Abstract
The retinoblastoma tumor suppressor, RB, assembles multiprotein complexes to mediate cell cycle inhibition. Although many RB binding partners have been suggested to underlie these functions, the validity of these interactions on the behavior of RB complexes in living cells has not been investigated. Here, we studied the dynamic behavior of RB by using green fluorescent protein-RB fusion proteins. Although these proteins were universally nuclear, phosphorylation or oncoprotein binding mediated their active exclusion from the nucleolus. In vivo imaging approaches revealed that RB exists in dynamic equilibrium between a highly mobile and a slower diffusing species, and genetic lesions associated with tumorigenesis increased the fraction of RB in a highly mobile state. The RB complexes dictating cell cycle arrest were surprisingly dynamic and harbored a relatively short residence time on chromatin. In contrast, this rapid exchange was attenuated in cells that are hypersensitive to RB, suggesting that responsiveness may inversely correlate with mobility. The stability of RB dynamics within the cell was additionally modified by the presence and function of critical corepressors. Last, the RB-assembled complexes present in living cells were primarily associated with E2F binding sites in chromatin. In contrast to RB, E2F1 consistently maintained a stable association with E2F sites regardless of cell type. Together, these results elucidate the kinetic framework of RB tumor suppressor action in transcriptional repression and cell cycle regulation.
The retinoblastoma tumor suppressor (RB) is a negative regulator of cellular proliferation that is functionally inactivated in the majority of human tumors through various distinct mechanisms (16, 39, 50, 51, 61). The Rb gene, initially identified based on biallelic inactivation in familial retinoblastoma, is mutated and lost in multiple sporadic cancers (50, 61). Alternatively, oncoproteins encoded by DNA tumor viruses (e.g., human papillomavirus type E7 in cervical cancer) bind to the RB protein and disrupt its function (39, 61). Last, aberrant phosphorylation of RB, as occurs through the amplification of cyclin-dependent kinase 4 (CDK4)/cyclin D1 or loss of p16ink4a, leads to the disruption of RB function (35, 50). Together, these different mechanisms of oncogenic inactivation target the ability of RB to mediate cell cycle inhibition and assemble transcriptional repressor complexes (37, 39, 50, 61).
RB limits cell cycle progression by assembling transcriptional repressor complexes that attenuate the expression of genes required for cellular proliferation (14, 16). A large number of proteins have been demonstrated to interact with RB, the majority of which are involved in transcriptional repression (37, 61). For example, RB binds to members of the E2F family of transcriptional activators (6, 16, 39). This binding not only attenuates the action of E2F in stimulating transcription but also serves to recruit a repressor module to E2F-responsive promoters (16, 62). This corepression function of RB likely involves a number of distinct factors. RB has been shown to recruit histone deacetylases, components of the mammalian SWI/SNF chromatin-remodeling complex, histone methyltransferases, heterochromatin proteins, DNA methyltransferases, and Polycomb group proteins to repress the transcription of genes required for cell cycle transitions (5, 8, 40, 45, 54, 69). Based on biochemical characterization of individual RB-interacting proteins, it has been hypothesized, but never unequivocally demonstrated, that RB must assemble large complexes involving multiple factors to mediate transcriptional repression.
During cell cycle progression, RB-assembled complexes are disrupted by CDK/cyclin-mediated phosphorylation (35, 61). Mitogenic signals trigger the upregulation of CDK4(6)/cyclin D complexes that initiate RB phosphorylation in mid-G1. Subsequent CDK2/cyclin E activity results in the complete hyperphosphorylation of RB. These two phosphorylation cascades disrupt RB-mediated cell cycle inhibition (35). This effect occurs through the disruption of RB-assembled repressor complexes, as virtually all RB-binding proteins fail to bind to the hyperphosphorylated form of RB (37, 61). Consistent with this idea, overexpression of wild-type RB protein in most cell types has little effect, as it is efficiently phosphorylated and inactivated by endogenous CDK/cyclins (23, 25, 30). The exception is SAOS-2 cells, which require the coexpression of ectopic cyclins to overcome RB-mediated arrest (17). However, mutants of RB that cannot be phosphorylated are potent inhibitors of both cell cycle progression and transcription in virtually all cells studied (4, 23, 25, 30).
While many studies have individually examined phosphorylation-dependent RB localization and binding, no prior study has analyzed the dynamics of RB and its assembled repressor complexes in vivo. From a number of in vitro experiments, it would be predicted that RB assembles a relatively stable repressor complex on target promoters to mediate cell cycle inhibition that is only displaced when RB is phosphorylated (16, 37, 61). However, in the case of other transcriptional regulators such as the estrogen or glucocorticoid receptor, in vivo behavior has been shown to be distinct from observations generated with in vitro systems (34, 53). In part, this can be due to weak or transient interactions that are not detectable in dilute cell lysates. Additionally, aberrant stabilization of complexes can occur through high concentrations of purified components to facilitate detection of specific complexes. Here, we utilized a combination of approaches to demonstrate that RB regulatory mechanisms result in disparate, measurable effects on RB mobility and function in living cells.
MATERIALS AND METHODS
Cell culture, adenoviral infections, and transfection.
Rat-1, SAOS-2, U2OS, SW13, and C33A cells were cultured and transfected as previously described (23, 54). A2-4 cells were cultured as previously described (49). MOLT-4 cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, and 100 U of penicillin-streptomycin/ml. Rat-1 cells stably expressing green fluorescent protein (GFP)-RB were cotransfected with pBABE-puro and selected with 2.5 μg of puromycin/ml. Rat-1 cells were infected with p16ink4a- or cyclin E-encoding adenovirus as described previously (2, 3).
Plasmids.
The human RB cDNA BamHI fragment was inserted in frame into the BamHI site of pEGFP-C2 (Clontech). The wild-type large pocket fragment of RB (WTLP) and a phosphorylation site mutant large pocket (7LP) were inserted in frame into the BamHI site of pEGFP-C1 (Clontech). Human E2F1 cDNA was subcloned in frame into pEGFP-C1. The cyclin E, large T antigen (T-Ag), pBABE-puro, p16ink4a, and H2B-GFP plasmids have been described elsewhere (23, 54). Hemagglutinin (HA)-tagged murine RB expression plasmid was provided by P. Hamel (University of Toronto, Toronto, Ontario, Canada). Wild-type and dominant-negative BRG-1 (wtBRG-1 and dnBRG-1, respectively) expression plasmids were provided by B. Weissman (Lineberger Comprehensive Cancer Center, Chapel Hill, N.C.). E2F-DB (E2F1, amino acids 1 to 374) expression plasmid was a gift from D. Dean (Washington University School of Medicine, St. Louis, Mo.).
Immunoblotting.
Immunoblotting was performed as described previously (2). The p300 (N-15), CDK2 (M2-G), and GFP (B-2) antibodies were from Santa Cruz. Rabbit polyclonal antibody against human E2F1 was from A. Yee (Tufts University School of Medicine, Boston, Mass.). The 851 antibody used to detect RB has been previously described (24). Monoclonal RB antibody (554136, clone G3-245) was from BD Transduction Labs.
Gel filtration.
The A2-4 cell line inducibly expresses PSM-RB under tetracycline control (49). Cells were cultured in the absence of doxycycline for 30 h to induce the expression of PSM-RB. Cells were subsequently lysed in NET-N (100 mM NaCl, 1 mM EDTA [pH 8.0], 20 mM Tris-HCl [pH 8.0], 0.5% Nonidet P-40) supplemented with protease and phosphatase inhibitors. Lysates were sonicated and clarified by centrifugation at 16,000 × g for 15 min. Clarified lysates were applied to a Superdex 200 column (Pharmacia), and 1-ml fractions were obtained. Fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and relevant proteins were detected by immunoblotting. The column was standardized under similar running conditions.
BrdU incorporation and immunofluorescence microscopy.
Bromodeoxyuridine (BrdU) staining was performed as previously described (23). For nucleophosmin (NPM), HA, and BRG-1 immunostaining, transfected cells were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.3% Triton X-100 in PBS. Monoclonal NPM antibody was provided by P.-K. Chan (Baylor College of Medicine, Houston, Tex.). HA (sc-805) and BRG-1 (sc-17796) antibodies were from Santa Cruz. For detection of endogenous RB, cells were fixed with ice-cold methanol for 5 min and blocked with 10% normal goat serum in PBS prior to staining. Monoclonal RB antibody (clone G3-245) used for staining was from BD Transduction Labs. Nuclei were counterstained with Hoechst dye.
E2F1 binding assay.
Glutathione S-transferase (GST)-E2F1 was expressed in bacteria and purified on glutathione-agarose beads as previously described (24). U2OS cells transfected with GFP, GFP-WTLP, or GFP-7LP were lysed with NET-N (100 mM NaCl, 1 mM EDTA [pH 8.0], 20 mM Tris-HCl [pH 8.0], 0.5% Nonidet P-40), clarified by centrifugation, and incubated with immobilized GST-E2F1 for 1 h at 4°C. Bound protein was recovered by centrifugation at 2,000 × g, washed 4 times in NET-N, solubilized in SDS sample buffer, and resolved by SDS-PAGE.
Reporter assay.
Rat-1 or C33A cells grown on 60-mm-diameter dishes were cotransfected with 0.5 μg of cytomegalovirus (CMV)-β-galactosidase, 0.5 μg of human −608cycA-LUC, and 7 μg (total) of either H2B-GFP, GFP-WTLP, GFP-7LP, GFP-7LP and BRG-1, or GFP-7LP and dnBRG-1. U2OS cells were grown on 60-mm-diameter dishes and cotransfected with 0.5 μg of CMV-β-galactosidase, 0.5 μg of 3XE2F-Luc, and 3 μg of GFP or GFP-E2F1. At 48 h posttransfection, cells were harvested and analyzed for luciferase activity and normalized by β-galactosidase activity as previously described (26).
FRAP.
Cells were seeded onto 25-mm-diameter coverslips and transfected as indicated. At 24 to 36 h posttransfection, coverslips were transferred to live-cell imaging chambers (Atto) in a water-jacketed stage incubator at 37°C. Fluorescent recovery after photobleaching (FRAP) was performed on a Zeiss LSM510 laser scanning confocal unit mated to a Zeiss Axiovert inverted microscope equipped with a C-Apochromat 63× 1.4 NA objective. A 2.9- by 2.9-μm area of the nucleus was photobleached with 100% transmission of 488-nm light from an argon laser running at 6.3 mW. Fluorescence intensity values of the bleached area and of a distal unbleached area of the nucleus of equal size were measured every 50 ms for the indicated lengths of time following the photobleaching. These values were compared to produce a relative fluorescence intensity to normalize for prebleach intensity. The data presented were collected from greater than 12 nuclei per condition from multiple independent experiments. The mean time for 50% recovery (t1/2) was determined from recovery curves by regression analysis with SigmaPlot.
FCS.
Rat-1 cells were seeded on 25-mm-diameter coverslips and transfected as indicated. At 24 to 36 h posttransfection, coverslips were transferred to live-cell imaging chambers (Atto). Fluorescence correlation microscopy (FCS) measurements were performed at room temperature with an LSM510 Confocor 2 microscope (Carl Zeiss) with a C-Apochromat 40× 1.2 NA objective. GFP fluorescence in cell nuclei was excited with 0.1% transmission of 488-nm light from an argon laser running at 13.25 mW and detected through a pinhole diameter of 70 μm and a 505-nm band-pass filter. Autocorrelation curves were derived from fluctuations of fluorescence intensity over time measured for 4-s intervals with Confocor software (Carl Zeiss). Autocorrelation curves were fit to a two-component model of free diffusion in two dimensions (48) by using Origin software to derive the translational diffusion time and number of molecules diffusing with Dfast and Dslow. Twenty measurements were performed per nucleus, and the data presented were collected from 12 nuclei per construct.
RESULTS
GFP-RB fusion proteins localize to the nucleus and possess cell cycle inhibitory activity.
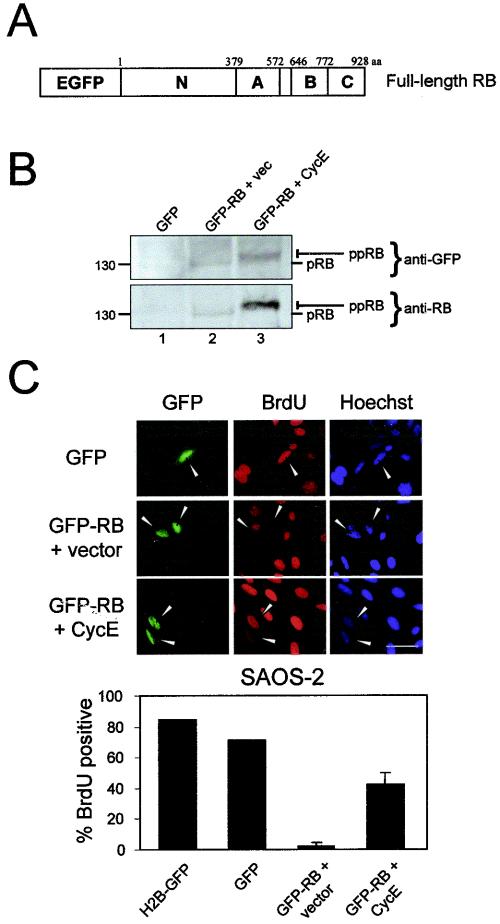
To investigate the in vivo behavior of RB, enhanced GFP was fused to the N terminus of wild-type human RB cDNA (Fig. 1A). To confirm expression, GFP-RB was initially transfected into SAOS-2 cells, an RB-deficient osteosarcoma cell line (Fig. 1B). The fusion protein migrated at approximately 130 kDa, consistent with its predicted molecular weight. Since SAOS-2 cells lack sufficient CDK/cyclin activity to phosphorylate ectopic RB (17, 43, 58), GFP-RB protein migrated as a single band (Fig. 1B, lane 2). To verify that GFP-RB could be phosphorylated and/or inactivated by CDK/cyclins, GFP-RB was cotransfected with human cyclin E (Fig. 1B, lane 3). Ectopic cyclin E facilitated the phosphorylation of GFP-RB, resulting in its slower migration as multiple bands (Fig. 1B, compare lanes 2 and 3).
FIG. 1.
GFP-RB fusion protein localizes to the nucleus and retains cell cycle-inhibitory activity. (A) Enhanced GFP was fused to the N terminus of wild-type RB. The N-terminal, A and B pocket, and C-terminal domains are indicated. aa, amino acids. (B) SAOS-2 cells were transfected with GFP (lane 1), GFP-RB and empty vector (vec) (lane 2), or GFP-RB and cyclin E (CycE) (lane 3). Immunoblotting was performed to detect either GFP or RB. The hyperphosphorylated (ppRB) or hypophosphorylated (pRB) bands are indicated on the right. Molecular masses (in kilodaltons) are indicated on the left. (C) SAOS-2 cells were transfected with GFP, GFP-RB and empty vector, or GFP-RB and cyclin E. (Upper panel) BrdU incorporation was detected by indirect immunofluorescent staining. Bar, 20 μm. (Lower panel) Quantitation of BrdU incorporation from two independent experiments with over 100 cells counted per experiment.
To confirm the retention of growth-inhibitory activity, GFP-RB was transiently transfected into SAOS-2 cells and BrdU incorporation was monitored (Fig. 1C, upper and lower panels). As expected, GFP expression alone did not elicit cell cycle arrest in SAOS-2 cells. In contrast, expression of GFP-RB in SAOS-2 cells significantly inhibited their ability to progress through the cell cycle. Importantly, inactivation of GFP-RB by coexpression of cyclin E was sufficient to overcome cell cycle arrest. Thus, the GFP-RB protein was active and subject to phosphoregulation.
Phosphorylation and oncoprotein binding regulate the subnuclear localization of RB.
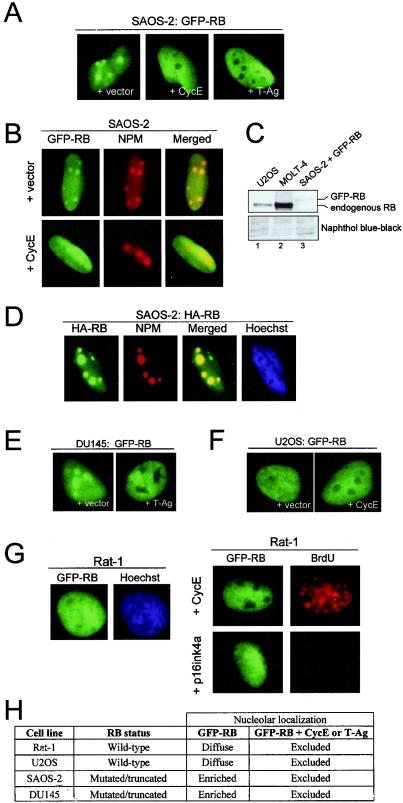
As cursory inspection only confirmed the presence of GFP-RB in the nucleus, its subnuclear localization was inspected in greater detail. Higher-resolution imaging of GFP-RB in SAOS-2 nuclei revealed its distribution into distinct foci (approximately 2 to 8 foci per nucleus) (Fig. 2A). Interestingly, cotransfection of GFP-RB with either cyclin E or simian virus 40 (SV40) T-Ag resulted in the exclusion of GFP-RB from these regions (Fig. 2A). To determine whether the observed regions were nucleoli, immunofluorescent staining for NPM was performed on SAOS-2 cells transfected with GFP-RB (Fig. 2B). As shown in Fig. 2B, the concentrated foci of GFP-RB colocalized with NPM-positive foci. Coexpression of cyclin E or T-Ag resulted in the exclusion of GFP-RB from NPM-positive nuclear regions (Fig. 2B and data not shown). Thus, the enrichment of GFP-RB in nucleoli can be regulated by phosphorylation or oncoprotein sequestration. The possibility existed that the nucleolar enrichment of GFP-RB was due to excessive protein within the nucleus. To determine the relative amount of GFP-RB protein expressed in RB-null SAOS-2 cells, endogenous RB levels from U2OS and MOLT-4 cells were compared to those from GFP-RB-transfected cells (Fig. 2C). Based upon the transfection efficiency achieved with SAOS-2 cells (∼40%), the relative levels were at or below the levels of endogenous RB in the other cell lines. Therefore, the nucleolar enrichment of GFP-RB was not due to disproportionate protein expression. To ensure that the GFP moiety was not altering the nuclear distribution of RB, an HA-tagged RB construct was expressed in SAOS-2 cells and immunofluorescent staining was performed to detect HA-RB and NPM (Fig. 2D). The HA staining was nuclear and enriched in distinct foci that colocalized with NPM. Therefore, the nucleolar association of RB in SAOS-2 cells was not attributable to a fused GFP moiety.
FIG. 2.
Hyperphosphorylated GFP-RB is excluded from the nucleolus. (A) SAOS-2 cells were transfected with GFP-RB and either vector, cyclin E (CycE), or SV40 T-Ag. (B) SAOS-2 cells as described for panel A were permeabilized, and immunofluorescent staining was performed with an antibody directed against NPM. GFP fluorescence, NPM staining, and merged images of nuclei are shown. (C) U2OS (lane 1), MOLT-4 (lane 2), and SAOS-2 cells transfected with GFP-RB (lane 3) were harvested, and whole-cell lysates were resolved by SDS-PAGE. RB was detected by immunoblotting. Naphthol blue-black staining is shown as a loading control. (D) SAOS-2 cells were transfected with HA-tagged RB. HA-RB was detected by indirect immunofluorescence. (E) DU145 cells were transfected with GFP-RB and either vector or T-Ag. (F) U2OS cells weretransfected with GFP-RB and either vector or cyclin E. (G) (Left panel) Rat-1 cells stably transfected with GFP-RB. (Right panel) Rat-1 cells stably expressing GFP-RB were infected with recombinant adenovirus encoding either p16ink4a or cyclin E for 16 h. BrdU incorporation was detected by indirect immunofluorescence. (H) Summary of GFP-RB localization in various cell lines. (A, B, and D to G) Representative photomicrographs taken at ×60 magnification.
To test the possibility that the nucleolar association was cell-type specific, the GFP-RB construct was expressed in multiple cell lines. Analysis of GFP-RB localization in other cell lines revealed that Rat-1 and U2OS cells exhibited diffuse nuclear fluorescence while SAOS-2 and DU145 cells exhibited nucleolar enrichment (Fig. 2E, F, and G). Interestingly, diffuse nuclear localization of GFP-RB correlated with the endogenous expression of functional, wild-type RB (Fig. 2H). To determine whether phosphorylation status or oncoprotein binding might still affect RB localization in these cell lines, T-Ag or cyclin E was coexpressed with GFP-RB in DU145 and U2OS cells (Fig. 2E and F). As shown in Fig. 2H, RB sequestration by T-Ag or hyperphosphorylation driven by cyclin E triggered the nucleolar exclusion of GFP-RB in all cell lines analyzed. Next, the effect of RB dephosphorylation and/or activation on its localization was investigated. The GFP-RB construct was stably integrated into an immortalized rat fibroblast cell line, Rat-1, and either p16ink4a or cyclin E was introduced by adenoviral infection (Fig. 2G, left and right panels). p16ink4a blocks CDK4/cyclin D and CDK2/cyclin E activity, leading to the accumulation of hypophosphorylated, active RB (52). To verify that p16ink4a elicited cell cycle arrest, BrdU incorporation was monitored concurrently. As shown in Fig. 2G, ectopic cyclin E expression resulted in the nucleolar exclusion of GFP-RB, as seen in SAOS-2 cells. However, p16ink4a-mediated activation of RB failed to stimulate enhanced association with nucleoli. Together, these findings suggest that the phosphorylation-mediated exclusion of RB from the nucleolus was common while the enrichment of active RB in the nucleolus was cell type dependent.
N terminus of RB enhances nucleolar localization but is not required for transcriptional repression and cell cycle inhibition.
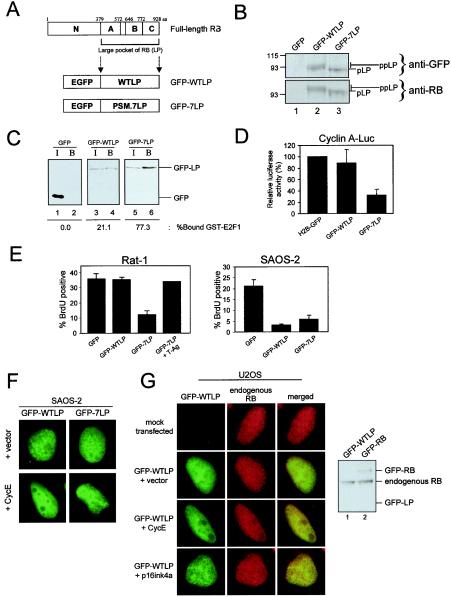
To determine whether the minimal transcriptional repressor region of RB exhibited nucleolar association, GFP-tagged RB alleles lacking the N-terminal domain were constructed (Fig. 3A). Previous studies have implicated the N terminus of RB in mediating interactions with nuclear lamina (9), whereas the LP fragment contains minimal functional domains required for transcriptional repression, cell cycle inhibition, and tumor suppression (Fig. 3A) (43, 67). To circumvent wild-type (GFP-WTLP) phosphorylation in cells with endogenous CDK/cyclin activity, we additionally GFP tagged a phosphorylation-site mutant of LP (GFP-7LP). The 7LP allele has been well characterized as a potent effector of transcriptional repression and cell cycle inhibition (23, 26).
FIG. 3.
The N-terminal domain of RB may contribute to nucleolar localization but is not required for cell cycle inhibition. (A) The critical growth inhibitory and tumor suppressive region of the retinoblastoma protein is comprised of an A and B pocket domain and the C terminus. These regions are termed the large pocket (LP) domain of RB. The cDNA encoding enhanced GFP (EGFP) was fused to wild-type LP (GFP-WTLP) and a phosphorylation site mutant LP (GFP-7LP). aa, amino acids. (B) C33A cells were transfected with either GFP (lane 1), GFP-WTLP (lane 2), or GFP-7LP (lane 3). Immunoblotting was performed to detect GFP (upper panel) or RB (lower panel). The hyperphosphorylated (ppLP) and hypophosphorylated (pLP) bands are indicated on the right. Molecular masses (in kilodaltons) are indicated on the left. (C) U2OS cells weretransfected with either GFP (lanes 1 and 2), GFP-WTLP (lanes 3 and 4), or GFP-7LP (lanes 4 and 6). Input (I) (lanes 1, 3, and 5) and GST-E2F1-bound (B) (lanes 2, 4, and 6) fractions were resolved by SDS-PAGE, and GFP was detected by immunoblotting. The percentage of GFP bound to E2F1 was determined by using Metamorph software. (D) Rat-1 cells were transfected with CMV-β-galactosidase, human −608Cyclin A-Luc reporter plasmid, and either H2B-GFP, GFP-WTLP, or GFP-7LP. Relative luciferase activity was determined and set to 100% for H2B-GFP. (E) Rat-1 cells (left panel) were transfected with either GFP, GFP-WTLP, or GFP-7LP and empty vector or GFP-7LP with SV40 T-Ag. SAOS-2 cells (right panel) were transfected with GFP, GFP-WTLP, or GFP-7LP. BrdU incorporation was determined by indirect immunofluorescence. Data shown are from over 200 cells counted. (F) SAOS-2 cells were transfected with GFP-WTLP or GFP-7LP and either empty vector or cyclin E. Representative photomicrographs were taken at a ×60 magnification. (G) (Left panel) U2OS cells were either mock transfected or transfected with GFP-WTLP and either vector, cyclin E (CycE), or p16ink4a. Endogenous RB was simultaneously detected by indirect immunofluorescence. Representative photomicrographs are shown at a magnification of ×54. (Right panel) U2OS cells transfected with either GFP-WTLP (lane 1) or GFP-RB (lane 2) were harvested, and whole-cell lysates were resolved by SDS-PAGE. Full-length RB, but not WTLP, was detected by immunoblotting with an N-terminal-specific antibody (G3-245).
To confirm the phosphorylation-dependent behavior of the GFP-LP proteins, their phosphorylation by endogenous CDK/cyclin activity was examined. As shown in Fig. 3B, GFP-LP proteins migrated at their predicted molecular mass (90 to 95 kDa) when expressed in C33A cells. Furthermore, GFP-WTLP, but not GFP-7LP, was subject to in vivo phosphorylation, as evidenced by its slower migration as a series of bands.
To substantiate that the fusion proteins were assembling repressor complexes, the relative binding of GFP, GFP-WTLP, and GFP-7LP to E2F was analyzed by an established pull-down assay (Fig. 3C) (24). As a negative control, GFP did not associate appreciably with GST-E2F1. In contrast, approximately 20% of the applied GFP-WTLP bound to GST-E2F1. However, with GFP-7LP, greater than 75% of the applied protein associated with GST-E2F1. Thus, phosphorylation by endogenous CDK/cyclin activity efficiently modulates the association of GFP-LP proteins with E2F1.
To demonstrate that the GFP-LP constructs could actively repress transcription in a phospho-dependent manner, Rat-1 cells were cotransfected with a cyclin A-LUC reporter plasmid and each of the GFP-LP constructs. As a control for nuclear GFP overexpression, a histone 2B-GFP (H2B-GFP) construct was utilized. As shown in Fig. 3D, expression of GFP-WTLP did not significantly reduce the activity of the cyclin A promoter compared to that of the H2B-GFP. In contrast, expression of GFP-7LP inhibited cyclin A promoter activity by 70% (Fig. 3D). Thus, GFP-7LP elicits strong transcriptional repression while GFP-WTLP is largely inactive in cells with significant CDK/cyclin activity.
Last, we sought to verify that the GFP-LP fusion proteins possessed biological activity. To confirm that these molecules effectively elicited cell cycle inhibition, transient transfections in both Rat-1 and SAOS-2 cells were performed and BrdU incorporation was monitored (Fig. 3E). Rat-1 cells were resistant to GFP and GFP-WTLP while GFP-7LP inhibited BrdU incorporation (Fig. 3E, left panel). To ensure that cell cycle arrest in response to GFP-7LP was due to complex formation, ectopic T-Ag expression was employed. It is known that T-Ag can disrupt the RB transcriptional repressor complexes via direct binding (16). Consistent with cell cycle inhibition resulting from the formation of repressor complexes, we observed that coexpression of T-Ag eliminated the cell cycle inhibition mediated by GFP-7LP. In contrast to Rat-1 cells, expression of either GFP-LP protein was sufficient to block cell cycle progression in SAOS-2 cells (Fig. 3E, right panel).
Initially, these fusion proteins were transiently transfected into SAOS-2 cells and their localization patterns were observed (Fig. 3F). Surprisingly, the GFP-WTLP and GFP-7LP fusions did not exhibit enriched nucleolar association in SAOS-2 cells. This observation indicated that the N-terminal domain of RB is required for the nucleolar enrichment observed in SAOS-2 cells. Since the localization of GFP-LP in SAOS-2 cells resembled that of GFP-RB in Rat-1 cells, we tested the hypothesis that phosphorylation of RB might regulate its localization. As shown in Fig. 3F, ectopic cyclin E led to the nucleolar exclusion of GFP-WTLP but not the phosphorylation-refractory GFP-7LP. Therefore, the direct cyclin E-mediated phosphorylation of GFP-RB or -WTLP altered its in vivo localization as opposed to an effect of CDK/cyclin on other nucleolar proteins. These data indicate that while the nucleolar association of RB was enhanced in SAOS-2 cells by the presence of the N terminus, this region is not essential for transcriptional repression or cell cycle inhibition. Furthermore, the minimal repressor fragment of RB retained the domain(s) required for nucleolar exclusion.
To determine whether the behavior of the GFP-LP proteins accurately recapitulated that of endogenous RB, indirect immunofluorescent detection of endogenous RB was performed (Fig. 3G). Using an antibody that recognizes an N-terminal epitope of RB, simultaneous detection of GFP-LP (lacking the N terminus) and endogenous RB was achievable. Importantly, endogenous RB was evenly distributed throughout the nucleus of U2OS cells, which was comparable to GFP-WTLP. Consistent with observations of GFP-RB alleles, nucleolar exclusion of endogenous RB was mediated by cyclin E while nucleolar enrichment was not induced by p16ink4a (Fig. 3G). To confirm that the N-terminal-specific antibody did not detect GFP-WTLP, U2OS cells transfected with either GFP-WTLP or GFP-RB were harvested and utilized for immunoblotting (Fig. 3G, left panel). As predicted, the antibody failed to detect GFP-WTLP by immunoblotting and the levels of GFP-RB achieved in U2OS cells (40% transfection efficiency) were below that of endogenous RB. Thus, the regulation of GFP-RB proteins is consistent with the regulation of endogenous RB protein.
FRAP analysis of GFP-RB reveals immobility in RB-sensitive cells.
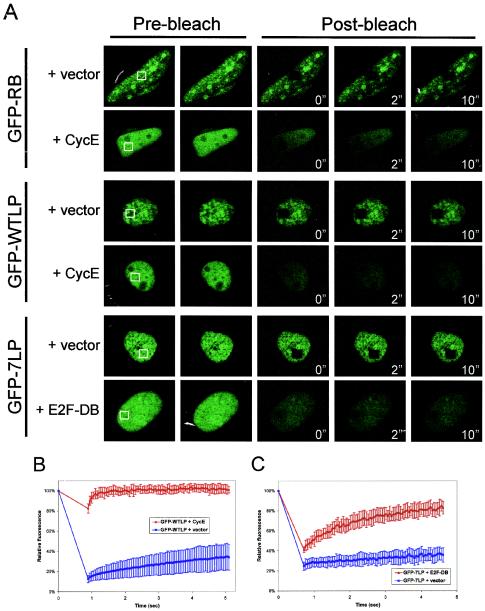
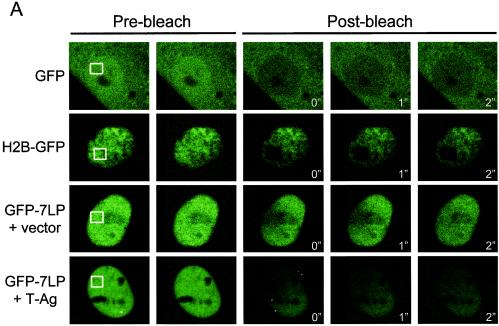
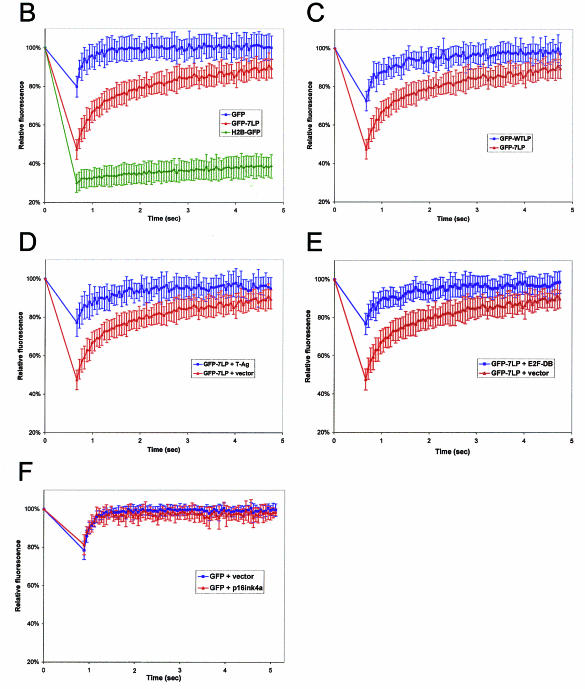
Since the GFP-fusions accurately recapitulated RB function and localization, we next investigated their behavior in living cells by FRAP. Because SAOS-2 is a commonly used model for the analysis of RB activity, we initially utilized this cell type to monitor the in vivo dynamics of RB in real time. Photobleaching was performed on a 2.9- by 2.9-μm area of the nucleus (Fig. 4A, pre-bleach). Sequential scanned images following photobleaching were utilized to determine the kinetics of fluorescence recovery (Fig. 4A, post-bleach). By measuring the fluorescence intensity of the bleached area and of a distal unbleached area of the nucleus of equal size every 50 ms, relative fluorescence intensity was determined. The rate and extent of recovery are dependent upon the mobility of the fluorescent molecule. From the recovery curves, the mean time for 50% recovery (t1/2) for each of the proteins can be determined. Lack of apparent mobility (i.e., higher t1/2 values) is indicative of higher-order complex formation or static interactions with fixed nuclear structures (e.g., chromatin) (29, 44). Quite unexpectedly, the fluorescence recovery of GFP-RB was minimal after photobleaching, suggesting that it was immobile in its active, repressive state (Fig. 4A, GFP-RB plus vector). This observation was in marked contrast to the observed behavior of other transcriptional regulators (28, 34, 53). FRAP analysis showed that GFP-RB in nucleolar foci exhibited comparable immobility to nucleoplasmic GFP-RB (data not shown). Since the N terminus of RB was not required for cell cycle inhibition in SAOS-2 cells, we next investigated the behavior of GFP-WTLP. Nuclear FRAP analysis revealed that GFP-WTLP behaved similarly to full-length GFP-RB (Fig. 4A and data not shown). In agreement with our qualitative observations, GFP-WTLP recovered to only 30% of its original intensity during the time course of the experiment (Fig. 4B, t1/2 >30 min). Thus, the apparent immobility of active RB in SAOS-2 cells was not attributable to nucleolar interactions mediated by the N terminus.
FIG. 4.
FRAP analysis reveals immobility of active GFP-RB in SAOS-2 cells. (A) SAOS-2 cells were transfected with either GFP-RB, GFP-WTLP, or GFP-7LP and either empty vector, cyclin E, or E2F-DB as indicated. At 24 h posttransfection, cells were subjected to nuclear FRAP analysis. (B and C) Recovery curves from SAOS-2 cells shown in panel A. Relative fluorescence intensities were determined by comparing the fluorescence intensity of a distal unbleached region of the nucleus to the photobleached area, and the results are plotted over time. The white boxes indicate the bleached areas (2.9 by 2.9 μm2).
RB repressor complex is mobilized by ectopic expression of cyclin E.
Phosphorylation of RB is presumed to disrupt essentially all of its protein-protein interactions, based on in vitro experimentation. However, the localization of RB to specific nuclear structures in vivo has been observed to occur in S-phase cells (19), wherein RB is hyperphosphorylated. These prior results suggest that the behavior of phosphorylated RB in living cells may not accurately recapitulate in vitro protein binding experiments. Therefore, to determine whether the static behavior of the RB complex was dependent on characterized protein-binding functions, we investigated the effect of phosphorylation on its mobility. Interestingly, the phosphorylation of either GFP-RB or GFP-WTLP by cyclin E converted it to a freely diffusible species (Fig. 4A and B). Therefore, these data indicate that the phosphorylation status of RB, not its presence alone, underlies its static behavior in SAOS-2 nuclei.
RB mobility is regulated by E2F site availability.
Active RB has been demonstrated to exhibit tight associations with the nuclear matrix (32, 36). However, whether this is due to specific nuclear protein binding or nonspecific chromatin association has not been determined. To elucidate the contribution of specific DNA interactions to RB mobility, we utilized a mutant allele of E2F1 (E2F-DB, amino acids 1 to 374) that lacks the domains required for RB binding and transactivation of E2F-dependent genes. Thus, E2F-DB retains its DNA-binding domain and is able to compete with RB-E2F repressor complexes for E2F sites in regulated promoters. Importantly, expression of this allele has been previously shown to alleviate RB-mediated transcriptional repression and overcome RB-mediated cell cycle arrest (31, 46, 70). When expressed in SAOS-2 cells, E2F-DB effectively mobilized GFP-7LP compared to the vector control (Fig. 4A and C). Thus, the static behavior of GFP-7LP was largely due to association with available E2F sites in chromatin.
Dynamic behavior of RB is cell type dependent and is regulated by endogenous CDK/cyclin phosphorylation and E2F-dependent chromatin association.
Since GFP-RB did not behave as a typical transcription factor in SAOS-2 cells, we next tested whether the repressor complex might exhibit different dynamics in another cell type. Initially, Rat-1 cells were transiently transfected with GFP-7LP for nuclear FRAP analysis. We observed a decrease in relative fluorescence intensity following photobleaching that recovered relatively rapidly in cells transfected with GFP-7LP (Fig. 5A and B). The t1/2 value calculated for GFP-7LP was 680 ms, which is comparable to the observed recovery of other transcription factors (34, 42, 53). Therefore, active GFP-7LP exists as a relatively mobile complex in Rat-1 cells. Because the behavior of GFP-7LP was less static in this cell type, we next utilized GFP and H2B-GFP fusion proteins as controls for freely diffusible and largely immobile species, respectively (Fig. 5A and B). In GFP-expressing cells, we detected negligible loss of relative fluorescence following nuclear photobleaching and a very small t1/2 value for recovery (<80 ms). These observations are consistent with a freely diffusible protein species in the nucleus (29). In contrast, a substantial decrease in relative fluorescence with less than 10% recovery during our time scale was observed in cells transfected with H2B-GFP (t1/2 > 100 min). This behavior is similar to that reported by other laboratories and is consistent with the stable tight association of H2B-GFP with chromatin (21, 42). The dynamics of GFP-7LP were intermediate compared to those of GFP and H2B-GFP. To investigate the regulation of complex assembly by phosphorylation, we then compared the behavior of GFP-7LP with GFP-WTLP (Fig. 5A and C). Similar to the dynamics of GFP, GFP-WTLP recovered significantly more rapidly than GFP-7LP after photobleaching. Importantly, the behavior of GFP-WTLP was quantitatively distinguishable from GFP-7LP. From the rate of recovery, a t1/2 value of 220 ms was obtained for GFP-WTLP (Fig. 5C). This analysis demonstrates that endogenous CDK/cyclin-mediated phosphorylation regulates the mobility of RB in vivo.
FIG. 5.
FRAP analysis uncovers dynamic regulation of GFP-LP by phosphorylation, oncoprotein binding, and E2F site availability. (A) Rat-1 cells were transfected as indicated. At 24 h posttransfection, cells were subjected to nuclear FRAP analysis. As a control, no recovery of fusion proteins was observed after bleaching of chemically fixed cells. The white boxes indicate the bleached areas (2.9 by 2.9 μm2). (B to F) Recovery curves, with relative fluorescence intensity plotted over time.
In contrast to phosphorylation-mediated RB inactivation, certain DNA tumor viruses produce oncoproteins that specifically target the active form of RB (39, 61). Disruption of RB repressor modules by these proteins has been inferred from exquisite biochemical analyses. To explicitly determine whether the perturbation of active RB complexes could be monitored in vivo, Rat-1 cells were cotransfected with GFP-7LP and either vector or T-Ag (Fig. 5A and D). Coexpression of T-Ag, but not vector, led to the mobilization of GFP-7LP (t1/2 dropped from 680 to 350 ms) (Fig. 5D). In fact, the behavior of GFP-7LP, in the context of T-Ag coexpression, was analogous to that of GFP-WTLP (compare Fig. 5D and C). Therefore, these data demonstrate that oncoprotein sequestration of RB serves to increase its dynamic behavior in living cells by preventing chromatin binding of the repressor.
Presumably, the influence of T-Ag on RB mobility could be attributed to the disruption of RB-E2F interactions. Therefore, we examined whether the immobility of active RB was dependent on the availability of free E2F sites. Cells were cotransfected with GFP-7LP and either empty vector or E2F-DB expression plasmid (Fig. 5A and E). GFP-7LP was largely converted to a freely diffusible species by the coexpression of E2F-DB (comparable to the direct binding of T-Ag). Collectively, these data indicate that the immobility of active RB repressor complexes in both SAOS-2 and Rat-1 cells is dependent on specific interactions with E2F binding sites in chromatin.
Several factors influence the observed behavior of molecules by FRAP analysis, including nuclear viscosity (i.e., hydrodynamic environment) and the formation of aggregates (29). To exclude the possibility that cell cycle arrest might alter these conditions (thereby impeding the diffusion of GFP-7LP), we investigated the dynamics of GFP in asynchronous and p16ink4a-arrested cells (Fig. 5F). Importantly, the induction of cell cycle arrest did not alter the apparent mobility of GFP, lending confidence to the interpretation that GFP-7LP was forming stable DNA interactions at E2F sites to elicit cell cycle inhibition.
E2F dynamic behavior is cell type independent and mediated by chromatin association.
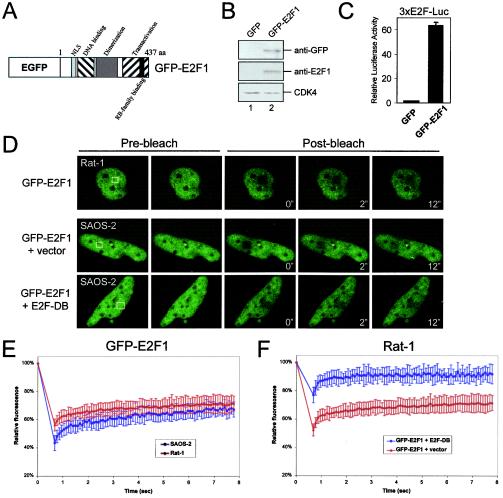
While the occupancy of E2F sites clearly affected RB mobility, the direct contribution of E2F dynamics to this behavior was unclear. To address this issue, human E2F1 was tagged with GFP at its N terminus (Fig. 6A). When expressed in U2OS cells, GFP-E2F1 protein migrated at the predicted molecular mass (∼75 kDa) and could be detected by immunoblotting with both E2F1- and GFP-specific antibodies (Fig. 6B). Furthermore, GFP-E2F1 significantly stimulated transactivation of an E2F-Luc reporter (Fig. 6C). To analyze the mobility of GFP-E2F1, nuclear photobleaching experiments were performed in asynchronously proliferating Rat-1 and SAOS-2 cells (Fig. 6D and E). As shown in Fig. 6D, GFP-E2F1 exhibited surprisingly slow dynamics, as the photobleached areas of both cell types displayed minimal recovery during the time frame of the experiments. Although GFP-7LP demonstrated cell-type-dependent in vivo dynamics, GFP-E2F1 behaved similarly in both Rat-1 and SAOS-2 nuclei (Fig. 6D and E). To verify that the immobility of GFP-E2F1 was, in fact, dependent on site-specific chromatin binding, the E2F-DB allele was coexpressed with GFP-E2F1. As shown in Fig. 6F, competition for available E2F sites significantly mobilized GFP-E2F1. Similar fluorescence recovery was observed for GFP-E2F1 in SAOS-2 cells upon E2F-DB coexpression (Fig. 6D and data not shown). Thus, GFP-E2F1 exchanges with available E2F sites in chromatin at a surprisingly slow rate that is independent of the cell type examined.
FIG. 6.
GFP-E2F1 dynamics are not cell type dependent and depend on E2F site occupancy. (A) Enhanced GFP (EGFP) was fused to the N terminus of human E2F1. The domain structure is indicated. aa, amino acids. (B) U2OS cells were transfected with GFP (lane 1) or GFP-E2F1 (lane 2), whole-cell extracts were resolved by SDS-PAGE, and the indicated proteins were detected by immunoblotting. (C) U2OS cells were transfected with CMV-β-galactosidase, 3XE2F-Luc reporter, and either GFP or GFP-E2F1. The relative luciferase activity was determined. The data shown are from two independent experiments. (D) Rat-1 or SAOS-2 cells were transfected with GFP-E2F1 alone or with either vector or E2F1-DB as indicated. At 24 h posttransfection, cells were subjected to nuclear FRAP analysis. The white boxes indicate the bleached areas (2.9 by 2.9 μm2). (E and F) Recovery curves for GFP-E2F1 from panel D are shown, with relative fluorescence intensity plotted over time.
Corepressor availability and function influence RB nuclear dynamics.
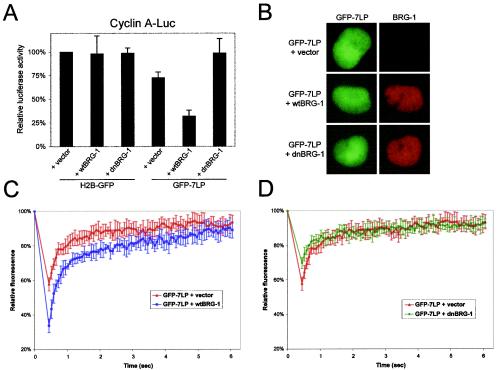
Next, we sought to determine whether specific transcriptional corepressors might contribute to the mobility of RB in living cells. Initially, we analyzed the consequence of inhibiting histone deacetylase (HDAC) activity through the use of trichostatin A (TSA). The pharmacological inhibition of HDAC, however, did not significantly alter the dynamics of RB complexes (data not shown). We then investigated the contribution of an additional RB corepressor. BRG-1 and BRM are the core catalytic ATPases of the SWI/SNF complex and are required for RB-mediated cell cycle arrest (22, 54, 55, 69). To elucidate the role of functional BRG-1 in the stability of RB complexes in vivo, we utilized the BRG-1- and BRM-deficient cell lines C33A and SW13. Initially, the ability of GFP-7LP to cooperate with BRG-1 to elicit transcriptional repression was determined by reporter analysis in C33A cells. As shown in Fig. 7A, GFP-7LP alone was incapable of substantially repressing cyclin A promoter activity compared to H2B-GFP controls. In contrast, coexpression of wtBRG-1 and GFP-7LP significantly attenuated transcriptional activity. To determine whether chromatin remodeling function was required for repression of the cyclin A promoter by GFP-7LP, dnBRG-1 was utilized. This allele is capable of binding RB and assembling SWI/SNF complexes but cannot remodel chromatin (8, 20). As shown in Fig. 7A, the ATPase mutant of BRG-1 failed to cooperate with GFP-7LP to repress cyclin A promoter activity. Therefore, GFP-7LP can cooperate with essential, functional corepressors to elicit transcriptional repression. To elucidate the contribution of BRG-1 to RB complex mobility, GFP-7LP and either empty vector, wtBRG-1, or dnBRG-1 were transfected into SW13 cells. As shown in Fig. 7B, immunofluorescent detection of the ectopically expressed BRG-1 proteins revealed their comparable accumulation and localization in SW13 nuclei. When analyzed by FRAP, coexpression of functional wtBRG-1 significantly diminished the fluorescence recovery of GFP-7LP after photobleaching (Fig. 7C). Since the dnBRG-1 allele cannot remodel chromatin, we also analyzed the contribution of enzymatic function to RB complex mobility. Surprisingly, coexpression of the dnBRG-1 allele did not appreciably affect the dynamics of GFP-7LP in SW13 cells (Fig. 7D). These data indicate that the presence and function of individual, requisite components of the active RB repressor complex can measurably alter its in vivo dynamics.
FIG. 7.
GFP-7LP recruitment of functional BRG-1 forms an active repressor module with distinct dynamics. (A) C33A cells were transfected with CMV-β-galactosidase, cyclin A-Luc reporter, H2B-GFP, or GFP-7LP and either vector, wtBRG-1, or dnBRG-1 as indicated. The relative luciferase activity was determined. The data shown are from three independent experiments. (B) SW13 cells were transfected with GFP-7LP and either vector, wtBRG-1, or dnBRG-1. BRG-1 was detected by indirect immunofluorescence. Representative photomicrographs were taken at a magnification of ×60. (C and D) SW13 cells transfected as described for panel B were utilized for nuclear FRAP analysis. Relative fluorescence intensity is plotted over time.
Direct in vivo analysis of RB complex dynamics by FCS.
In contrast to FRAP, which determines diffusion kinetics from recovery into a photobleached region, FCS directly monitors the diffusion of fluorescent species within a confocal volume smaller than 1 fl. As such, it enables higher-order dissection of the behavior of specific molecular complexes. Utilizing FCS, GFP was present exclusively as a single species with an approximate τD of 215 μs, which translates to an apparent molecular mass of 40 kDa (determined by assuming free diffusion in two dimensions and using a calculated nuclear viscosity of 1.5 g/m·s) (Fig. 8A, upper panel). This observation is consistent with the actual molecular mass of monomeric GFP (27 kDa). In contrast, H2B-GFP was virtually immobile. As such, it was not amenable to FCS analysis, which is dependent on a degree of mobility through the analytical volume. Likewise, the relative immobility of active RB complexes in SAOS-2 cells required the utilization of Rat-1 cells for FCS analysis.
FIG. 8.
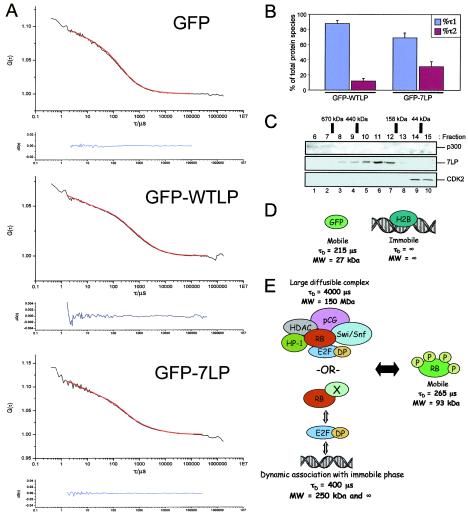
FCS analysis of RB complex dynamics. (A) Rat-1 cells expressing either GFP, GFP-WTLP, or GFP-7LP were subjected to nuclear FCS analysis. Shown are mean autocorrelation curves (upper plots) derived from fluctuations of fluorescence intensity over time measured for 4-s intervals (lower plots). Autocorrelation curves were fit to a two-component model of free diffusion in two dimensions with Origin software to derive the translational diffusion time and number of molecules diffusing through the confocal volume with Dfast and Dslow. (B) The data obtained from FCS identified that GFP-LP diffused as two distinct species: a mobile species with rapid diffusion rate (τ1) and a 10- to 20-fold-more-slowly diffusing complex (τ2). The percentages of total protein species diffusing through the confocal volume with τ1 and τ2 are shown. (C) A2-4 cells expressing 7LP were harvested, whole-cell lysate was subjected to gel filtration, and eluted fractions were resolved by SDS-PAGE. The indicated proteins were detected by immunoblotting. Molecular mass standards are indicated. (D and E) Proposed models for the behavior of GFP, H2B, and RB in vivo. RB can potentially interact with multiple cellular proteins, forming a large diffusible complex. Data presented here support a model wherein RB and essential transcriptional corepressors dynamically exchange with E2F-binding sites in chromatin.
In the case of GFP-LP, two distinct species were identified for both GFP-WTLP and GFP-7LP (Fig. 8A and B). The first species exhibited a rapid diffusion time (τD of 265 μs) and a calculated average molecular mass of 76 kDa, similar to the monomeric molecular mass of the GFP-LP fusion proteins (93 kDa). The second species diffused 10- to 20-fold more slowly (τD of 2,000 to 6,000 μs), indicative of a relatively immobile complex. Consistent with the FRAP analysis, it was apparent by FCS that GFP-WTLP was largely present in the rapidly diffusing species, with only 10% in the less mobile complex (Fig. 8B). In contrast, GFP-7LP was more frequently associated with the less mobile complex (greater than 30% of the species detected) (Fig. 8B). Such a finding is consistent with the phosphoregulation of RB-assembled complexes and suggests that the slower diffusing complex represents the active form of RB that mediates cell cycle inhibition and transcriptional repression.
As a biochemical complement to our live-cell imaging, we sought to elucidate the size of transcriptional repressor complexes assembled by active RB in vivo. Utilizing cells that inducibly express the 7LP fragment of RB, soluble proteins were subjected to gel filtration through a Superdex 200 column that separates complexes from 30 to 700 kDa (Fig. 8C). Specific monomeric markers were utilized to calibrate the column (data not shown). Verifying the calibration, p300 (known to assemble 10-MDa complexes) (12) preferentially eluted in the void volume (fraction 6). In contrast, 7LP eluted as a somewhat broad peak, with the majority eluting at approximately 400 to 200 kDa. As a control for smaller protein complexes, we also analyzed the fractions for CDK2, which was detectable in fractions corresponding to 100 to 20 kDa. These in vitro data indicate that an active, growth-inhibitory allele of RB forms stable multiprotein complexes with an approximate size of 250 kDa. Collectively, these data provide the basis for a model of phosphorylation-mediated RB repressor complex dynamics (Fig. 8D and E and see below).
DISCUSSION
Extensive analysis of RB function has suggested that it assembles large multiprotein complexes to mediate transcriptional repression and suppress tumorigenesis. Over 100 different RB binding proteins have been identified (37); however, their relative contribution to RB action and the actual existence of these complexes in vivo have not been explored. Here, we utilized GFP-RB fusion proteins to investigate the dynamic localization of RB and the behavior of RB-assembled complexes in living cells. We show that these GFP fusions retain RB activities and can be regulated by phosphorylation, validating their utility as models of RB function. We find that RB is enriched in the nucleolus in specific cell types and is universally excluded from the nucleolus following its CDK2/cyclin E-catalyzed phosphorylation. By nuclear FRAP analysis, we documented the mobility of the active RB repressor module in living cells. RB complex dynamics can be modulated by oncoproteins and CDK/cyclin-dependent phosphorylation. Furthermore, we provide the first direct demonstration that the majority of RB repressor complexes primarily target and associate with E2F DNA-binding sites. Interestingly, the behavior of RB exhibited cell type-dependent variance, while E2F1 dynamics were relatively consistent. Last, the mobility of a functional RB complex is dependent on the availability and/or function of critical corepressors. Together, these data provide the first detailed analysis of RB action in real time and illuminate the dynamic interactions of this critical cell cycle-regulatory molecule.
Subnuclear localization of RB.
Following its initial characterization, RB was determined to be a nuclear protein, which harbored a classical bipartite nuclear localization signal (27, 68). As evidence for the inactivation of RB by phosphorylation or oncoprotein binding was revealed, analysis of RB localization under these conditions was undertaken. Regardless of cell cycle phase or the presence of oncoproteins, RB remained confined to the nucleus. We consistently observed both GFP-RB and HA-tagged RB proteins in the nucleus under all conditions of study. Interestingly, prior studies have suggested that RB interacts with a myriad of subnuclear structures. This observation was initially documented by immunofluorescence microscopy and biochemical extraction in SAOS-2 cells, wherein hypophosphorylated RB was retained in an insoluble nuclear fraction (36). In our hands, RB enrichment was detected in the nucleolus only in specific cell lines. Importantly, the enhanced localization in this subnuclear region required the N terminus of RB, although the enrichment in the nucleolus was not requisite for cell cycle inhibition.
In contrast to the cell-specific enrichment of RB in the nucleolus, hyperphosphorylated RB was demonstrably eliminated from the nucleolus in all cell systems studied. This exclusion was dependent on phosphorylation occurring in the large pocket domain of RB and was observed with both full-length and large pocket fragments of RB. This finding was consistent with previously published results showing the dissociation of phosphorylated RB from nucleolar regions (13). Although the precise molecular mechanisms determining this exclusion are at present unknown, RB has been shown to interact with the nucleolus-associated proteins NPM and MDM2 (57, 66). Two intriguing possibilities exist for the function of nucleolar enrichment of RB observed in SAOS-2 and DU145 cells. First, this association could represent a negative regulatory mechanism to sequester RB during the cell cycle, as occurs with cdc14 in Saccharomyces cerevisiae (60). Alternatively, enrichment of active RB in the nucleolus could represent a means of attenuating RNA polymerase I/III transcription (65). An unexpected extension of our results is that RB cannot be expected to behave identically in all cells under study. It will be interesting to explore the localization of GFP-RB in primary cells versus immortalized cell lines, wherein discrepancies have been previously noted (19). Thus, these observations could explain some of the inconsistencies regarding RB localization in different cell types (1, 7, 19, 33, 57).
RB complex dynamics—regulation by viral oncoproteins and phosphorylation.
The critical role of RB in controlling cell division was supported by the observation that RB is the target of specific protein products of DNA tumor viruses to deregulate proliferation (18, 50, 61). Similarly, the phosphorylation status of RB was shown to correlate with the growth status of cell populations: RB was phosphorylated in proliferating cells and unphosphorylated in quiescent or senescent cells (18, 50, 61). As the tumor-suppressive role of RB could be bypassed by viral oncoproteins or deregulated phosphorylation, it has been predicted that these events displace essential proteins from RB in vivo. Many such complexes (including the RB-E2F interaction) are disrupted by viral oncoprotein binding to RB. Consistent with this idea, T-Ag efficiently mobilized GFP-RB proteins (Fig. 5). However, not all RB-associated proteins mobilized GFP-RB; for example, BRG-1 expression stabilized RB (as discussed below) while the expression of the androgen receptor (37) had no effect on RB mobility (data not shown). Thus, we provide the first demonstration that viral oncoproteins mobilize active RB complexes, as opposed to promoting static sequestration within the nucleus. Based on biochemical analysis, phosphorylation is known to disrupt the association of RB with most of its binding partners (35, 37, 61). However, specific exceptions to this phenomenon have been documented (37). By FRAP analysis, we observed a clear effect of phosphorylation on RB behavior. Wild-type RB is not phosphorylated in SAOS-2 cells, but its phosphorylation can be driven by the ectopic expression of cyclins (17). Under these conditions, cyclin E completely mobilized RB from its static behavior in SAOS-2 cells (Fig. 4). Second, a significant difference between the behavior of phospho-regulated GFP-WTLP and constitutively active GFP-7LP was observed in Rat-1 cells, which harbor endogenous RB kinase activity. From analysis of the recovery curves after nuclear photobleaching, GFP-WTLP and GFP-7LP had t1/2 values of 220 and 680 ms, respectively (Fig. 5C). These observations were confirmed by direct measurement of RB mobility by FCS, demonstrating the presence of two distinct species of RB whose vastly different behaviors are phosphorylation dependent. These in vivo observations suggest that phosphorylated and/or inactivated RB becomes highly mobile and does not stably associate with structural components of the nucleus nor is subject to other potential sequestration mechanisms.
Chromatin association of active RB.
Previous studies have led to the hypothesis that RB assembles a large transcriptional repressor complex to attenuate transcription and mediate cell cycle inhibition (15). Active RB interacts simultaneously with transcription factors such as E2F/DP and a myriad of corepressors (37). Additionally, RB interacts with more than 20 other transcription factors (37). Based on these associations, one might predict that RB is principally involved in chromatin binding to regulate transcription. However, several studies have reported interactions of hypophosphorylated RB with the nuclear matrix or nuclear lamina (9, 32, 33, 41), suggesting that the majority of active RB is associated with these proteinaceous components of the nucleus. Here, we sought to elucidate the contribution of E2F target gene repression to RB function in living cells. In contrast to direct phosphorylation (i.e., cyclin E) or direct binding (i.e., T-Ag), the E2F-DB allele indirectly competes with RB repressor modules for available E2F binding sites in chromatin. Using this allele, we demonstrate that RB complex mobility is largely dependent on the accessibility of E2F-regulated promoters, indicating that the primary targets for RB in living cells are E2F binding sites. This finding is supported by a recent study by Wells et al. demonstrating that chromatin-associated RB is primarily found at the same promoter regions as E2F proteins (64).
Interestingly, analysis of active RB in Rat-1 and SAOS-2 cells revealed cell type-specific mobility differences, potentially representing enhanced recruitment and/or stability of RB at E2F binding sites. While there has been some controversy concerning the presence of RB at E2F-regulated promoters by chromatin immunoprecipitation (56, 63), recent data have shown that RB-mediated senescence correlates with the stable repression of E2F target genes and the detectable association of RB at E2F promoter regions (38). Therefore, the rapid induction of a senescent phenotype in SAOS-2 cells arrested by RB may in fact be a consequence of the relative strength of RB-chromatin interactions. Taken together, however, these results cast doubt on the extent to which RB interacts with non-E2F promoter elements or large proteinaceous components of the nucleus.
Although numerous studies have investigated the presence of E2F family members at target promoters by chromatin immunoprecipitation (56, 63, 64), the dynamic behavior of E2F family transcription factors has not been analyzed in living cells. Intriguingly, GFP-E2F1 did not exhibit cell type-specific behavior and displayed relatively stable behavior in both Rat-1 and SAOS-2 cells. Similar results were obtained with GFP-E2F2 (data not shown). Ultimately, differences in E2F mobility do not apparently underlie the observed discrepancy of RB behavior in SAOS-2 versus Rat-1 nuclei. These data support a model wherein RB repressor complexes do not stably sequester free E2F; rather, the dynamics of the active repressor module may be dictated by interactions with relatively stable E2F-chromatin complexes (Fig. 8E). Recent reports have indicated that the promoter specificity of individual E2F proteins occurs through the coupled chromatin binding of specific E2F family members and associated transcription factors (e.g., YY1/RYBP and TFE3) (11, 47). It will be interesting to further explore the dynamic behavior of E2F proteins as a function of RB association, cell cycle phase, or varied genetic backgrounds (e.g., deficiency of specific coactivators).
RB mobility and transcriptional corepressors.
In addition to the direct binding to E2F, RB mediates repression through the recruitment of essential corepressors to chromatin (16). These corepressors themselves exist in very large complexes. For example, the SWI/SNF complex is approximately 2 MDa and the pCG complex is approximately 2 MDa (10, 22, 59). Based only on interactions with these three partners, RB would assemble a complex of greater than 4 MDa. It is apparent that RB binds to additional large corepressor complexes (e.g., DNMT-1, Sin3/HDAC, and SUV39H1/HP-1), suggesting a possible molecular mass for RB and all of its associated proteins approaching 10 MDa (Fig. 8E) (5, 40, 45). However, the manner in which RB associates to mediate repression is not completely understood. To address this question, we initially explored the contribution of HDAC function to the behavior of active RB. We have recently utilized the HDAC inhibitor TSA to disrupt RB-mediated repression of specific target genes (52a). The addition of TSA under these conditions had no detectable effect on the mobility of active RB, indicating that HDAC function is not required for the relative stability of RB in a living cell.
RB is known to require SWI/SNF subunits to elicit transcriptional repression and cell cycle inhibition (54, 69). Presumably, the function of SWI/SNF could be to stabilize RB on promoters, as is observed for SBF in S. cerevisiae (10). Alternatively, SWI/SNF association with RB could result in subsequent modification of target promoters (10, 22). Therefore, we assessed the action of functional SWI/SNF on RB in living cells. In cells deficient in SWI/SNF activity due to the coordinate loss of the ATPase core subunits (BRG-1 and BRM), we found that GFP-7LP dynamics were specifically slowed by functional wtBRG-1 and not an ATPase-deficient dominant-negative allele. This finding suggested that the availability of SWI/SNF chromatin remodeling function permits the establishment of a retained RB repressor complex. Since SWI/SNF is a large complex (2 MDa) that is associated with additional heteromeric complexes, it is formally possible that the enhanced retention simply represents the corresponding increase in mass due to association with the multiprotein complex. However, we could eliminate this possibility since the dnBRG-1 utilized does bind to RB (8) and assemble large SWI/SNF complexes (20) but failed to influence RB behavior. This result supports the idea that SWI/SNF acts upstream of active RB, facilitating its stable association with promoter elements.
In summary, RB exists in dynamic equilibrium between two species. We propose that this dynamic nature of RB is an essential feature of its function as a tumor suppressor. Phosphorylation or viral oncoprotein expression increase the fraction of RB in a highly mobile state, indicating a lack of association with chromatin. Furthermore, the loss of critical corepressor molecules, suspected to act as tumor suppressor proteins (e.g., BRG-1), renders RB a more mobile nuclear factor. These observations indicate that the activity state of RB in living cells can be accurately assessed by complex mobility. RB complexes dictating cell cycle arrest are typically dynamic and harbor a relatively short residence time on chromatin. In contrast, this rapid exchange is attenuated in cells that are hypersensitive to RB, suggesting that the biological response to RB correlates with chromatin residence time. Last, the vast majority of RB-assembled complexes were associated with E2F promoter elements, indicating that the bulk of RB action is directed at E2F-dependent gene regulation. Together, these results provide the first description of the kinetic framework of RB tumor suppressor action in transcriptional repression and cell cycle regulation.
FIG. 5—Continued.
Acknowledgments
We are grateful to Karen Knudsen, Christin Petre, and the members of the Knudsen laboratories for thought-provoking discourse. We thank Wallace Ip, Robert Brackenbury, David Gilbert, and Tom Misteli for suggestions and technical advice. Doug Dean, Bernard Weissman, and Paul Hamel generously provided plasmids. We also thank Dan Wiginton for providing MOLT-4 cells. Zeiss kindly provided the FCS instrument used in these studies. All imaging was performed at the UC Center for Biological Microscopy.
S.P.A. is supported by the Albert J. Ryan Foundation. E.S.K. is supported by the National Cancer Institute (CA82525).
S.P.A. and D.A.S. contributed equally to this work.
REFERENCES
- 1.Alcalay, M., L. Tomassoni, E. Colombo, S. Stoldt, F. Grignani, M. Fagioli, L. Szekely, K. Helin, and P. G. Pelicci. 1998. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 18:1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus, S. P., A. F. Fribourg, M. P. Markey, S. L. Williams, H. F. Horn, J. DeGregori, T. F. Kowalik, K. Fukasawa, and E. S. Knudsen. 2002. Active RB elicits late G1/S inhibition. Exp. Cell Res. 276:201-213. [DOI] [PubMed] [Google Scholar]
- 3.Angus, S. P., L. J. Wheeler, S. A. Ranmal, X. Zhang, M. P. Markey, C. K. Mathews, and E. S. Knudsen. 2002. Retinoblastoma tumor suppressor targets dNTP metabolism to regulate DNA replication. J. Biol. Chem. 277:44376-44384. [DOI] [PubMed] [Google Scholar]
- 4.Chew, Y. P., M. Ellis, S. Wilkie, and S. Mittnacht. 1998. pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene 17:2177-2186. [DOI] [PubMed] [Google Scholar]
- 5.Dahiya, A., S. Wong, S. Gonzalo, M. Gavin, and D. C. Dean. 2001. Linking the Rb and polycomb pathways. Mol. Cell 8:557-569. [DOI] [PubMed] [Google Scholar]
- 6.DeGregori, J. 2002. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim. Biophys. Acta 1602:131-150. [DOI] [PubMed] [Google Scholar]
- 7.Dimitrova, D. S., and R. Berezney. 2002. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115:4037-4051. [DOI] [PubMed] [Google Scholar]
- 8.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 9.Durfee, T., M. A. Mancini, D. Jones, S. J. Elledge, and W. H. Lee. 1994. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J. Cell Biol. 127:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 11.Giangrande, P. H., T. C. Hallstrom, C. Tunyaplin, K. Calame, and J. R. Nevins. 2003. Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol. Cell. Biol. 23:3707-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 13.Hannan, K. M., B. K. Kennedy, A. H. Cavanaugh, R. D. Hannan, I. Hirschler-Laszkiewicz, L. S. Jefferson, and L. I. Rothblum. 2000. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene 19:3487-3497. [DOI] [PubMed] [Google Scholar]
- 14.Harbour, J. W., and D. C. Dean. 2000. Chromatin remodeling and Rb activity. Curr. Opin. Cell Biol. 12:685-689. [DOI] [PubMed] [Google Scholar]
- 15.Harbour, J. W., and D. C. Dean. 2001. Corepressors and retinoblastoma protein function. Curr. Top. Microbiol. Immunol. 254:137-144. [DOI] [PubMed] [Google Scholar]
- 16.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed] [Google Scholar]
- 17.Hinds, P. W., S. Mittnacht, V. Dulic, A. Arnold, S. I. Reed, and R. A. Weinberg. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70:993-1006. [DOI] [PubMed] [Google Scholar]
- 18.Kaelin, W. G., Jr. 1997. Alterations in G1/S cell-cycle control contributing to carcinogenesis. Ann. N. Y. Acad. Sci. 833:29-33. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy, B. K., D. A. Barbie, M. Classon, N. Dyson, and E. Harlow. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14:2855-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 23.Knudsen, E. S., C. Buckmaster, T. T. Chen, J. R. Feramisco, and J. Y. Wang. 1998. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 12:2278-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen, E. S., and J. Y. Wang. 1996. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J. Biol. Chem. 271:8313-8320. [DOI] [PubMed] [Google Scholar]
- 25.Knudsen, E. S., and J. Y. Wang. 1997. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 17:5771-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudsen, K. E., A. F. Fribourg, M. W. Strobeck, J. M. Blanchard, and E. S. Knudsen. 1999. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J. Biol. Chem. 274:27632-27641. [DOI] [PubMed] [Google Scholar]
- 27.Lee, W. H., J. Y. Shew, F. D. Hong, T. W. Sery, L. A. Donoso, L. J. Young, R. Bookstein, and E. Y. Lee. 1987. The retinoblastoma susceptibility gene encodes a nuclear phosphoprotein associated with DNA binding activity. Nature 329:642-645. [DOI] [PubMed] [Google Scholar]
- 28.Lillemeier, B. F., M. Koster, and I. M. Kerr. 2001. STAT1 from the cell membrane to the DNA. EMBO J. 20:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 30.Lukas, J., T. Herzinger, K. Hansen, M. C. Moroni, D. Resnitzky, K. Helin, S. I. Reed, and J. Bartek. 1997. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 11:1479-1492. [DOI] [PubMed] [Google Scholar]
- 31.Lukas, J., B. O. Petersen, K. Holm, J. Bartek, and K. Helin. 1996. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol. Cell. Biol. 16:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mancini, M. A., B. Shan, J. A. Nickerson, S. Penman, and W. H. Lee. 1994. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc. Natl. Acad. Sci. USA 91:418-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markiewicz, E., T. Dechat, R. Foisner, R. A. Quinlan, and C. J. Hutchison. 2002. Lamin A/C binding protein LAP2alpha is required for nuclear anchorage of retinoblastoma protein. Mol. Biol. Cell 13:4401-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNally, J. G., W. G. Muller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 35.Mittnacht, S. 1998. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 8:21-27. [DOI] [PubMed] [Google Scholar]
- 36.Mittnacht, S., and R. A. Weinberg. 1991. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell 65:381-393. [DOI] [PubMed] [Google Scholar]
- 37.Morris, E. J., and N. J. Dyson. 2001. Retinoblastoma protein partners. Adv. Cancer Res. 82:1-54. [DOI] [PubMed] [Google Scholar]
- 38.Narita, M., S. Nunez, E. Heard, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 39.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 41.Ozaki, T., M. Saijo, K. Murakami, H. Enomoto, Y. Taya, and S. Sakiyama. 1994. Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene 9:2649-2653. [PubMed] [Google Scholar]
- 42.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 43.Qin, X. Q., T. Chittenden, D. M. Livingston, and W. G. Kaelin, Jr. 1992. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 6:953-964. [DOI] [PubMed] [Google Scholar]
- 44.Reits, E. A., and J. J. Neefjes. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3:E145-E147. [DOI] [PubMed] [Google Scholar]
- 45.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 46.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Peeper. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell 2:55-65. [DOI] [PubMed] [Google Scholar]
- 47.Schlisio, S., T. Halperin, M. Vidal, and J. R. Nevins. 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21:5775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwille, P. 2001. Fluorescence correlation spectroscopy and its potential for intracellular applications. Cell Biochem. Biophys. 34:383-408. [DOI] [PubMed] [Google Scholar]
- 49.Sever-Chroneos, Z., S. P. Angus, A. F. Fribourg, H. Wan, I. Todorov, K. E. Knudsen, and E. S. Knudsen. 2001. Retinoblastoma tumor suppressor protein signals through inhibition of cyclin-dependent kinase 2 activity to disrupt PCNA function in S phase. Mol. Cell. Biol. 21:4032-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherr, C., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 51.Sherr, C. J. 2001. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 52.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 52a.Siddiqui, H., D. A. Solomon, R. W. Gunawardena, Y. Wang, and E. S. Knudsen. 2003. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol. Cell. Biol. 23:7719-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stenoien, D. L., K. Patel, M. G. Mancini, M. Dutertre, C. L. Smith, B. W. O'Malley, and M. A. Mancini. 2001. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol 3:15-23. [DOI] [PubMed] [Google Scholar]
- 54.Strobeck, M. W., K. E. Knudsen, A. F. Fribourg, M. F. DeCristofaro, B. E. Weissman, A. N. Imbalzano, and E. S. Knudsen. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 97:7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strobeck, M. W., D. N. Reisman, R. W. Gunawardena, B. L. Betz, S. P. Angus, K. E. Knudsen, T. F. Kowalik, B. E. Weissman, and E. S. Knudsen. 2002. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 277:4782-4789. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 57.Takemura, M., F. Ohoka, M. Perpelescu, M. Ogawa, H. Matsushita, T. Takaba, T. Akiyama, H. Umekawa, Y. Furuichi, P. R. Cook, and S. Yoshida. 2002. Phosphorylation-dependent migration of retinoblastoma protein into the nucleolus triggered by binding to nucleophosmin/B23. Exp. Cell Res. 276:233-241. [DOI] [PubMed] [Google Scholar]
- 58.Templeton, D. J., S. H. Park, L. Lanier, and R. A. Weinberg. 1991. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc. Natl. Acad. Sci. USA 88:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Lohuizen, M. 1999. The trithorax-group and polycomb-group chromatin modifiers: implications for disease. Curr. Opin. Genet. Dev. 9:355-361. [DOI] [PubMed] [Google Scholar]
- 60.Visintin, R., and A. Amon. 2000. The nucleolus: the magician's hat for cell cycle tricks. Curr. Opin. Cell Biol. 12:372-377. [DOI] [PubMed] [Google Scholar]
- 61.Wang, J. Y., E. S. Knudsen, and P. J. Welch. 1994. The retinoblastoma tumor suppressor protein. Adv. Cancer Res. 64:25-85. [DOI] [PubMed] [Google Scholar]
- 62.Weintraub, S. J., K. N. Chow, R. X. Luo, S. H. Zhang, S. He, and D. C. Dean. 1995. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 375:812-815. [DOI] [PubMed] [Google Scholar]
- 63.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wells, J., P. S. Yan, M. Cechvala, T. Huang, and P. J. Farnham. 2003. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene 22:1445-1460. [DOI] [PubMed] [Google Scholar]
- 65.White, R. J. 1997. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem. Sci. 22:77-80. [DOI] [PubMed] [Google Scholar]
- 66.Xiao, Z. X., J. Chen, A. J. Levine, N. Modjtahedi, J. Xing, W. R. Sellers, and D. M. Livingston. 1995. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature 375:694-698. [DOI] [PubMed] [Google Scholar]
- 67.Yang, H., B. O. Williams, P. W. Hinds, T. S. Shih, T. Jacks, R. T. Bronson, and D. M. Livingston. 2002. Tumor suppression by a severely truncated species of retinoblastoma protein. Mol. Cell. Biol. 22:3103-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zacksenhaus, E., R. Bremner, R. A. Phillips, and B. L. Gallie. 1993. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol. Cell. Biol. 13:4588-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, H. S., A. A. Postigo, and D. C. Dean. 1999. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell 97:53-61. [DOI] [PubMed] [Google Scholar]