions of very rapidly exchanging H1, which would facilitate this process (22). H1 depletion correlated positively with H3 K9 methylation, which is associated with repressive chromatin.
The transcribed region.
Our results with the NF-E2 mutant HS2 indicate that in the absence of a functional enhancer, the ɛ-globin coding region is unmarked by H3 and H4 modification. H3 hypoacetylation in the coding region in vivo results in transcriptional inhibition (27). In contrast, in the actively transcribed coding region, H3 was highly modified by acetylation, consistent with the association of H3 acetylation with euchromatic chromatin (39). Methylation on H3 K4 was also highest in the coding region. Recent data indicate that H3 K4 trimethylation may be a particular mark of transcribed regions (45). The anti-dimethylated H3 antibody we used for our ChIP assays cross-reacts with the trimethyl modification, and therefore the segregation of the di- and trimethyl modification in our model locus is unclear and warrants further study. Nucleosomes were also mobilized in the actively transcribed coding region. Both remodeling and covalent histone modification could facilitate RNA polymerase II elongation, and recent evidence shows that RNA polymerase II holoenzyme can contain remodeling complexes (40).
In our study, although nucleosome mobilization extended through the transcribed sequences of the gene, it was notably not associated with increased restriction enzyme accessibility. Differences in activity of the restriction enzymes used could explain this result. However, we do not favor this explanation, because in the coding sequences the AvaII and NcoI sites are clearly inaccessible, while in the promoter they are accessible (even in the inactive promoter). Possibly, transcription per se may cause these alterations. Alternatively, a second remodeling complex, recruited directly to the promoter together with RNA polymerase II, could be responsible for the coding sequence alterations. One candidate is an ISWI-type remodeling complex. ISWI complexes mobilize nucleosomes but do not bring about the conformational changes in nucleosomes that are typical of SWI/SNF and that result in increased restriction enzyme accessibility (1). Underlying this observation may be fundamental differences in the mechanism of action of BRG1, the catalytic ATPase of SWI/SNF, and of SNF2H, the catalytic ATPase of the human ISWI complex (2). The above alternatives will be experimentally addressed in future work.
Acknowledgments
We thank Gary Felsenfeld and Michael Litt for use of the ABI Prism 7700 and for many useful discussions. We thank David Clark, Craig Peterson, and Jane Little for critical comments on the manuscript.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does “chromatin remodeling” mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Aalfs, J. D., G. J. Narlikar, and R. E. Kingston. 2001. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem. 276:34270-34278. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 4.Bender, M. A., M. Bulger, J. Close, and M. Groudine. 2000. β-Globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol. Cell 5:387-393. [DOI] [PubMed] [Google Scholar]
- 5.Bulger, M., T. Sawado, D. Schubeler, and M. Groudine. 2002. ChIPs of the beta-globin locus: unraveling gene regulation within an active domain. Curr. Opin. Genet. Dev. 12:170-177. [DOI] [PubMed] [Google Scholar]
- 6.Carter, D., L. Chakalova, C. S. Osborne, Y. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, W. L., S. D. Briggs, and C. D. Allis. 2000. Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 12:326-333. [DOI] [PubMed] [Google Scholar]
- 8.Davie, J. K., and S. Y. Dent. 2002. Transcriptional control: an activating role for arginine methylation. Curr. Biol. 12:R59-R61. [DOI] [PubMed] [Google Scholar]
- 9.Elefant, F., N. E. Cooke, and S. A. Liebhaber. 2000. Targeted recruitment of histone acetyltransferase activity to a locus control region. J. Biol. Chem. 275:13827-13834. [DOI] [PubMed] [Google Scholar]
- 10.Elefant, F., Y. Su, S. A. Liebhaber, and N. E. Cooke. 2000. Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO J. 19:6814-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming, A. B., and S. Pennings. 2001. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 20:5219-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsberg, E. C., and E. H. Bresnick. 2001. Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 23:820-830. [DOI] [PubMed] [Google Scholar]
- 13.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 15.Gong, Q. H., J. C. McDowell, and A. Dean. 1996. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the ɛ-globin gene in vivo by 5′ hypersensitive site 2 of the β-globin locus control region. Mol. Cell. Biol. 16:6055-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui, C. Y., and A. Dean. 2001. Acetylation of a specific promoter nucleosome accompanies activation of the epsilon-globin gene by beta-globin locus control region HS2. Mol. Cell. Biol. 21:1155-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gui, C. Y., and A. Dean. 2003. A major role for the TATA box in recruitment of chromatin modifying complexes to a globin gene promoter. Proc. Natl. Acad. Sci. USA 100:7009-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 19.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 20.Hebbes, T. R., A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 1994. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 13:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrera, J. E., K. L. West, R. L. Schiltz, Y. Nakatani, and M. Bustin. 2000. Histone H1 is a specific repressor of core histone acetylation in chromatin. Mol. Cell. Biol. 20:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn, P. J., L. M. Carruthers, C. Logie, D. A. Hill, M. J. Solomon, P. A. Wade, A. N. Imbalzano, J. C. Hansen, and C. L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263-267. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, K. D., and E. H. Bresnick. 2002. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 26:27-36. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 25.Kim, A. R., and V. Murray. 2001. Chromatin structure at the 3′-boundary of the human beta-globin locus control region hypersensitive site-2. Int. J. Biochem. Cell Biol. 33:1183-1192. [DOI] [PubMed] [Google Scholar]
- 26.Kim, Y., and D. J. Clark. 2002. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. USA 99:15381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kristjuhan, A., J. Walker, N. Suka, M. Grunstein, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2002. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol. Cell 10:925-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krumm, A., L. Madisen, X. J. Yang, R. Goodman, Y. Nakatani, and M. Groudine. 1998. Long-distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc. Natl. Acad. Sci. USA 95:13501-13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laybourn, P. J., and J. T. Kadonaga. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254:238-245. [DOI] [PubMed] [Google Scholar]
- 31.Li, Q., S. Harju, and K. R. Peterson. 1999. Locus control regions: coming of age at a decade plus. Trends Genet. 15:403-408. [DOI] [PubMed] [Google Scholar]
- 32.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 33.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 35.Louie, M. C., H. Q. Yang, A. H. Ma, W. Xu, J. X. Zou, H. J. Kung, and H. W. Chen. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. USA 100:2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madisen, L., A. Krumm, T. R. Hebbes, and M. Groudine. 1998. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol. Cell. Biol. 18:6281-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 38.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 39.Noma, K., and S. I. Grewal. 2002. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16438-16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471-475. [DOI] [PubMed] [Google Scholar]
- 41.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 42.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 43.Postnikov, Y. V., V. V. Shick, A. V. Belyavsky, K. R. Khrapko, K. L. Brodolin, T. A. Nikolskaya, and A. D. Mirzabekov. 1991. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 19:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7:529-538. [DOI] [PubMed] [Google Scholar]
- 45.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 46.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 47.Schubeler, D., M. Groudine, and M. A. Bender. 2001. The murine beta-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc. Natl. Acad. Sci. USA 98:11432-11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 49.Spicuglia, S., S. Kumar, J. H. Yeh, E. Vachez, L. Chasson, S. Gorbatch, J. Cautres, and P. Ferrier. 2002. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol. Cell 10:1479-1487. [DOI] [PubMed] [Google Scholar]
- 50.Stamatoyannopoulos, G., and F. Grosveld. 2001. Hemoglobin switching, p. 135-182. In G. Stamatoyannopoulos, P. W. Majerus, R. M. Perlmutter, and H. Varmus (ed.), The molecular basis of blood diseases. W. B. Saunders, Philadelphia, Pa.
- 51.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 52.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 53.Travers, A. 1999. Chromatin modification by DNA tracking. Proc. Natl. Acad. Sci. USA 96:13634-13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vignali, M., D. J. Steger, K. E. Neely, and J. L. Workman. 2000. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and Nu4A complexes. EMBO J. 19:2629-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]