Abstract
The murine ZnT3 gene was cloned by virtue of its homology to the ZnT2 gene, which encodes a membrane protein that facilitates sequestration of zinc in endosomal vesicles. ZnT-3 protein is predicted to have six transmembrane domains and shares 52% amino acid identity with ZnT-2, with the homology extending throughout the two sequences. Human ZnT-3 cDNAs were also cloned; the amino acid sequence is 86% identical to murine ZnT-3. The mouse ZnT3 gene has 8 exons and maps to chromosome 5. Northern blot and reverse transcriptase–PCR analyses demonstrate that murine ZnT-3 expression is restricted to the brain and testis. In situ hybridization reveals that within the brain, ZnT-3 mRNA is most abundant in the hippocampus and cerebral cortex. Antibodies raised against the C-terminal tail of mouse ZnT-3 react with the projections from these neurons and produce a pattern similar to that obtained with Timm’s reaction, which reveals histochemically reactive zinc within synaptic vesicles. We propose that ZnT-3 facilitates the accumulation of zinc in synaptic vesicles.
Cytoplasmic zinc concentration is maintained within a narrow range in mammalian cells (1). Zinc is required for the maintenance and activity of numerous metalloproteins where it plays either a structural role (e.g., in zinc-finger proteins) or catalytic function as part of the active site of various metalloenzymes (2). Cells can tolerate slight increases in zinc over the amount required to fulfill metalloprotein needs, but beyond that excess, zinc becomes toxic unless cells can induce protective mechanisms (1). One protective mechanism involves the induction of metallothioneins that can sequester the excess zinc (3). However, another important mode of zinc regulation is likely to be at the level of transporters that facilitate zinc influx during deficiency and efflux during excess. Some of the molecules involved in zinc transport have been cloned recently. ZRT1 is a yeast protein with eight predicted transmembrane domains that mediates high-affinity uptake of zinc and is inducible by zinc limitation (4). A related low-affinity zinc uptake transporter (ZRT2) has also been cloned from yeast (5). Zinc efflux in mammalian cells is mediated by ZnT-1, a protein predicted to span the membrane six times. ZnT-1 is activated by excess zinc (1).
Zinc can also be concentrated in organelles in some cell types. Vesicular zinc has been visualized in pancreas, salivary gland, testis, and brain with the Timm’s sulfide–silver staining procedure (6), as well as with the fluorescent probes 6-methoxy-8-p-toluene sulfonamide quinoline (TSQ) and zinquin (7, 8). These probes do not react appreciably with cytoplasmic zinc, suggesting that vesicular zinc is not tightly complexed with proteins. In pancreatic beta cells and male salivary glands, the detectable zinc is in secretory granules (7, 8), whereas in the brain the staining is predominantly in synaptic vesicles of neuronal axons (7). Neurons containing histochemically detectable zinc are particularly abundant in the hippocampal formation and cerebral cortex. The most intense Timm’s or TSQ staining is seen in the “mossy fibers” that project from the granule cells of the dentate gyrus to the CA3 region of the hippocampus (9–12). Other regions of the hippocampus, cortex, and amygdala are also reactive. These neurons are thought to be excitatory glutamatergic neurons (7). Specific transporters are probably essential to concentrate zinc in various vesicular compartments.
We cloned a gene encoding a zinc transporter (ZnT-2) that facilitates accumulation of zinc in endosomal vesicles and confers resistance to zinc toxicity by complementation of a zinc-sensitive baby hamster kidney (BHK) cell line (13). This transporter is homologous to ZnT-1, but it is localized on endosomal vesicles instead of the plasma membrane. BHK cells expressing ZnT-2 accumulate detectable levels of zinc in endosomes only when they are exposed to elevated levels of extracellular zinc.
In the process of screening a mouse genomic λ library with rat ZnT-2 cDNA, a homologous gene was cloned. Herein we characterize that gene, which we call ZnT3, and show that its mRNA is abundant in the hippocampus and cortex. The ZnT-3 protein is detected immunologically in the mossy fibers, where zinc-containing vesicles are most abundant. Overall, the pattern of ZnT-3 expression resembles that observed with the Timm’s stain or TSQ. Because the zinc in synaptic vesicles is coreleased with glutamate in response to high-frequency stimulation (14, 15) and can modulate the activity of various receptors in vitro, including ionotropic glutamate receptors and γ-aminobutyric acid receptors (16–23), zinc has been postulated to function as a neuromodulator (7, 24, 25). We propose that ZnT-3 is an essential component of the complex that sequesters zinc in synaptic vesicles, enabling it to serve as a neuromodulator.
MATERIALS AND METHODS
Cloning and Sequencing.
A mouse (strain 129) genomic λ library was screened using rat ZnT-2 cDNA as a probe at reduced stringency. Eight clones were purified; four overlapping clones corresponded to mouse ZnT-2, whereas the other four overlapping clones represented a related gene. Most of the hybridization to the related gene was confined to a 2-kb EcoRI fragment that was isolated and sequenced. That EcoRI fragment was then used as a probe in a screen of a mouse brain cDNA library (Stratagene). The clone with the longest insert was sequenced. An oligonucleotide probe based on the 5′ end of the cDNA sequence was used to find the corresponding exon in the genomic DNA, which was then subcloned and sequenced. DNA sequencing was by the dideoxynucleotide method, using 7-deaza-dATP and 7-deaza-dGTP when necessary to resolve ambiguities. The cDNA was sequenced on both strands but the genomic DNA was only sequenced on one strand. The human ZnT-3 cDNA was isolated from a brain temporal cortex library (Stratagene) and sequenced on both strands.
Immunological Techniques.
DNA corresponding to the C-terminal 93 amino acids of ZnT-3 was amplified by PCR and inserted, in-frame, downstream of the maltose binding protein in pMAL-CR1 (New England Biolabs). The fusion protein was purified from Escherichia coli on amylose resin and used to immunize two rabbits (R & R Rabbitry, Stanwood, WA). The immunoglobulin fraction was isolated by ammonium sulfate precipitation and affinity-purified by passing it through a column containing the same C-terminal tail of ZnT-3 fused to a biotinylated peptide (PinPoint vector, Promega). The concentration of the purified antibody was 65 μg/ml.
For Western blots, cell pellets or solid tissues were homogenized in 8 vol of sample buffer (2% sodium dodecyl sulfate/5% 2-mercapoethanol/50 mM Tris·HCl, pH 7/10% glycerol/0.01% bromophenol blue) and boiled, and aliquots were electrophoresed on a 0.1% sodium dodecyl sulfate/10% polyacrylamide gel. The proteins were electrophoretically transferred to Hybond-C (Amersham). The nitrocellulose was soaked in 5% Blotto [PBS (138 mM NaCl/2.7 mM KCl/10 mM phosphate, pH 7.4) containing 5% nonfat powdered milk and 0.1% Tween-20] overnight at 4°C and then exposed to the purified antibody (diluted 1:100 in 1% Blotto) overnight at 4°C. The filter was washed three times in 1% Blotto and then incubated 1 hr with peroxidase-linked donkey anti-rabbit IgG (Amersham) that was diluted 1:3000 in 1% Blotto. This was followed by three washes in PBS/0.1% Tween-20, and the bound antibody was visualized using the Renaissance Western Blot Chemiluminescence reagents (DuPont/NEN) and exposed to X-Omat AR film (Eastman Kodak).
For immunocytochemistry, tissues were quickly removed from CO2-asphyxiated C57BL/6 mice into ice-cold isopentane for 20 sec and then stored at −80°C prior to cutting 10-μm sections. The sections were air-dried, fixed in 4% paraformaldehyde in PBS, rinsed twice in PBS, and then boiled for 8 min in 10 mM citric acid using a microwave oven. The sections were returned to PBS and exposed to 3% H2O2 for 15 min, washed in PBS, and incubated overnight at 4°C with the primary antibody (1:100 dilution) in PBS containing 1% bovine serum albumin and 3% goat serum. After washing, the sections were incubated for 1 hr at 4°C with biotinylated anti-rabbit IgG, diluted 1:200 in PBS (Vector Laboratories). The reaction product was detected with streptavidin-peroxidase (1:20 dilution) using the AEC reagents from Zymed.
Timm’s Staining.
A modification of the Timm’s staining procedure (26) was performed as described by Masters et al. (27).
RNA Detection.
Total RNA was isolated from various organs of adult C57BL/6 mice using TRIzol reagent (BRL/GIBCO). Complementary DNA was prepared from 2 μg of total RNA using reverse transcriptase with oligo(dT) as a primer. Aliquots of the cDNA were then amplified by 35 cycles of PCR (94°C for 30 sec; 72°C for 90 sec) using the following oligonucleotide primers from genomic ZnT-1 sequence (1) [5′ primer, GGCCAACACCAGCAATTCCAACG (1011); 3′ primer, AAGGCATTCACGACCACGATCACG (2470)], from the mouse ZnT-2 cDNA sequence (13) [5′ primer, GTTGAGCTGGCTGTCCAGAGCAACC (133); 3′ primer, GCCGAAGTTCATGGTCTTGGTGGC (411)], from the mouse ZnT-3 cDNA sequence (this paper) [5′ primer, GGCCAACACCAGCAATTCCAACG (371); 3′ primer, AAGGCATTCACGACCACGATCACG (821)]. As a control for RNA integrity, primers for elongation factor 1α (EF1α) cDNA (28) were used at 94°C for 30 sec, 55°C for 30 sec, and 72°C for 60 sec [5′ primer, ACATTAAGAAAATTGGCTAC (589); 3′ primer, ATTGAAGCCCACATTGTCCC (992)]. The number brackets refers to the location of the first base of the oligonucleotide on the published sequence. All of the primer pairs lie in separate exons to obviate any amplification due to DNA contamination. The products were visualized by ethidium bromide staining after electrophoresis through 0.8% agarose/2% NuSieve GTG agarose (FMC).
In Situ Hybridization.
In situ hybridization was performed essentially as described by Marks et al. (29). Tissues from C57BL/6 mice were prepared as described above. Frozen coronal sections (10 μm) were cut and placed on Superfrost/Plus microscope slides (Fisher Scientific). The sections were heated to 50°C for 2 min, air-dried 30 min, and stored at −80°C. The dried sections were fixed in 4% paraformaldehyde in PBS for 10 min at 4°C. After rinsing for two 5-min periods in PBS, the brain sections were dehydrated through a graded ethanol series to 70% ethanol, delipidated in chloroform for 5 min, rehydrated through the graded ethanol series, and rinsed in PBS for 5 min. Testis sections were not delipidated. To decrease nonspecific binding, the sections were treated with 0.25% acetic anhydride in 0.1 M triethanolamine (pH 9.5) for 10 min. After a brief rinse in PBS, the sections were incubated in 100 μl of hybridization mixture [50% formamide/1× Denhardt’s solution/10% dextran sulfate/0.6 M NaCl/10 mM NaOAc/0.1% Tween 20/50 mM dithiothreitol (DTT)/heparin sodium (Fisher Scientific) (50 μg/ml)/yeast RNA (0.5 mg/ml)/herring sperm DNA (0.1 mg/ml)] for 3 hr at 50°C under a coverslip.
For the production of RNA probes, 1 μg of linearized template DNA [ZnT3 cDNA (1956 bp) in Bluescript (Stratagene)] was added to a 20-μl reaction mixture containing 2 μl of 10× transcription buffer (Boehringer Mannheim), 500 μM ATP, 500 μM GTP, 500 μM CTP, 5 μl (3 μM) of [33P]UTP (Andotek, Irvine, CA), 40 units of RNAguard (Pharmacia), and 50 units of T3 or T7 RNA polymerase and incubated for 3 hr at 37°C. To destroy the template, the transcription reaction was diluted to 40 μl with H2O and incubated with 1 μl (10 units) RNase-free DNase I (Boehringer Mannheim) for 30 min at 37°C. RNA was precipitated with 0.8 M LiCl and 3 vol of ethanol and resuspended in 50 μl of 0.2× SET (2 mM Tris/1 mM EDTA/0.2% SDS). This procedure gave a yield of 15,000 cpm/ng of RNA. The probe was diluted into fresh hybridization mixture (2 ng of RNA per μl). The coverslips were floated off the sections in PBS and rinsed briefly in PBS, 25 μl of the probe solution was applied, and a coverslip was sealed in place with a 1:1 mixture of petroleum ether and rubber cement. The sections were incubated for ≈16 hr at 50°C, after which the sealant was removed, the coverslips were floated off in 4× SSC (1× SSC = 150 mM NaCl/15 mM sodium citrate, pH 7.5), rinsed for 10 min in 2× SSC with 2 mM DTT, then treated with RNase A solution (RNase A at 20 μg/ml in 0.5 M NaCl/10 mM Tris, pH 8.0/10 mM DTT) for 30 min at 37°C. This was followed by a series of washes: 2× SSC/50% formamide/10 mM DTT at 60°C for 30 min; 1× SSC/50% formamide/10 mM DTT at 60°C (for brain) or 50°C (for testis) for 30 min; 0.1× SSC at 37°C for 30 min. The sections were then dehydrated through a graded ethanol series, air-dried for about 16 hr, and coated with emulsion (NTB-3, Eastman Kodak). Coated slides were exposed to emulsion for various times (4–17 days) at 4°C. After developing the sections, silver grains were visualized and photographed in dark field, using a Nikon Microphot FX microscope.
Chromosomal Mapping.
ZnT-3 was mapped to chromosome 5 as described (13). The approved designation for its gene locus is ZnT3. The chromosomal map location is available on the World Wide Web at http://www.jax.org/resources/documents/cmddata.
Expression of ZnT-3 in BHK Cells.
The ZnT-3 cDNA was inserted into the cloning site of pcDNA1 that had been modified to carry a dihydrofolate reductase gene driven by simian virus 40 promoter/enhancer. This construct was transfected into a zinc-sensitive BHK cell line (3286–8-8) and cells resistant to methotrexate were cloned (1). A clone with the highest level of ZnT-3 mRNA was selected for further studies. A derivative, ZnT-3:GFP, with the green fluorescent protein (containing alanine at position 65) fused at the C terminus of ZnT-3 was also prepared in this vector.
RESULTS
Cloning and Characterization of Mouse ZnT3 Gene and cDNA.
Four overlapping mouse genomic clones were isolated from a λ library by screening with rat ZnT-2 cDNA. Restriction mapping revealed that most of the hybridzation signal corresponded to a 2-kb EcoRI fragment that was subcloned and sequenced. It contained several putative exons with homology to ZnT-2. The EcoRI fragment was then used to screen a mouse brain cDNA library. The longest cDNA clone was sequenced; it contained an open reading frame encoding a protein of 388 amino acids that is 52% identical to rat ZnT-2 (Fig. 1). We call the new protein ZnT-3 (Zinc Transporter-3) because of its similarity to ZnT-2. The two proteins are predicted to have similar membrane topology with six transmembrane domains and both N and C termini on the cytoplasmic side of the membrane.
Figure 1.
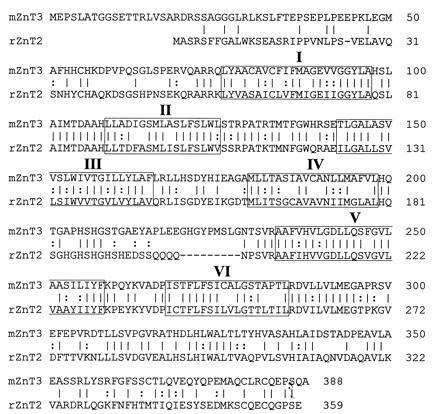
Comparison of mouse ZnT-3 protein sequence with rat ZnT-2. Amino acids are designated by one-letter code; identical amino acids are connected by lines; similar amino acids are connected by dots; the six predicted transmembrane domains are boxed with Roman numerals.
Seven exons encoding most of ZnT-3 were located in the λ clones and sequenced. About 800 nt corresponding to the 3′ end of the cDNA were not included in any of the λ clones; however, PCR of mouse genomic DNA with opposing primers from each end of the missing 800 nt gave a product with the same size as that obtained from the cDNA, indicating that they reside within one exon. This exon, number 8, which includes the C-terminal 50 amino acids and 3′ untranslated region, is located ≈1.05 kb downstream of exon 7, as determined by restriction mapping. A composite map of the mouse ZnT3 gene is shown in Fig. 2B; the locations of the introns are indicated in Fig. 2A. Because the genomic sequence upstream of the 5′ end of the cDNA is G+C-rich and lacks potential splice acceptor sites, we assume that this is the first exon; however, no obvious TATA box was discerned and the transcription start site has not been mapped. Although the mouse ZnT2 gene has not been completely characterized, its genomic organization appears to be similar to that of ZnT3; both of these genes are more complex than ZnT1, which has only two exons.
Figure 2.

Sequence of mouse ZnT-3 cDNA, comparison of mouse and human ZnT-3 protein sequences, and mouse ZnT-3 genomic organization. (A) Mouse ZnT-3 cDNA sequence with amino acid sequence in three-letter code above. Differences between human and mouse ZnT-3 are indicated above the mouse protein sequence. Arrows indicate the location of introns and numbers designate the flanking exons. (B) A map of the mouse ZnT-3 genomic DNA showing the location of 8 exons. The regions that have been sequenced are indicated with a dotted line.
Cloning Human ZnT-3 cDNA.
The human counterpart of mouse ZnT-3 was cloned from a human temporal cortex cDNA library in a λ ZAP vector. Twenty-four clones were identified by hybridization to mouse ZnT-3 at low stringency; sequencing revealed that they all represented parts of the same cDNA. Two overlapping clones were sequenced to produce the complete open reading frame, which encodes a protein of 388 amino acids that is 86% identical to mouse ZnT-3; the amino acid differences are shown in Fig. 2A.
Chromosomal Mapping of ZnT-3.
Genomic DNAs from C57BL/6J and Mus spretus were digested with 10 different enzymes, electrophoresed on agarose, and transferred to nitrocellulose, and the blots were probed with the mouse ZnT-3 cDNA. Polymorphisms were apparent for BglII, EcoRI, and MspI. The BglII polymorphism was chosen to screen 94 segregants of the Jackson Laboratory BSS backcross panel (30) using Southern blot filters provided by the Jackson Laboratory BC Panel Map Service. This analysis indicated that ZnT3 cosegregates with Nos3, Pep1b, and Plk on mouse chromosome 5 near the engrailed (En) locus. Human ZnT-3 was not mapped, but the syntenic region is on human chromosome 7. ZnT3 is on a different chromosome than ZnT2, which resides on mouse chromosome 4 (10).
Expression of ZnT-3 in BHK Cells.
Because the ZnT3 gene was cloned by virtue of its homology to ZnT2, it was tested for its ability to confer resistance to metals in zinc-sensitive BHK cells. The ZnT-3 cDNA was inserted into an expression vector with a DHFR-selectable gene, transfected into BHK cells and a stable clones were isolated after growth in 20 μM methotrexate. Solution hybridization with an oligonucleotide complementary to ZnT-3 mRNA was used to identify clones with the highest levels of ZnT-3 mRNA. Western blots revealed a protein of the expected size in these cells (see below). Fluorescence with zinquin was not observed in the transfected BHK cells regardless of whether they were incubated in normal medium or in medium containing up to the maximum amount of zinc they could tolerate. These cells were also compared with the parental cells for their resistance to zinc, cadmium, cobalt, and copper toxicity. No resistance to the toxic effects of any of these metals was observed (data not shown). We conclude that expression of ZnT-3 in BHK cells has no discernible effect on their phenotype, in contrast to the pronounced effects of ZnT-1 or ZnT-2 expression in these cells (1, 13).
Expression of ZnT-3 in Vivo.
To determine where ZnT-3 is expressed in vivo, total RNA was isolated from various mouse organs and converted to cDNA with reverse transcriptase using oligo(dT) as a primer. Aliquots were then subjected to PCR using primers in exons 2 and 5 (see Fig. 2). A product was detected by ethidium bromide staining with RNA from brain and testis but not the other organs indicated in Fig. 3. In contrast, ZnT-1 is expressed in most organs and ZnT-2 is expressed in intestine, kidney, seminal vesicles, and testis.
Figure 3.
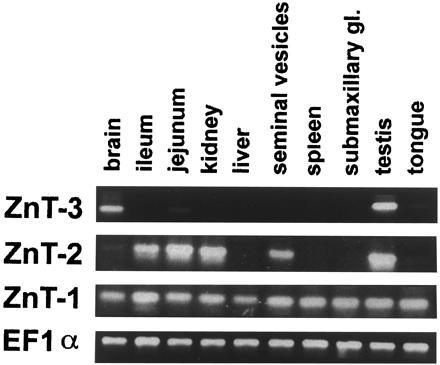
Organ distribution of ZnT-3, ZnT-2, and ZnT-1 mRNAs. Total RNA from the indicated organs was prepared and subjected to reverse transcription followed by PCR. The products were electrophoresed through 2.8% agarose and stained with ethidium bromide.
In situ hybridization with antisense RNA probes to mouse ZnT-3 was used to determine which cells in brain and testis express ZnT-3. Fig. 4A shows a coronal section of a mouse brain, revealing hybridization to the hippocampal formation, piriform cortex, and lateral amygdala, as well as several layers of the neocortex. Hybridization was also detectable in the paraventricular thalamic nucleus and the zona inserta. In the hippocampus (Fig. 4B), ZnT-3 mRNA is evident in granule cell neurons of the dentate gyrus and in pyramidal cells in the CA3 and CA1 regions. This hybridization pattern corresponds closely to the cell bodies of neurons that sequester zinc in synaptic vesicles (compare Fig. 4 D and F). Examination of sagittal sections of mouse brain revealed the presence of ZnT-3 mRNA in the entorhinal cortex as well, whereas there was none in olfactory bulb, cerebellum, or hypothalamus (data not shown). In the testis, hybridization was confined to germ cells and was most intense in pachytene spermatocytes and round spermatids (data not shown). No specific hybridization to either brain or testis sections was observed with a ZnT-3 sense probe (data not shown).
Figure 4.
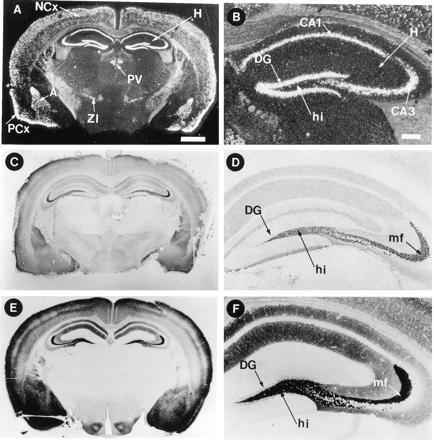
Localization of ZnT-3 mRNA, protein, and histochemically reactive zinc in the brain. Coronal sections of mouse brain were subjected to in situ hybridization with a 33P-labeled probe complementary to mouse ZnT-3 mRNA (A and B), immunocytochemistry with affinity-purified antisera raised against the C-terminal tail of ZnT-3 (C and D), and Timm’s staining procedure for vesicular zinc (E and F). (B, D, and F, ×2.8; A, C, and E, ×0.7). In the hippocampus, ZnT-3 mRNA is present in the cell bodies of dentate granule cells, and ZnT-3 protein is abundant in the zinc-rich mossy fiber projections emanating from these neurons. A, amygdala (lateral nucleus); H, hippocampus; NCx, neocortex; PCx, piriform cortex; PV, paraventricular thalamic nucleus; ZI, zona inserta; GC, granule cell neurons of the dentate gyrus; hi, hilus; mf, mossy fibers. [Bars = 1000 μm (A) and 200 μm (B).]
ZnT-3 Protein Is Present in the Mossy Fibers of the Hippocampus That Sequester Zinc in Synaptic Terminals.
A rabbit polyclonal antibody was raised against the C-terminal cytoplasmic tail of mouse ZnT-3 and purified by affinity chromatography. The specificity of this antibody was assessed by Western blots of total cell proteins from control BHK cells and BHK cells that were stably transfected with constructs expressing ZnT-1 (1), ZnT-2 (12), or ZnT-3 (described above). Only BHK cells expressing ZnT-3 reacted with this antibody, indicating that it does not react with the homologous proteins (Fig. 5). The specific band detected in the BHK cells has an apparent molecular weight of ≈40 kDa. An immunoreactive protein of similar size was detected in brain samples, but no protein of this size was detected in either the kidney or testis samples (Fig. 5). A faint band of lower molecular weight was evident in testis after longer development times.
Figure 5.
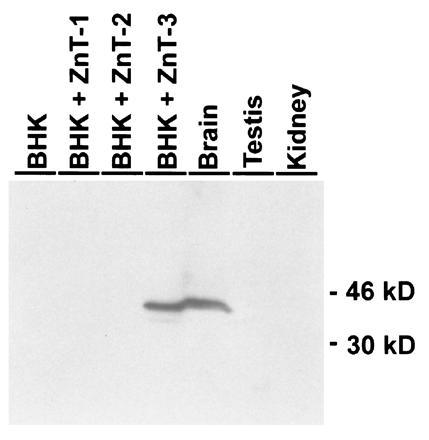
Western blot analysis of ZnT-3 protein. Lanes 1–4 represent equivalent amounts of protein isolated from confluent BHK cells: untransfected cells (lane 1), cells expressing ZnT-1 (lane 2), cells expressing ZnT-2 (lane 3), and cells expressing ZnT-3 (lane 4). Lanes 5–7 represent total protein from brain (lane 5), testis (lane 6), and kidney (lane 7). The amount of protein in all lanes was similar by Coomassie blue 250 staining of a replicate gel. Affinity-purified antibody to ZnT-3 was detected with a peroxidase-linked secondary antibody by chemiluminescence.
When this antibody was applied to brain sections, the most intense immunostaining was observed over the mossy fiber projections emanating from the granule cell neurons in the dentate gyrus (Fig. 4 C and D). Projections from pyramidal cells of the hippocampus and in the cortex also stained with this antibody but not with preimmune serum (data not shown). The antibody staining pattern in the brain is very similar to the histochemical localization of zinc by the Timm’s reaction, which detects loosely bound synaptic zinc (compare Fig. 4 C and E with D and F). The only exception being the dendritic field of the dentate granule cells that react with the antibody but not with Timm’s staining procedure (compare Fig. 4 D and F).
Neither Western blots of total testis proteins (Fig. 5) nor immunohistochemistry on sections of adult testis using the purified antibody (data not shown) gave a reaction under conditions that worked well for detection of ZnT-3 protein in the brain.
DISCUSSION
We have cloned mouse and human ZnT-3 cDNAs encoding membrane proteins that are homologous to the mammalian zinc transporters, ZnT-1 and ZnT-2, as well as two yeast proteins, ZRC and COT1, that confer resistance to zinc and cobalt, respectively. All of these proteins are predicted to have six transmembrane domains with the N and C termini located in the cytoplasm (1). The cytoplasmic loop connecting the transmembrane domains IV and V is relatively short in ZnT-3 and lacks the characteristic histidines of ZnT-1 and ZnT-2. The transmembrane domains and C-terminal tail of ZnT-3 (≈150 amino acids) are similar to ZnT-2. Furthermore, the exon–intron structures of the ZnT-2 and ZnT-3 genes are similar, but unlike ZnT-1, which has only one intron. The low-stringency screen of the human brain cDNA library only produced the human counterpart of mouse ZnT-3, suggesting that if there are other members of this gene family, they are either too divergent from ZnT-3 to hybridize or are not expressed in temporal cortex.
The best clues for the function of ZnT-3 arise from its expression pattern and its homology to ZnT-2, which facilitates accumulation of zinc into vesicles in BHK cells. ZnT-3 mRNA is detected only in brain and testis when measured by reverse transcriptase–PCR or Northern blot analysis. ZnT-3 mRNA is readily detected in the hippocampus and cortex by in situ hybridization. Within the hippocampal formation, the cell bodies of the granule cells in the dentate gyrus produce the most intense hybridization signal, while pyramidal cells in the CA3 and CA1 regions also give strong signals. All of these glutamatergic neurons have processes containing vesicular zinc as revealed by a variety of histochemical techniques (9–12). The mossy fiber projections of granule cells produce the most intense staining for histochemically reactive zinc with the sulfide/silver or TSQ fluorescence methods. The mossy fibers also give the most intense reaction with the antibody against ZnT-3, suggesting that ZnT-3 is transported down the axons of granule cells in association with zinc. We also note in situ hybridization signals and immunostaining in many regions of the cerebral cortex that resemble the Timm’s staining pattern; however, because the cellular architecture of the cortex is complex, we cannot be certain that ZnT-3 is localized exclusively in neurons that sequester zinc.
Because ZnT-3 is homologous to ZnT-2, which has been localized to endosomes of BHK cells and facilitates accumulation of zinc in those vesicles (13), we predict that ZnT-3 might also be localized to the membranes of a cellular organelle. An attractive possibility is that ZnT-3 is one component of a complex involved in transport of zinc into synaptic vesicles. In the hippocampus, the ZnT-3 protein is most abundant where zinc-laden synaptic vesicles reside, which is consistent with it being a membrane component of those vesicles. However, it is also possible that other organelles are transported down the axons of those neurons and ZnT-3 may be associated with them. Electron microscopy of immunogold-labeled sections after Timm’s staining should answer the question of whether ZnT-3 is on the same vesicles that accumulate zinc in the mossy fibers and elsewhere in the brain.
We have no direct evidence that ZnT-3 transports zinc; however, its homology to other zinc transporters and its association with vesicular zinc in the brain lends credence to this hypothesis. Unlike ZnT-2 (13), expression of ZnT-3 in BHK cells does not protect against zinc toxicity, does not result in accumulation of zinc, and does not produce zinquin fluorescence when the cells are grown in normal or zinc-supplemented medium. We tested whether ZnT-3 might provide resistance to other heavy metals (copper, cobalt, or cadmium) in cell culture, but none was detected. The mechanism of metal transport by members of this gene family are unknown. Presumably other components must be involved because these proteins do not have obvious ATP-binding domains. Thus, one possible explanation for the lack of ZnT-3 function in BHK cells may be that it normally functions as part of a protein complex and critical components of that complex may be missing in BHK cells. BHK cells expressing ZnT-3 were transfected with brain-derived cDNA expression libraries, followed by selection for zinc resistance; however, no cDNAs that could complement ZnT-3 were identified in this screen.
ZnT-3 mRNA, as detected by Northern blot and reverse transcriptase–PCR, is more abundant in adult testis than in brain and in situ hybridization reveals that ZnT-3 mRNA is in developing germ cells. However, neither Western blots nor immunocytochemistry of testis sections revealed any ZnT-3 protein. Northern blots of polysomal preparations from testis indicated that the ZnT-3 transcripts were abundant on ribonucleoprotein particles that sedimented slower than monoribosomes, rather than in the polysome region, suggesting that this mRNA may not be translated efficiently (data not shown). A small amount of translation at a late stage in spermiogenesis might be undetectable by the techniques used.
The cloning of the mouse ZnT-3 gene provides an opportunity to inactivate it by homologous recombination. This could provide definitive evidence regarding the role of this gene product in zinc sequestration. If inactivation of ZnT-3 prevents the accumulation of zinc in synaptic vesicles, as we predict, it will provide a unique opportunity to explore the function of this vesicular pool of zinc in neuronal function, learning, and memory, as well as its contribution to various disease processes.
Acknowledgments
We thank Glenda Froelick for preparing the histological sections, Mark Fajardo for sharing the testis polysome Northern blots, and our colleagues for valuable discussions during the course of this work and preparation of the manuscript.
Footnotes
Abbreviations: TSQ, 6-methoxy-8-p-toluene sulfonamide quinoline; BHK, baby hamster kidney.
Data deposition: The sequences reported in this paper have been deposited in the GenBank data base (accession nos. U76007U76007 for mouse ZnT3 cDNA, U76008U76008 and U76009U76009 for mouse ZnT3 genomic, and U76010U76010 for human ZnT3 cDNA sequences).
References
- 1.Palmiter R D, Findley S D. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallee B L, Falchuk K H. Physiol Rev. 1993;73:79–117. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 3.Palmiter, R. D. (1987) Experientia 52, Suppl., 63–80. [DOI] [PubMed]
- 4.Zhao H, Eide D. Proc Natl Acad Sci USA. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Eide D. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 6.Timm F. Dtsch Z Ges Gericht Med. 1958;46:706–711. [PubMed] [Google Scholar]
- 7.Frederickson C J, Moncrieff D D. Biol Signals. 1994;3:127–139. doi: 10.1159/000109536. [DOI] [PubMed] [Google Scholar]
- 8.Zalewski P D, Millard S H, Forbes I J, Kapaniris O, Slavotinek A, Betts W H, Ward D, Lincoln S F, Mahadevan I. J Histochem Cytochem. 1994;42:877–884. doi: 10.1177/42.7.8014471. [DOI] [PubMed] [Google Scholar]
- 9.Danscher G. Histochemistry. 1981;71:1–16. doi: 10.1007/BF00592566. [DOI] [PubMed] [Google Scholar]
- 10.Frederickson C J. Int Rev Neurobiol. 1989;31:145–238. doi: 10.1016/s0074-7742(08)60279-2. [DOI] [PubMed] [Google Scholar]
- 11.Frederickson C J, Danscher G. Prog Brain Res. 1990;83:71–84. doi: 10.1016/s0079-6123(08)61242-x. [DOI] [PubMed] [Google Scholar]
- 12.Slomianka L. Neuroscience. 1992;48:325–352. doi: 10.1016/0306-4522(92)90494-m. [DOI] [PubMed] [Google Scholar]
- 13.Palmiter R D, Cole T B, Findley S D. EMBO J. 1996;15:1784–1791. [PMC free article] [PubMed] [Google Scholar]
- 14.Assaf S, Chung S. Nature (London) 1984;308:734–738. doi: 10.1038/308734a0. [DOI] [PubMed] [Google Scholar]
- 15.Howell G A, Welch M G, Frederickson C J. Nature (London) 1984;308:736–738. doi: 10.1038/308736a0. [DOI] [PubMed] [Google Scholar]
- 16.Westbrook G L, Mayer M L. Nature (London) 1987;328:640–643. doi: 10.1038/328640a0. [DOI] [PubMed] [Google Scholar]
- 17.Aniksztejn L, Charton G, Ben-Ari Y. Brain Res. 1987;404:58–64. doi: 10.1016/0006-8993(87)91355-2. [DOI] [PubMed] [Google Scholar]
- 18.Mayer M L, Vyklicky L, Jr, Westbrook G L. J Physiol (London) 1989;415:329–350. doi: 10.1113/jphysiol.1989.sp017724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christine C W, Choi D W. J Neurosci. 1990;10:108–116. doi: 10.1523/JNEUROSCI.10-01-00108.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rassendren F A, Lory P, Pin J P, Nargeot J. Neuron. 1990;10:943–954. doi: 10.1016/0896-6273(90)90199-p. [DOI] [PubMed] [Google Scholar]
- 21.Xie X, Smart T G. Nature (London) 1991;349:521–524. doi: 10.1038/349521a0. [DOI] [PubMed] [Google Scholar]
- 22.Smart T G. J Physiol (London) 1992;447:587–625. doi: 10.1113/jphysiol.1992.sp019020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollman M, Boulter J, Mason C, Beasly L, Sullivan J, Pechet G, Heinemann S. Neuron. 1993;4:733–739. doi: 10.1016/0896-6273(93)90209-a. [DOI] [PubMed] [Google Scholar]
- 24.Smart T G, Xie X, Krishek B J. Prog Neurobiol. 1994;42:393–441. doi: 10.1016/0301-0082(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 25.Harrison N L, Gibbons S J. Neuropharmacology. 1994;33:935–952. doi: 10.1016/0028-3908(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 26.Sloviter R S. Brain Res Bull. 1982;8:771–774. doi: 10.1016/0361-9230(82)90104-6. [DOI] [PubMed] [Google Scholar]
- 27.Masters B A, Quaife C J, Erickson J C, Kelly E J, Froelick G J, Zambrowicz B P, Brinster R L, Palmiter R D. J Neurosci. 1994;14:5844–5857. doi: 10.1523/JNEUROSCI.14-10-05844.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy N S, Roth W W, Bragg P W, Wahba A J. Gene. 1988;70:231–243. doi: 10.1016/0378-1119(88)90195-3. [DOI] [PubMed] [Google Scholar]
- 29.Marks D L, Wienman J N, Burton K A, Lent K L, Clifton D K, Steiner R A. Mol Cell Neurosci. 1992;3:395–401. doi: 10.1016/1044-7431(92)90051-3. [DOI] [PubMed] [Google Scholar]
- 30.Rowe L B, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S H, Thurston S J, Birkenmeier E H. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]


