Abstract
Substrates of the ubiquitin-dependent N-end rule pathway include proteins with destabilizing N-terminal residues. UBR1−/− mice, which lacked the pathway's ubiquitin ligase E3α, were viable and retained the N-end rule pathway. The present work describes the identification and analysis of mouse UBR2, a homolog of UBR1. We demonstrate that the substrate-binding properties of UBR2 are highly similar to those of UBR1, identifying UBR2 as the second E3 of the mammalian N-end rule pathway. UBR2−/− mouse strains were constructed, and their viability was found to be dependent on both gender and genetic background. In the strain 129 (inbred) background, the UBR2−/− genotype was lethal to most embryos of either gender. In the 129/B6 (mixed) background, most UBR2−/− females died as embryos, whereas UBR2−/− males were viable but infertile, owing to the postnatal degeneration of the testes. The gross architecture of UBR2−/− testes was normal and spermatogonia were intact as well, but UBR2−/− spermatocytes were arrested between leptotene/zygotene and pachytene and died through apoptosis. A conspicuous defect of UBR2−/− spermatocytes was the absence of intact synaptonemal complexes. We conclude that the UBR2 ubiquitin ligase and, hence, the N-end rule pathway are required for male meiosis and spermatogenesis and for an essential aspect of female embryonic development.
Proteolysis by the ubiquitin (Ub)-proteasome system controls the concentrations of many regulatory proteins (31, 36, 52). A substrate of the Ub system is conjugated to Ub through the action of three enzymes, E1, E2, and E3. The selectivity of ubiquitylation is determined by E3, which recognizes a substrate's degradation signal, or degron (18, 34). The term Ub ligase designates either an E2-E3 complex or its E3 component. One Ub-dependent proteolytic pathway, called the N-end rule pathway, targets proteins bearing destabilizing N-terminal residues (Fig. 1A) (24, 53). The corresponding degron, called the N-degron, consists of two determinants: a destabilizing N-terminal residue and an internal Lys residue of a substrate (48). The Lys residue is the site of formation of a substrate-linked poly-Ub chain. A ubiquitylated substrate is degraded by the 26S proteasome (18, 38). The N-end rule pathway is present in all organisms examined, from mammals and plants to fungi and prokaryotes (23, 24, 26, 53).
FIG.1.
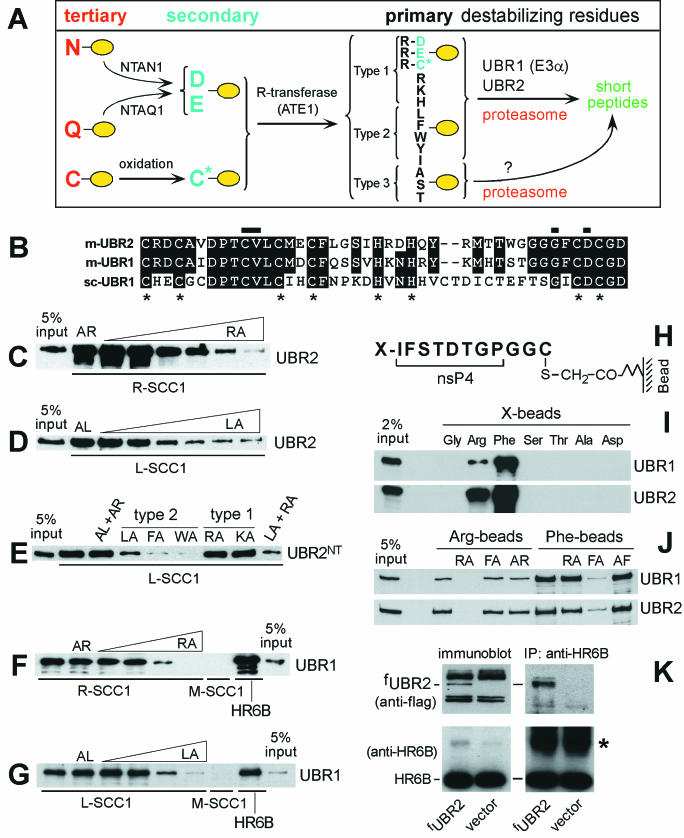
Mouse UBR2 is the ubiquitin ligase of the N-end rule pathway. (A) N-end rule pathway in mammals. N-terminal residues are indicated by single-letter abbreviations for amino acids. The yellow ovals designate the rest of a protein substrate. (B) Alignment of the sequences of the UBHC (Ub, His, and Cys) domain (12, 25) of m-UBR2 (residues 112 to 153), m-UBR1, and sc-UBR1. The asterisks indicate conserved Cys and His. Horizontal bars indicate residues important for the binding of type 1 destabilizing N-terminal residues by sc-UBR1 (see Results). (C to G) GST pull-down assays. Equal amounts of an extract from S. cerevisiae containing either m-fUBR1 (E3α), m-fUBR2, or its 1,041-residue N-terminal fragment (m-fUBR21-1041) were incubated with glutathione-Sepharose beads preloaded with X-SCC1-GST (X = Arg [R], Leu [L], or Met [M]) or with GST-HR6B. The incubations were performed in the presence or absence of the indicated dipeptides: RA, Arg-Leu; AR, Ala-Arg; LA, Leu-Ala; AL, Ala-Leu; FA, Phe-Ala; WA, Trp-Ala; KA, Lys-Ala. The bound proteins were eluted, fractionated by SDS-PAGE, and immunoblotted with anti-Flag antibody. The 5% input lanes refer to a directly loaded sample of yeast extract that corresponded to 5% of the amount of extract used in GST assays. The concentrations of a competitor peptide were 1 μM, 10 μM, 0.1 mM, 0.5 mM, 1 mM, and 2 mM for the results shown in panels C and D, and 1 mM in for the results shown in panels E, F, and G. (H to K) Pull-down assay with peptide-linked microbeads. (H) Bead-linked 12-mer peptides. X = Arg, Phe, Gly, Ser, Thr, Ala, or Asp. (I) m-UBR1 and m-UBR2 bind to Arg (type 1) and Phe (type 2) destabilizing N-terminal residues but not to the other tested N-terminal residues. (J) Binding competition assays with m-UBR1 and m-UBR2, the 12-mer peptides bearing N-terminal Arg or Phe, and competitor dipeptides. (K) m-UBR2 binds to the HR6B E2 enzyme. Extracts (0.1 mg of protein) from control NIH 3T3 cells and NIH 3T3 cells stably expressing m-fUBR2 from the PCMV promoter were fractionated by SDS-12% PAGE (left panel) followed by immunoblotting with anti-Flag (top panel) or anti-HR6B antibody (bottom panel). Extracts from the same cell lines (1 mg of protein) were immunoprecipitated with anti-HR6B (right panel) followed by SDS-12% PAGE and immunoblotting with anti-Flag (top panel) or anti-HR6B antibody (bottom panel). The asterisk designates the band of light chain immunoglobulin G.
In the yeast Saccharomyces cerevisiae, two substrate-binding sites of the 225 kDa, RING domain Ub ligase UBR1 (sc-UBR1) recognize primary destabilizing N-terminal residues of two types, basic (type 1: Arg, Lys, and His) and bulky hydrophobic (type 2: Phe, Leu, Tyr, Trp, and Ile) (Fig. 1A) (12). Several other N-terminal residues function as tertiary (Asn and Gln) and secondary (Asp and Glu) destabilizing residues in that they are recognized by sc-UBR1 after their enzymatic conjugation to Arg, a primary destabilizing residue (53). In the case of N-terminal Asn and Gln (as well as Cys in metazoans), the conjugation of Arg is preceded by other enzymatic modifications (Fig. 1A) (23, 24). sc-UBR1 also contains a third substrate-binding site that targets proteins through their internal (non-N-terminal) degrons (12).
The established functions of the N-end rule pathway include the regulation of peptide import in yeast, through degradation of the repressor, CUP9, which controls the expression of a peptide transporter (50); the maintenance of chromosome stability, through the degradation of a fragment of SCC1, a subunit of cohesin, at the metaphase-anaphase transition (37); an essential role in mammalian cardiovascular development (24); and the regulation of apoptosis in Drosophila melanogaster, through the degradation of DIAP1, an inhibitor of apoptosis (11, 52).
Several other proteins have also been shown to be degraded by the N-end rule pathway, including the GTPase-activating proteins RGS4 and RGS16 (8), the RNA polymerases of alphaviruses (9), the integrase of the human immunodeficiency virus (30), p60, a protein secreted by the bacterium Listeria monocytogenes into the host cell's cytosol (46), 3C protease of encephalomyocarditis virus (27), and a subset of γ2 subunits of mammalian G proteins (15). The functions of the N-end rule pathway in controlling the levels of these proteins remain to be understood. In addition, the results of studies in which dipeptides with destabilizing N-terminal residues were used to perturb the N-end rule pathway suggested its involvement in cell differentiation (32), turnover of muscle proteins (47), and limb regeneration in newts (49).
In mammals, the tertiary destabilizing residues Asn and Gln are deamidated by two distinct N-terminal amidases, yielding the secondary destabilizing residues Asp and Glu (Fig. 1A). NTAN1−/− mice, which lacked the Asn-specific N-terminal amidase but retained the arginylation step of the N-end rule pathway, exhibited abnormal behaviors but appeared to be wild type in other respects (23). In contrast, ATE1−/− mice, which lacked the ATE1-encoded R-transferases and thus lacked the pathway's deamidation-arginylation branch (Fig. 1A), died as embryos, with defects in angiogenic remodeling and heart development (24). N-terminal Cys, a stabilizing residue in fungi, is a tertiary destabilizing residue in metazoans, in that it is arginylated in mammalian cells after its (apparently) enzymatic oxidation (Fig. 1A) (24).
The Ub ligase of the mammalian N-end rule pathway, termed E3α, was characterized biochemically in rabbit reticulocyte extracts (18). Its cloned mouse gene, termed mouse UBR1 (m-UBR1), encoded a 200-kDa protein that was similar to sc-UBR1 not only in sequence but also in the presence and arrangement of specific domains and in the substrate-binding properties as well (25). UBR1−/− mice were constructed and were found to be viable but different from their +/+ littermates in several ways that remain to be understood mechanistically (26). Although UBR1−/− mice lacked the m-UBR1 Ub ligase, the N-end rule pathway was still active at least in embryonic fibroblasts (EFs) from these mice, indicating the presence of a complementing Ub ligase(s) (26).
The present work describes the cloning and characterization of m-UBR2, a homolog of m-UBR1, demonstrates high similarity of their substrate-binding properties, and describes the gender-specific phenotypes of UBR2−/− strains. In addition to showing that UBR2 and, hence, the N-end rule pathway are required for an essential aspect of the female's embryonic development, we also demonstrate that the N-end rule pathway is required for male meiosis and spermatogenesis.
MATERIALS AND METHODS
Cloning and chromosomal mapping of UBR2 cDNA.
To amplify m-UBR2 cDNA fragments, preparations of poly(A)+ RNA from mouse EFs were subjected to reverse transcription (RT)-PCR with the forward and reverse primers 5′-TAATGTGAAATGCAGACGTGAGATG and 5′-GATCCATGCCATTCTCTTCTGTACATG, specific, respectively, for the human and mouse expressed sequence tag (EST) clones (accession no. T62713 and W78536), the only UBR2 ESTs available at the time. The same approach was used to amplify human UBR2 (h-UBR2) cDNA fragments by RT-PCR with RNA from human 293 cells (data not shown). The resulting 2.4-kb h- and m-UBR2 cDNA fragments encoded amino acid sequences similar to that of m-UBR1 (E3α) (25). The 2.4-kb m-UBR2 cDNA fragment was used as a probe to screen the λgt10 mouse cDNA library from MEL-C19 cells (Clontech, Palo Alto, Calif.), and this screening was followed by a second screening with a probe specific for the 5′ end of a cDNA fragment from the first screening. To isolate the 5′ end of the full-length m-UBR2 cDNA, 5′ rapid amplification of cDNA ends PCR (2) was carried out with poly(A)+ RNA from mouse L cells and a primer derived from a cDNA fragment produced from the second screening described above. h-UBR2 is on chromosome 6p11-21, whereas m-UBR2 is in the middle of mouse chromosome 17, as determined by radiation hybrid mapping and fluorescence in situ hybridization. In contrast, h- and m-UBR1 are located, respectively, on the human and mouse chromosomes 15 and 2 (25, 26). Lymphocytes isolated from mouse spleen were cultured, synchronized, grown to subconfluence, harvested, and deposited on slides for in situ hybridization (17). Mouse bacterial artificial chromosome (BAC) DNA containing UBR2 was biotinylated (BioNick kit; GIBCO, Frederick, Md.) and followed by fluorescence in situ hybridization, 4′-6-diamino-2-phenylindole (DAPI) staining, and image analysis as described previously (17). m-UBR2 was also mapped by using a mouse-hamster radiation hybrid panel (Research Genetics, Huntsville, Ala.). The primers derived from the mouse UBR2 cDNA, 5′-AGTGACACCTACTACTGCATGCTG and 5′-GTAGACTTGGTTCAATAGCATTGGC, yielded 730- and 850-bp PCR fragments with mouse and hamster DNAs, respectively. The data were analyzed by using Jackson Lab's server. The intracellular localization of m-UBR2 was assayed by using a fusion of m-UBR2 and green fluorescent protein; the bulk of UBR2-green fluorescent protein was present in the nucleus (A. Kashina, Y. T. Kwon, and A. Varshavsky, unpublished data). Northern analyses, with RNA from whole mouse embryos of different ages, indicated that the overall levels of m-UBR1 and m-UBR2 mRNAs were approximately constant during embryogenesis (data not shown).
Construction and analyses of UBR2−/− mouse strains.
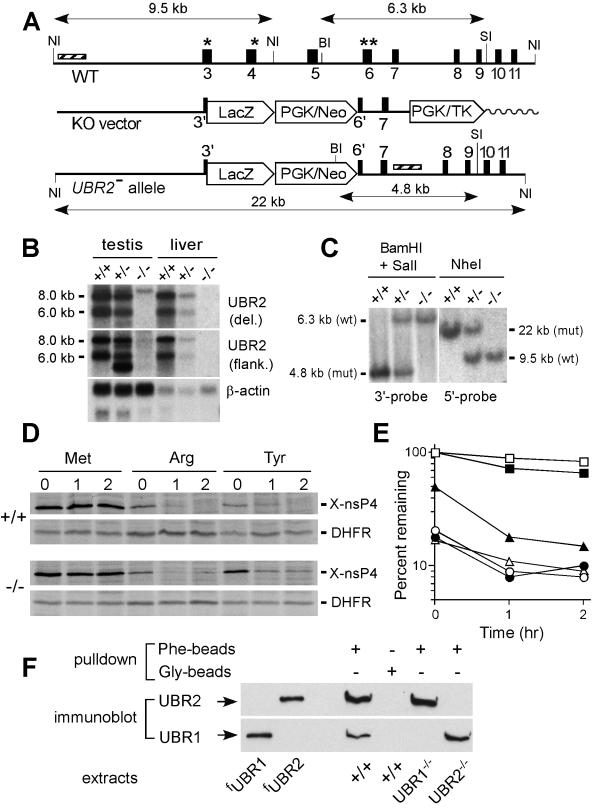
m-UBR2 was isolated by screening, with a fragment of the UBR2 cDNA (nucleotides [nt] 732 to 1674), a BAC DNA library (Genome Systems) from 129SvImJ embryonic stem (ES) mouse cells. The exon-intron organization of the first ∼30 kb of UBR2 was determined by using exon-specific PCR primers to produce genomic DNA fragments flanked by exons, as described previously (26). Details of the targeting vector construction (Fig. 2A) are available upon request. The vector was linearized with NsiI and electroporated into CJ7 ES cells, and this was followed by standard procedures (23, 24, 26) to select for and identify the correctly targeted UBR2+/− ES cell clones, which were further selected for clones with the apparently normal karyotype. Homologous recombination in ES cells resulted in the replacement of genomic DNA from codon 7 (Cys) of exon 3 to codon 16 (His) of exon 6 with the nuclear localization signal (NLS)-lacZ marker gene through in-frame fusion into codon 7 of exon 3. No β-galactosidase activity derived from the UBR2− allele could be detected histologically during embryogenesis or in adult testes, probably because of abnormal splicing of RNA transcribed from the UBR2−/− allele. Standard techniques were used to produce males chimeric for UBR2+/− cells and to derive from them, through backcrosses and intercrosses, the UBR2−/− mouse strains. In experiments that involved embryos of a specific age, successful matings were identified by the presence of a vaginal plug on the morning after mating, which was designated embryonic day 0.5 (E0.5). PCR-mediated genotyping of embryos and pups, RT-PCR, and Southern and Northern hybridizations were carried out as described previously (26). The phenotypes described in this paper were observed with UBR2−/− mouse strains produced from at least two independently derived UBR2+/− ES clones. Northern analysis with an m-UBR2 cDNA probe that contained exclusively the region deleted in the UBR2− allele did not detect UBR2-specific transcripts in the testes and livers of UBR2−/− mice (Fig. 2B, top panel). Low levels of UBR2-specific transcripts in UBR2−/− tissues could be detected with an m-UBR2 probe that encompassed both the deleted region and the 3′-flanking (undeleted) region of m-UBR2 cDNA (Fig. 2B, middle panel).
FIG. 2.
Construction of UBR2−/− mouse strains. (A) Targeting vector. A map of the ∼30-kb 5′-proximal region of the ∼96-kb m-UBR2 gene, the targeting vector, and the deletion disruption UBR2−/− allele. Exons are designated by vertical rectangles. Solid and wavy horizontal lines designate, respectively, the mouse and plasmid DNAs. The directions of gene transcription are indicated as well. Exons 3 and 4, marked by single asterisks, and exon 6, marked by a double asterisk, contain conserved residues that are essential for integrity of the substrate-binding sites of sc-UBR1. NI, NheI; BI, BamHI; SI, SalI. Southern hybridization probes are indicated by striped rectangles. PGK, phosphatidylglycerol kinase; TK,thymidine kinase. (B) Northern analysis of m-UBR2 expression with total RNA from the testes and livers of adult +/+, UBR+/−, and UBR2−/− mice. The probes used were either the 350-bp UBR2 cDNA fragment (nt 383 to 731) that was deleted in the UBR2−/− allele (data not shown) or the 2.3-kb UBR2 cDNA fragment (nt 863 to 3227) adjacent to the deleted region (upper panel) or the human β-actin cDNA fragment (lower panel). The wild-type 8.0- and 6.0-kb UBR2 mRNAs are indicated on the left. (C) Southern analysis of BamHI/SalI-cut (3′ probe) and NheI-cut (5′ probe) mouse tail DNA. The 760-bp BamHI/SalI 3′ probe detected 6.3- and 4.8-kb UBR2 fragments for the wild-type (wt) and mutant (mut) UBR2 alleles, respectively. The 1.2-kb NheI 5′ probe detected 9.5-kb (wild type) and 22-kb (mutant) fragments. (D) Pulse-chase analysis with +/+ and UBR2−/− EF cell lines. EFs were transfected with plasmids expressing fDHFRh-UbR48-X-nsP4f, which yielded the fDHFRh-UbR48 reference protein (designated DHFR), and an X-nsP4f (X-nsP4-Flag) test protein (X = Met, Arg, or Tyr) (designated X-nsP4). Cells were labeled for 10 min with [35S]methionine and chased for 1 and 2 h. (E) Quantitation of the patterns in shown in panel D with PhosphorImager. For each time point, the ratio of 35S in X-nsP4f to 35S in the Met-DHFR-UbR48 reference protein at the same time point was plotted as the percentage of that ratio relative to that for Met-nsP4f (which bore a stabilizing N-terminal residue) at time zero (the beginning of chase). Open and closed symbols designate the results with +/+ and UBR2−/− EFs, respectively. Squares, Met-nsP4f; circles, Arg-nsP4f; triangles, Tyr-nsP4f. (F) Extracts from +/+, UBR1−/−, and UBR2−/− EFs were assayed (see Results) by using peptide pull-down assays and immunoblotting with anti-UBR1 and anti-UBR2 antibodies. fUBR1 and fUBR2 designate S. cerevisiae extracts expressing Flag-tagged m-UBR1 and m-UBR2.
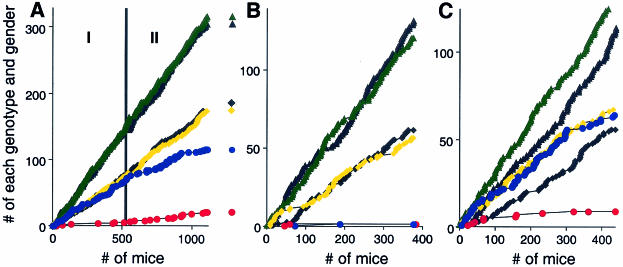
Genotyping of stage I 129SvImJ/C57BL/6J (designated 129/B6) offspring mice (n = 510) yielded 74 +/+ males, 76 +/+ females, 142 UBR2+/− males, 144 UBR2+/− females, and 67 UBR2−/− males but only 6 UBR2−/− females (Table 1 and Fig. 3). No disproportionate increase in the number of UBR2−/− females was observed among ∼15 1-day-old pups that were found dead in the course of stage I 129/B6 UBR2+/− intercrosses, indicating that most UBR2−/− females died in utero. When the embryos from UBR2+/− intercrosses were isolated, characterized, and genotyped at various times of gestation, 6 of 13 examined female UBR2−/− embryos appeared to be arrested and otherwise deformed at E7.5 to E10.5 (examples are shown in Fig. 4C, panels b, d, f) while the other 7 UBR2−/− embryos were at most growth retarded (data not shown), suggesting that female UBR2−/− embryos do not have a specific terminal phenotype (Fig. 4C and data not shown). The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining of sectioned female UBR2−/− embryos showed increased apoptosis in severely arrested E9.5 and E11.5 embryos but little or no excessive apoptosis in those female UBR2−/− embryos whose development was perturbed less strongly. Histological examination of female UBR2−/− embryos of the latter class revealed no obvious abnormalities in the major organs, including hearts, lungs, livers, brains, blood vessels, and kidneys. The rare surviving stage I UBR2−/− females of the 129/B6 background (Table 1) exhibited, by 2 months of age, an ∼20% growth retardation. These rare UBR2−/− females were fertile with both +/+ and UBR2+/− 129/B6 males but less so than +/+ females: the average litter sizes of UBR2−/− females (n = 6) mated with UBR2+/− (n = 3) and +/+ males (n = 3) were 3.4 ± 1.9 and 5.3 ± 2.1, respectively, in contrast to the corresponding litter sizes of 7.4 ± 2.3 and 8.2 ± 1.7 for +/+ 129/B6 females. Genotyping of stage II 129/B6 offspring mice (n = 596) yielded 99 +/+ males, 98 +/+ females, 174 UBR2+/− males, 161 UBR2+/− females, 49 UBR2−/− males, and 15 UBR2−/− females (compared with 6 females in stage I, n = 510) (Fig. 3A and Table 1).
TABLE 1.
Genotypes and genders of 3-week-old offspring of UBR2+/− intercrosses in two genetic backgrounds, inbred (129) and mixed (129/B6)a
| Genotype | Sex | No. of mice of background:
|
|||
|---|---|---|---|---|---|
| 129SvJ/ (C57BL/6J) (mixed) in phase:
|
129SvJ/SvImJ (inbred) | 129SvJ/CD1 (mixed) | |||
| I | II | ||||
| +/+ | Male | 74 | 99 | 19 | 67 |
| Female | 76 | 98 | 21 | 56 | |
| +/− | Male | 142 | 174 | 43 | 130 |
| Female | 144 | 161 | 39 | 115 | |
| −/− | Male | 67 | 49 | 1 | 64 |
| Female | 6 | 15 | 1 | 9 | |
| Total | 510 | 596 | 124 | 441 | |
The significantly underrepresented progeny types are shown in boldface type.
FIG. 3.
Viability and other phenotypes of UBR2−/− mice depend on gender and genetic background. (A) Tabulation of progeny from matings between UBR2+/− mice of the 129SvImJ/(C57BL/6J) hybrid background, designated 129/B6 below. Green and black triangles, UBR2+/− males and females, respectively; yellow and black squares, +/+ males and females, respectively; blue and red circles, UBR2−/− males and females, respectively. The numbers on the abscissa refer to 1,106 offspring of UBR2+/− intercrosses. These numbers are temporally ordered, in that the tabulation of mice produced from earlier UBR2+/− intercrosses precede, on the abscissa, the tabulation of mice produced from later intercrosses. The accumulated numbers of intercross progeny of each genotype (+/+, UBR2+/−, or UBR2−/−) of a given gender are plotted on the ordinate. To produce 129/B6 UBR2−/− mice, the founder chimeras were mated with +/+ C57BL/6 females, yielding UBR2+/− 129/B6 mice followed by intercrosses. The aging breeding pairs of initial UBR2+/− mice were replaced, in the course of these experiments, with F2 generation UBR2+/− 129SvJ/(C57BL/6J) (129/B6) mice. For the actual numbers of progeny with specific genotypes, see Table 1. Phase I and phase II are described in Results. (B) Same as in panel A, except that the offspring were from UBR2+/− intercrosses in the 129SvImJ (inbred) background. (C) Same as in panel A, except that the offspring were from UBR2+/− intercrosses in the 129SvJ/CD1 background.
FIG. 4.
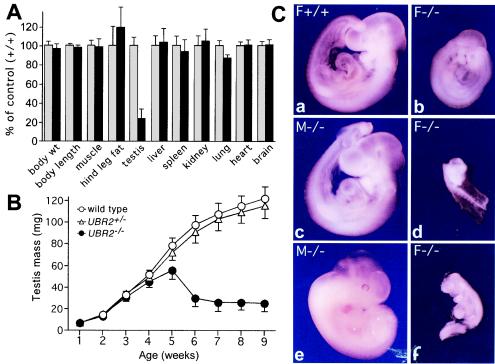
Testis degeneration and female lethality in UBR2−/− mice. (A) Weights of organs and tissues in UBR2−/− male mice [129SvJ/(C57BL/6J)] expressed as percentages of the corresponding weights for age-matched +/+ mice. The comparison involved 24 pairs of 2-month-old male mice (13 +/+, 11 UBR2+/−, and 24 UBR2−/−) produced through UBR2+/− intercrosses. Muscle, hind leg muscle; hind leg fat, fat pad of the hind leg. (B) Testis weights of +/+, UBR2+/−, and UBR2−/− mice [129SvJ/(C57BL/6J)] as a function of postnatal age. The total body weights of UBR2−/− mice were approximately equal to those of their +/+ littermates. (C) Whole-mount views of unstained +/+ and UBR2−/− embryos of the 129SvJ/(C57BL/6J) background. (a and b) E9.5 +/+ female embryo (a) and its UBR2−/− female littermate (b). (c and d) E9.5 UBR2−/− male embryo (c) and its UBR2−/− female littermate (d). (e and f) E11.5 UBR2−/− male embryo (e) and its UBR2−/− female littermate (f).
We observed a significantly decreased fertility of heterozygous UBR2+/− males: the average litter sizes of 129/B6 UBR2+/− males mated with UBR2+/− and +/+ females were 5.6 ± 1.3 and 7.4 ± 2.3, respectively. The time until the first plug formation in matings between UBR2+/− mice was 21.2 ± 7.3 days, whereas this time was reduced to 5.3 ± 3.7 days for matings between UBR2+/− males and +/+ females. Furthermore, 3 UBR2+/− testes, out of 12 examined, weighed ∼30% less than +/+ testes. The epididymal sperm count in UBR2+/− testes (n = 11) was, on average, 30% lower than in the testes of +/+ littermates.
Given the results with UBR2+/− intercrosses in the mixed (129/B6) background, we also produced UBR2+/− mice in the inbred (129SvImJ) background and carried out intercrosses. Genotyping of 376 offspring (at ∼4 weeks of age) from 129SvImJ UBR2+/− intercrosses yielded 57 +/+ males, 62 +/+ females, 121 UBR2+/− males, and 132 UBR2+/− females but only 2 UBR2−/− females and also, strikingly, only 2 UBR2−/− males (Fig. 3B and Table 1). Thus, the UBR2−/− genotype was nearly completely lethal to both genders in the inbred (129SvImJ) background, in contrast to the 129/B6 background, where the male UBR2−/− mice were born at nearly the Mendelian frequency. The 2 viable strain 129 UBR2−/− males were sterile and exhibited severe testis degeneration. Strikingly, the 2 surviving strain 129 UBR2−/− females, while growth retarded, were apparently normal otherwise and, in addition, fertile, similar to the above-described rare viable UBR2−/− females of the 129/B6 background. We also produced UBR2+/− mice in a different mixed background, 129SvImJ/CD1 (designated 129/CD1 below), and carried out intercrosses. Genotyping of 441 offspring from UBR2+/− 129/CD1 intercrosses yielded 67 +/+ males, 56 +/+ females, 130 UBR2+/− males, 115 UBR2+/− females, 64 UBR2−/− males, and 9 UBR2−/− females (Fig. 3C and Table 1). Thus, the mixed 129/CD1 background was the most permissive, among the three backgrounds tested, for viability of UBR2−/− females, but the lethality of the UBR2−/− female genotype was still high even in this background. UBR2−/− males of the 129/CD1 background exhibited less severe testis degeneration than stage I UBR2−/− males of the 129/B6 background (note that most UBR2−/− males of the inbred, 129-only, background died in utero) (Fig. 3B).
At 8 weeks, the testes of stage I UBR2−/− males of the 129/B6 background weighed uniformly ∼4-fold less than the testes of their +/+ littermates (Fig. 4A and B). In contrast, the testes of stage II UBR2−/− males of the same background (Fig. 3A and description above) varied in mass from 30 to 70% of that of the +/+ controls and exhibited a correspondingly less-severe testis degeneration. Of the 8 tested stage II 129/B6 UBR2−/− males, 4 of them contained low but detectable amounts of spermatozoa in their epididymides, on average 13% of the amount in +/+ littermates, in contrast to the virtual absence of spermatozoa from stage I UBR2−/− males (data not shown). However, closer examination showed that ∼90% of spermatozoa in stage II UBR2−/− epididymides were overtly abnormal, with aberrant head morphology, reduced motility, and often absent head (see Fig. 6G), consistent with the infertility of both stage I and II UBR2−/− males.
FIG. 6.
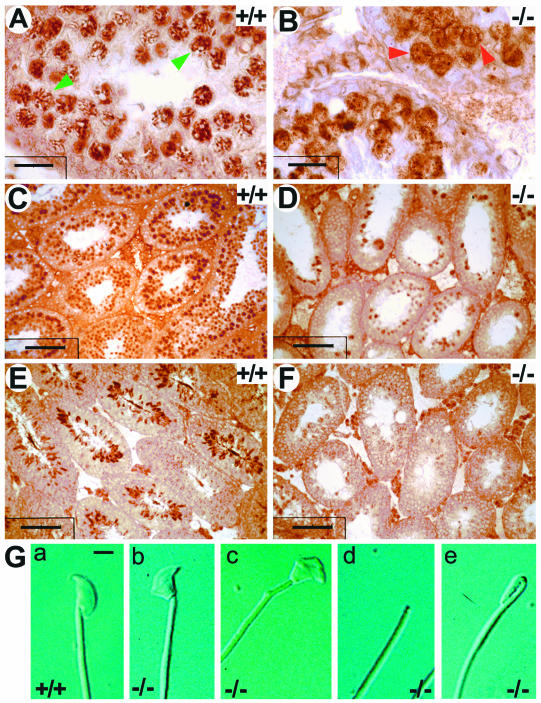
Perturbed spermiogenesis in UBR2−/− mice. (A and B) Testis sections of 2-week-old +/+ (A) and UBR2−/− (B) littermates stained with anti-SCP3 antibody. Green arrowheads in panel A, typical pachytene spermatocytes; red arrowheads in panel B, arrested UBR2−/− spermatocytes. (C to F) Testis sections from 4-week-old +/+ (C and E) and UBR2−/− (D and F) littermates were stained with anti-tH2B (C and D) and anti-TP2 (E and F) antibodies. Scale bars, 20 μm (A and B), 7 μm (C), and 60 μm (D to F). (G) Appearance of +/+ (a) and UBR2−/− (b to e) spermatozoa.
Genotyping of embryos and mice.
A three-primer PCR assay specific for UBR2 was carried out as previously described (26) with the primers 5′-CTACTGCATGCTGTTTAATGATGAG (for UBR2, forward), 5′-CCAGCTCATTCCTCCCACTCATGATC (for neo, forward), and 5′-GGAGGTAGAAACATGCAAATCTCTG (for UBR2, reverse). The sex of an embryo was determined with the primers 5′-AGAGCCACAAGCTAACCATTAAGAC (forward) and 5′-TCAGGAGACAGATGCCACACTTCAG (reverse), which were specific for ZFY1, located on the Y chromosome, and the primers 5′-CTGCCTGCACCATTCAAATTGGCAAG (forward) and 5′-AGGCGGTGGCTCGAGTTGTTTGCAG (reverse), specific for ZFX1, located on the X chromosome.
Cell lines from EFs and pulse-chase analysis.
Cultures of primary EFs were established from E13.5 UBR2−/− and littermate +/+ embryos produced through intercrosses of 129/B6 UBR2+/− mice (23, 24, 26). Primary EFs were immortalized through repeated splitting of the culture over ∼3 months. For pulse-chase analysis, the UBR2−/− and +/+ EF cell lines were transiently transfected, by using Lipofectamine-Plus (GIBCO), with plasmids pcDNA3flagDHFRhaUbXnsP4flag (X = Met, Arg, Tyr, Asp, Glu, or Cys), which expressed the Ub fusion proteins fDHFR (dihydrofolate reductase)h-UbR48-X-nsP4f from the PCMV promoter (26). The superscripts f and h designate, respectively, the hemagglutinin and Flag epitopes (2). The nsP4 moiety was the 69-kDa Sindbis virus RNA polymerase (9). An fDHFRh-UbR48-X-nsP4f fusion is cotranslationally cleaved in vivo at the Ub-protein junction, yielding the reference protein fDHFRh-UbR48 (DHFR-Ub) and a test protein X-nsP4f (X = Met, Arg, Tyr, Asp, Glu, or Cys) (23, 26, 54). EFs ∼24 h after transfection were labeled with [35S]methionine/cysteine (35S-EXPRESS; New England Nuclear, Boston, Mass.) for 10 min at 37°C, followed by a chase for 0, 1, and 2 h in the presence of cycloheximide, preparation of extracts, immunoprecipitation with monoclonal anti-Flag antibody (Sigma, St. Louis, Mo.), sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE), autoradiography, and quantitation by PhosphorImager.
Cell lines expressing Flag-m-UBR2.
NIH 3T3 cells were transfected with either the plasmid pcDNA3flagUBR2, expressing Flag-tagged m-UBR2 (m-fUBR2) from the PCMV promoter, or with the pcDNA3 vector (Invitrogen, Carlsbad, Calif.). In the N-terminal sequence of m-UBR2f, MDYKDDDDKGASM, the underlined part was the Flag tag and the last Met was the first residue of the m-UBR2 moiety. Stable transformants were selected with G418 at 0.4 mg/ml for 10 to 14 days, propagated further in the presence of 0.2 mg of G418/ml, and examined for expression of m-fUBR2 by immunoblotting with anti-Flag antibody (Sigma).
In situ hybridization and Northern analyses.
Freshly removed mouse testes were rapidly frozen in liquid N2 and embedded in TissueTek OCT embedding medium (Sakura Finetek, Torrance, Calif.). Ten-micrometer-thick tissue sections, prepared by using a cryostat, were collected onto poly-l-lysine-coated microscope slides. The plasmid pYK118 was constructed by subcloning the 1.4-kb UBR2 cDNA fragment (nt 984 to 2376) into XbaI-XhoI-cut pBluescript KS+. The plasmid pMR24 was constructed similarly with the 1.2-kb XbaI-EcoRI fragment of the mouse UBR1 cDNA. Digoxigenin-labeled, strand-specific RNA probes were transcribed from linearized pYK118 or pMR24, with either T3 RNA polymerase (for the sense strand) or T7 RNA polymerase (for the antisense strand). In situ hybridization was performed as described previously (2), with tissue sections prefixed with 4% HCHO (freshly prepared from paraformaldehyde) in 0.1 M Na phosphate buffer, pH 8.0. For Northern analyses, 10 μg of total RNAs from +/+, UBR2+/−, and UBR2−/− testes of 6-week-old mice were electrophoresed on an agarose gel and probed with 32P-labeled cDNA fragments of several mouse genes (see Fig. 8B to H) prepared by RT-PCR.
FIG. 8.
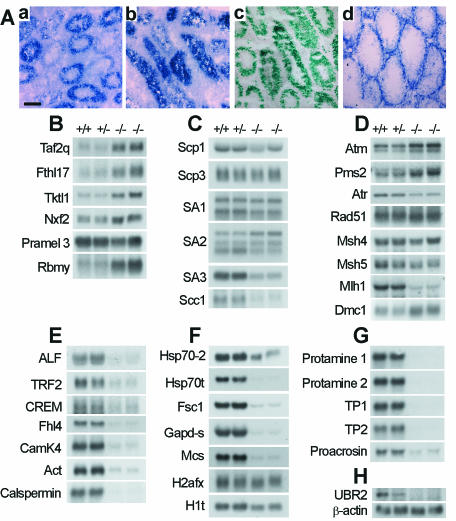
Northern and in situ hybridization analyses of UBR2−/− testes. (A) Distribution of lacZ-containing transcripts, derived from the UBR2−/− allele, in testis. Sections of 3-week-old testes from UBR2+/− (a) and UBR2−/− (b) mice were hybridized with antisense RNA probes specific for lacZ, which marked the deletion and/or disruption of the UBR2− allele. A UBR2-specific probe yielded a weak signal, hence the use of the lacZ-specific probe. No significant hybridization was detected with sections from these testes with a sense RNA probe (data not shown). (c) Testis sections from 3-week-old ATE1+/− mice which were heterozygous for the NLS-lacZ-marked deletion-disruption ATE1− allele (24) probed for lacZ expression as in panel a. (d) Distribution of UBR1 RNA in a 6-week-old UBR2+/− testis. Scale bar, 60 μm. (B to H) Northern analysis of RNA from +/+, UBR2+/−, and UBR2−/− testes of 6-week-old mice, with cDNA probes for the indicated genes.
Histology and staining for LacZ.
Either testes or whole embryos were fixed in 10% buffered formalin or Bouin's fixative, treated with 70% ethanol for 2 to 3 days, dehydrated, and embedded in paraffin wax. Isolated testes were punctured to facilitate fixation, followed by preparation of 10-μm-thick sections and their staining with hematoxylin-eosin or periodic acid-Schiff. For LacZ (β-galactosidase) staining (24), the isolated testes were frozen and thereafter embedded in the OCT reagent (Sakura Finetek). The sections were stained for 6 to 12 h at 37°C in 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) buffer (1.3 mg of potassium ferrocyanide/ml, 1 mg of potassium ferricyanide/ml, 0.2% Triton X-100, 1 mM MgCl2, and 1 mg of X-Gal/ml in phosphate-buffered saline [PBS] [pH 7.2]).
Immunohistochemistry and TUNEL assay.
Rabbit polyclonal antibodies to m-UBR2 were raised against the synthetic peptides RGPNPFPPLKEDT [UBR2 residues 51 to 63; antibody UBR2(3-1)], GSIHRDHQYRMTTWG [UBR2 residues 130 to 144; antibody UBR2(4-1)], and PEGFRPDFYPRNPY [UBR2 residues 1285 to 1298; antibody UBR2(5-1)] cross-linked to keyhole limpet hemocyanin. The sequence of the second peptide [antibody UBR2 (4-1)] was in the region of m-UBR2 that was absent from the UBR2− allele. The antibodies were affinity purified by using the Sepharose-linked initial peptides. A rabbit polyclonal antibody to rat TP2 (1) was a gift from W. Baarends (Erasmus University, Rotterdam, The Netherlands). The testis-specific histone H2B (tH2B) protein was detected with a mouse monoclonal immunoglobulin G raised against tyrosine hydroxylase (TH) (Boehringer, Mannheim, Germany), which is known to bind to tH2B because of sequence similarity near the N-termini of TH and tH2B (51). Mouse testis and epididymal sections were mounted on slides coated with 3-aminopropyltrietoxysilane (Sigma) and incubated at 60°C overnight. The sections were dewaxed in xylene, and the endogenous peroxidase was blocked through a 20-min incubation with 3% H2O2 in methanol. For anti-TP2 staining (but not for anti-tH2B staining), an antigen retrieval step was then performed by heating the sections in 0.01 M Na citrate (pH 6.0) in a microwave oven for 10 min. Nonspecific binding was blocked with goat serum (Vector Laboratories, Burlingame, Calif.) diluted 1:10 in 5% (wt/vol) bovine serum albumin (BSA) in PBS (pH 7.4). The sections were incubated at 4°C overnight with the primary antibody (diluted 1:10,000 for anti-TP2 and 1:100 for anti-TH in 5% BSA [wt/vol] in PBS). Immunostaining was carried out by using biotinylated goat anti-rabbit immunoglobulin and diaminobenzidine substrate. The sections were counterstained for 15 s with Mayer's hematoxylin. Control sections were incubated with 5% BSA (wt/vol) in PBS without primary antibody. To detect synaptonemal complexes (SCs), spreads of spermatocyte chromosomes from +/+ and UBR2−/− testes were prepared (19) and processed for immunofluorescence microscopy (29). The two primary antibodies were a rabbit antiserum (diluted 1:1,000) to rat SCP3, recognizing mouse SCP3, a component of SC (a gift from Peter Moens, York University, Toronto, Canada), and a human CREST antiserum (diluted 1:2,000), recognizing centromere proteins (a gift from Christer Höög, Karolinska Institutet, Stockholm, Sweden) (33, 56). Secondary antibodies conjugated with fluorescein isothiocyanate or rhodamine (goat anti-mouse and goat anti-rabbit) were from Jackson Immunoresearch Laboratories. Cells were counterstained with 0.5 μg of DAPI/ml to visualize the nuclei. Images were produced with a confocal microscope (Zeiss). For the nuclear DNA fragmentation assay (TUNEL), the testes were fixed with 4% HCHO in PBS and then embedded in paraffin. Sections (7 μm thick) were mounted on slides, dewaxed, and treated with proteinase K (Sigma). Apoptotic cells were detected by TUNEL labeling with fluorescein-dUTP (cell death detection kit; Boehringer). The cells were counterstained with propidium iodide before fluorescence microscopy.
Coimmunoprecipitation of UBR2 and HR6B.
Mouse NIH 3T3 fUBR2 cells stably expressing the N-terminally Flag-tagged full-length m-UBR2 (m-fUBR2) and NIH 3T3 pCDNA3 cells carrying the integrated empty vector pCDNA3 were harvested at subconfluence and lysed in buffer A (0.2% Triton X-100, 50 mM NaCl, 50 mM Na-HEPES [pH 7.5], 0.2 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, and 5 μg of aprotinin/ml). Cell extracts were diluted fourfold with Triton-free buffer A, and dilution was followed by immunoprecipitation with anti-HR6B antiserum (a gift from H. P. Roest, Erasmus University), SDS-12% PAGE, and immunoblotting with either anti-Flag monoclonal antibody (Sigma) or anti-HR6B antibody. These antibodies were also used to assess, through immunoblotting, the expression of Flag-UBR2 and HR6B in NIH 3T3 fUBR2 cells.
GST pull-down assay.
sc-fUBR1, m-fUBR1, m-fUBR2, and its 1,041-residue N-terminal fragment (m-fUBR21-1041) were expressed in S. cerevisiae SC295 (MAT a GAL4 GAL80 ura3-52 leu2-3,112 reg1-501 gal1 pep4-3) from the PADH1 promoter in a high-copy-number vector, and expression was followed by preparation of yeast extracts by the liquid N2 method (12). Three X-SCC1-glutathione S-transferase (GST) fusion proteins (X = Arg, Leu, or Met) were produced in Escherichia coli BL21(DE3) and modified with the intein-based IMPACT system (New England Biolabs, Beverly, Mass.). The S. cerevisiae SCC1269-566 moiety of these proteins corresponded to the separase-produced 33-kDa C-terminal fragment of SCC1 (37). The presence of desired N-terminal residues in the purified X-SCC1-GST proteins was verified by N-terminal sequencing through Edman degradation. GST-HR6B, a fusion of GST and m-HR6B, was overexpressed in E. coli BL2(DE3) from the plasmid pGEX-HR6B (26). Construction details are available upon request. Purified X-SCC1-GST (X = Arg, Leu, or Met) (∼2 μg) was diluted into 250 μl of the loading buffer (10% glycerol, 1% Triton X-100, 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.4]) and incubated with 15 μl (bed volume) of glutathione-Sepharose (Amersham-Pharmacia, Piscataway, N.J.) for 1 h at 4°C. The beads were washed once with 1 ml of the loading buffer and twice with 1 ml of the binding buffer (0.1% Nonidet P-40, 10% glycerol, 0.2 M KCl, 5 mM EDTA, 1 mM dithiothreitol, 50 mM K-HEPES [pH 7.5]). The total protein concentration in UBR-containing yeast extracts, produced as described in reference 12, was 7 mg/ml. Eighty microliters of the extract (containing either m-fUBR1 or m-fUBR2) was added to 170 μl of the binding buffer containing the protease inhibitor tablet (Roche, Indianapolis, Ind.) and 50 μM bestatin (Sigma), and this was followed by the addition of washed beads containing X-SCC1-GST, in the presence or absence of dipeptides (Sigma), as described previously (12). In the case of yeast extract containing m-fUBR21-1041, 60 μl of the extract was added to 190 μl of the binding buffer, because m-fUBR21-1041 was expressed in yeast to a higher level than full-length UBR proteins. After 1 h of incubation at 4°C, the beads were washed three times with 150 μl of the binding buffer containing dipeptides at the same concentrations. The beads were then suspended in 20 μl of SDS-PAGE loading buffer and heated at 100°C for 5 min, and this was followed by SDS-10% PAGE, electrophoretic transfer of proteins onto a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.), and immunoblotting with anti-Flag antibody.
Pull-down assay with peptide microbeads.
A 12-mer peptide (1.5 mg) (Fig. 1H) was cross-linked, via its C-terminal Cys residue, to 1 ml (packed volume) of UltraLink Iodoacetyl beads (Pierce) according to the manufacturer's protocol. S. cerevisiae extract expressing m-fUBR1 or m-fUBR2 was diluted by the lysis buffer (12) to 4 mg/ml (total protein) and was incubated in either the absence or the presence of added dipeptide for 15 min at 0°C. When present, each of the dipeptides Arg-Ala, Ala-Arg, Phe-Ala, or Ala-Phe (Sigma) was at a concentration of 10 mM in addition to the aminopeptidase inhibitor bestatin (Sigma), which was present at a concentration of 50 μM. An incubated sample (0.3 ml) was transferred to a new tube containing 5 μl (packed volume) of a bead-linked 12-mer peptide, and transfer was followed by gentle mixing, through tube rotation, for 1 h at 4°C. The beads were pelleted by a brief centrifugation and then washed three times, for 2 min each, with the lysis buffer either containing or lacking the same dipeptides, at 10 mM each. The beads were then suspended in 20 μl of SDS-PAGE loading buffer and heated at 95°C for 5 min, and this was followed by a brief centrifugation in a microcentrifuge, SDS-8% PAGE of the supernatant, and detection of m-fUBR1 or m-fUBR2 by immunoblotting with anti-Flag M2 antibody. To detect endogenous m-UBR1 or m-UBR2 in mouse cells, the extracts of +/+, UBR2+/−, and UBR2−/− EFs were incubated with specific peptide-linked microbeads for 12 h (Fig. 2F) and the retained proteins were fractionated by SDS-8% PAGE followed by immunoblotting with anti-UBR1 or anti-UBR2 antibodies.
Nucleotide sequence accession number.
The sequence of mouse UBR2 cDNA was submitted to GenBank and assigned accession number AY280958.
RESULTS
The m-UBR2 cDNA and gene.
Given the retention of the N-end rule pathway in UBR1−/− mice that lacked the previously described Ub ligase of this pathway (26), we identified several mouse ESTs that encoded a putative homolog of m-UBR1. Standard methods (25, 26) were used to clone and characterize the corresponding gene, termed m-UBR2, which encoded a 200-kDa protein 47% identical and 68% similar to m-UBR1. The residues that were previously identified as essential for the integrity of the type 1 and 2 substrate-binding sites of sc-UBR1 (A. Webster, M. Ghislain, and A. Varshavsky, unpublished data) were also present in m-UBR1 and m-UBR2. m- and h-UBR1 and -UBR2 were also similar to sc-UBR1 in containing the previously described UBHC, BRR, RING-H2, and UBLC domains (12, 25, 26). Through characterization of BAC DNA clones containing m-UBR2 (Fig. 2A), and using public databases as well, we determined that m-UBR2 encompasses ∼98 kb and contains 48 exons (see Materials and Methods for details of m-UBR2 mapping). The exon-intron junctions of at least the first 12 exons were identical between m-UBR1 and m-UBR2 (data not shown).
Binding specificities of m-UBR2 and -UBR1 define them as ubiquitin ligases of the N-end rule pathway.
We have previously shown (26) that m-UBR1 (E3α), the previously characterized Ub ligase of the mouse N-end rule pathway, could partially complement this pathway in ubr1Δ S. cerevisiae in the presence of either m-HR6A or m-HR6B, the cognate mouse E2 enzymes. S. cerevisiae RAD6, the E2 enzyme of the yeast N-end rule pathway that interacts with sc-UBR1 (55), could not substitute for mouse HR6A/HR6B in this assay. In contrast to m-UBR1 (25, 26), m-UBR2 could not rescue the N-end rule pathway in ubr1Δ S. cerevisiae (data not shown). This result was essentially neutral in regard to the function of m-UBR2, given the evolutionary distance between mammals and fungi. To determine whether m-UBR2 interacts with m-HR6B, we constructed NIH 3T3 fUBR2, a mouse cell line that stably expressed m-fUBR2. Extracts from 3T3 cells expressing or lacking m-fUBR2 were immunoprecipitated with anti-HR6B antibody (a gift from H. P. Roest, Erasmus University), and immunoprecipitation was followed by SDS-PAGE and immunoblotting with anti-Flag or anti-HR6B antibodies. As shown in Fig. 1K (right panel), m-HR6B was coimmunoprecipitated with m-fUBR2, strongly suggesting their direct interaction in vivo, similar to the interaction between the sc-RAD6 E2 enzyme and sc-UBR1 (55).
We also examined m-UBR2 for its binding to the type 1 and/or type 2 destabilizing N-terminal residues of test substrates. This binding specificity is, operationally, a prerequisite for a Ub ligase to be an E3 of the N-end rule pathway (12, 53). The test proteins were fusions of GST to the C terminus of a fragment of S. cerevisiae SCC1, a subunit of cohesin. This 33-kDa SCC1 fragment, which normally bears N-terminal Arg, is produced in yeast through cleavage by the separase ESP1 and is degraded by the N-end rule pathway (37). The purified X-SCC1-GST proteins (X = Arg, Leu, or Met) bearing either type 1 (Arg), type 2 (Leu), or stabilizing (Met) N-terminal residues were conjugated to glutathione-Sepharose, and GST pull-down assays were carried out with S. cerevisiae extracts that contained m-fUBR1 or m-fUBR2. Similar to the findings with sc-UBR1 (12), the results (Fig. 1C, D, F, and G) showed that both m-fUBR2 and m-fUBR1 bound to Arg-SCC1 and Leu-SCC1 and also that neither m-fUBR2 nor m-fUBR1 bound to Met-SCC1. The binding of m-fUBR2 and m-fUBR1 to Arg-SCC1 could be inhibited by the type 1 dipeptide Arg-Ala but not by Ala-Arg (Fig. 1C and F). Similarly, the binding of m-fUBR2 and m-fUBR1 to Leu-SCC1 could be inhibited by the type 2 dipeptide Leu-Ala but not by Ala-Leu (Fig. 1D and G). Previous work (12) showed that the N-terminal half of the 225-kDa sc-UBR1 contained all three of its substrate-binding sites. The m-fUBR21-1041 N-terminal fragment was found to bind to Leu-SCC1. This binding could be specifically inhibited by type 2 dipeptides (Leu-Ala, Phe-Ala, or Trp-Ala) but not by type 1 dipeptides (Arg-Ala or Lys-Ala) or by control dipeptides with N-terminal Ala (Fig. 1E).
The binding specificity of m-UBR2 was also examined by a different pull-down assay (12), which utilized a set of otherwise identical 12-mer peptides, X-Ile-Phe-Ser-Thr-Asp-Thr-Gly-Pro-Gly-Gly-Cys, that bore different N-terminal residues (X = Arg, Phe, Asp, Gly, Ser, Thr, or Ala) (Fig. 1H to J). The peptides were cross-linked to microbeads through their C-terminal Cys residue (12). As shown in Fig. 1I, m-fUBR2 bound to 12-mer peptides bearing N-terminal Arg (type 1 residue) or Phe (type 2 residue) but did not bind to the otherwise identical peptides bearing either N-terminal Gly (a stabilizing residue), or Asp (a secondary destabilizing residue), or Ser, Thr, and Ala (type 3 destabilizing residues). In addition, the binding of m-fUBR2 to either Arg- or Phe-bearing peptides could be selectively inhibited by dipeptides bearing, respectively, a type 1 or type 2 destabilizing N-terminal residue (Fig. 1J), in agreement with the results of GST pull-down assays (Fig. 1C to G). The substrate-binding properties of m-UBR2 are thus similar if not identical to those of m-UBR1 and sc-UBR1, indicating that m-UBR2 is the second Ub ligase of the mouse N-end rule pathway.
Mouse UBR2−/− strains.
In the deletion allele of m-UBR2, exons 3 to 6 were replaced by a reading frame encoding the NLS-LacZ marker followed by the independently expressed neo gene (Fig. 2A). The deleted segment encompassed the region of high sequence conservation between sc-UBR1, m-UBR1, and m-UBR2, including positions that were previously found to be essential for the integrity of the type 1 binding site (Gly147 and Asp150) and the type 2 binding site (Asp233 and His236) of sc-UBR1 (Webster et al., unpublished data). Of the ∼500 ES cell clones resistant to both G418 and 1-(2′-deoxy,2′-fluoro-β-d-arabinofluranosyl)-5-iodouracil, ∼85 clones contained the expected deletion and/or disruption allele (Fig. 2A) (data not shown). Ten of these correctly targeted 129SvImJ ES cell clones were used to generate male chimeras, and in six of them, the UBR2− allele was transmitted through the germ line. Male chimeras were mated with either C57BL/6, CD1, or 129/SvEv females, yielding UBR2+/− heterozygotes; the intercrosses of heterozygotes produced UBR2−/− mice in three different genetic backgrounds (Fig. 3) (see Materials and Methods for details of this extensive analysis).
To verify the absence of m-UBR2 from UBR2−/− cells, the extracts from UBR2−/−, UBR1−/−, and +/+ EFs were added to microbead-linked 12-mer peptides (Fig. 1H) bearing either N-terminal Phe (type 1) or N-terminal Gly (stabilizing) residues, and this was followed by pelleting and washing of the beads, solubilization of coprecipitated proteins, SDS-PAGE, and immunoblotting with either anti-UBR1 or anti-UBR2 antibodies (Fig. 2F). This two-step assay (used to increase the sensitivity of detection) indicated that both m-UBR1 and m-UBR2 were present in +/+ extracts, that only m-UBR2 was present in UBR1−/− extracts, and that only m-UBR1 was present in UBR2−/− extracts. The control assay, with Gly peptide instead of Phe peptide, did not detect either m-UBR1 or m-UBR2, even in +/+ extracts (Fig. 2F).
To examine the N-end rule pathway in UBR2−/− and +/+ EFs, they were converted into permanent cell lines and transiently transfected with plasmids expressing X-nsP4 test proteins (X = Met, Arg, or Tyr). The latter were a part of the Ub protein reference-based fusions fDHFRh-UbR48-X-nsP4f, whose cotranslational in vivo cleavage yielded the reference protein fDHFRh-UbR48 and the X-nsP4f test proteins (48, 54). Arg-nsP4 (type 1) and Tyr-nsP4 (type 2) N-end rule substrates were found to be short-lived not only in +/+ EFs but in UBR2−/− EFs as well, owing, presumably, to the presence of m-UBR1 in UBR2−/− EFs (Fig. 2D and E). Note, however, that the initial (during pulse) degradation (48) of Tyr-nsP4 was significantly and reproducibly reduced in UBR2−/− EFs in comparison to +/+ EFs. Specifically, ∼17 versus ∼50% of the initial amount of Tyr-nsP4 remained after a 10-min pulse in +/+ versus UBR2−/− EFs, respectively (Fig. 2D and E).
Female-specific or general lethality of the UBR2−/− genotype as a function of genetic background.
In the intercrosses of UBR2+/− mice, UBR2+/− heterozygotes were produced at the approximately Mendelian frequency and were phenotypically wild type, except for a reduced fertility, in comparison to +/+ mice (see below). The initially produced UBR2+/− mice were of the mixed 129SvImJ/C57BL/6J background (designated 129/B6 below), which resulted from mating of the founding chimeras (containing cells derived from ES cells of the 129SvImJ background) with +/+ C57BL/6J mice. The gender-specific viability of UBR2−/− mice produced in 129/B6 UBR2+/− intercrosses is illustrated in Fig. 3A and Table 1. Among the first 510 total offspring of these intercrosses (Fig. 3A), the male UBR2−/− mice survived to at least 1.5 years at nearly the expected frequency (nearly equal to that of +/+ males) but exhibited degeneration of the testes (see below). In striking contrast to UBR2−/− males, the female UBR2−/− mice exhibited severe prenatal lethality (Fig. 3A and Table 1) (see Materials and Methods for details). The aging breeding pairs of initial UBR2+/− mice were eventually replaced with F2 generation UBR2+/− 129/B6 mice. The resulting temporal evolution of the viability of UBR2−/− mice in the 129/B6 background can be viewed, in the first approximation, as a two-stage process. During stage II, which encompassed the later 596 offspring, UBR2−/− males exhibited consistent but less severe testis degeneration than stage I UBR2−/− males and the prenatal lethality of UBR2−/− females decreased as well (Fig. 3A; Table 1) (see Materials and Methods). These statistically robust patterns suggested a selection for modifier genes that enhanced the viability of female UBR2−/− progeny in UBR2+/− intercrosses.
Female UBR2−/− embryos, most of which died in utero, did not have a specific terminal phenotype (Fig. 4C) (see Materials and Methods). The rare surviving stage I UBR2−/− females of the 129/B6 background (Table 1) exhibited, by 2 months of age, an ∼20% growth retardation but were fertile. Given the results with UBR2+/− intercrosses in the mixed (129/B6) background, we also produced UBR2+/− mice in the inbred (strain 129) background and carried out intercrosses. Genotyping of 376 offspring (at ∼4 weeks of age) from strain 129 UBR2+/− intercrosses yielded 57 +/+ males, 62 +/+ females, 121 UBR2+/− males, and 132 UBR2+/− females but only 2 UBR2−/− females and also, strikingly, only 2 UBR2−/− males (Fig. 3B and Table 1). Thus, the UBR2−/− genotype was nearly completely lethal to both genders in the inbred (strain 129) background, in contrast to the 129/B6 background, where the male UBR2−/− mice were born at nearly the Mendelian frequency (Fig. 3; Table 1) (see Materials and Methods). That the choice of a strain background can have a dramatic effect on the severity of a mutant phenotype has been demonstrated for several mouse genes (for examples, see reference 6).
Infertility of UBR2−/− males and reduced fertility of UBR2+/− males.
In contrast to UBR2−/− females of the 129/B6 background, most of which died in utero (Fig. 3 and 4), the 129/B6 UBR2−/− males were born at nearly the expected (Mendelian) frequency, were of normal size and weight, oriented to sound, and their limb movements and behavior appeared to be normal. UBR2−/− males copulated normally with +/+ females, as indicated by the formation of vaginal plugs, but none of mated females became pregnant. No defects were observed in the external genitalia and nonreproductive organs of UBR2−/− males, but their testes, at 8 weeks of age, weighed ∼4 times less than +/+ testes (Fig. 4A and B and data not shown). UBR2−/− testes developed similarly to +/+ ones until ∼3 weeks of age and then degenerated over the next 5 to 6 weeks (Fig. 4A and B). Caudal epididymides in all 16 of the examined 8-week-old UBR2−/− males were devoid of sperm (data not shown). We also observed a significantly decreased fertility of heterozygous UBR2+/− males (see Materials and Methods for details). It would be, therefore, interesting to determine whether the UBR2+/−genotype is overrepresented, relative to the male population at large, in human males that seek medical assistance because of reduced fertility.
Arrest and apoptosis of UBR2−/− spermatocytes.
Examination of testis sections with the TUNEL assay detected occasional, rare apoptotic cells in +/+ testes of 4 to 9 weeks of age, mainly in the spermatogonial layer next to the basal lamina of seminiferous tubules (Fig. 5G and I and data not shown). In UBR2−/− testes, the fraction of TUNEL-positive apoptotic cells, while already significant by 4 weeks of age, increased sharply between 4 and 8 weeks (Fig. 5H, J, and K), resulting, by 8 weeks, in hollowed-out, vacuolized tubules containing little or no spermatocytes, in striking contrast to the tubules of sexually mature +/+ males of the same age (Fig. 5E, I, and K). The apoptosis of UBR2−/− spermatocytes apparently did not extend to the spermatogonial layer (Fig. 5H and J; also data not shown).
FIG. 5.
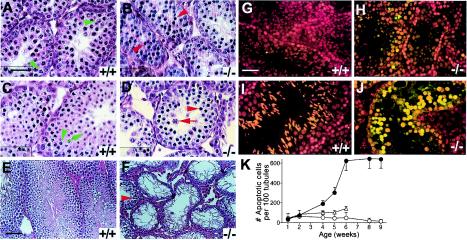
Arrest and apoptosis of spermatocytes in UBR2−/− mice. (A to F) Hematoxylin-eosin staining of testis sections from +/+ mice (A, C, and E) and UBR2−/− littermates (B, D, and F) of the 129SvJ/(C57BL/6J) background at 2 weeks (A and B), 3 weeks (C and D), and 8 weeks (E and F). Green arrowheads in panels A and C, pachytene spermatocytes; green arrow in panel C, round spermatid; red arrowheads in panels B, D, and F, apparently arrested spermatocytes; red arrow in panel D, aberrant pachytene spermatocyte. (G and H) Fluorescent TUNEL staining (and counterstaining with propidium iodide) of testis sections from 5-week-old +/+ male mice (G) and UBR2−/− littermates (H). (I and J) Same as panels G and H, respectively, but with 8-week-old littermates. (K) Quantitation of apoptosis in +/+ (○), UBR2+/− (▵), and UBR2−/− (•) testes as a function of postnatal age. The plotted numbers of apoptotic cells per 100 seminiferous tubules were derived from the examination of ∼400 tubule sections. Scale bars, 20 μm (A to D and G to J) and 60 μm (E and F).
Histological analyses of 8- to 10-day-old UBR2−/− testes showed normally developed Sertoli cells as well as preleptotene/leptotene spermatocytes (data not shown), suggesting that the proliferation and differentiation of spermatogonia to at least the preleptotene stage did not require UBR2. The first abnormality was detected at ∼2 weeks, the time of appearance of the first pachytene stage spermatocytes in +/+ tubules (Fig. 5A). The tubules of 2-week-old UBR2−/− males contained much lower amounts of pachytene stage spermatocytes, which in addition appeared aberrant (Fig. 5B). By 3 weeks, most +/+ tubules contained both pachytene spermatocytes and round spermatids (Fig. 5C), surrounded by 1 to 2 layers of leptotene/zygotene spermatocytes. In contrast, the tubules of littermate UBR2−/− males were almost completely devoid of round spermatids; small amounts of obviously abnormal pachytene-like spermatocytes were present as well (Fig. 5D), surrounded by leptotene/zygotene spermatocytes (Fig. 5D). By 4 weeks, when the first wave of spermiogenesis produced mature sperm in the lumen of +/+ tubules, they were filled with round spermatids, elongating and condensing and/or condensed spermatids, and mature spermatozoa; most spermatocytes in 4-week-old +/+ tubules appeared to be in pachytene and were surrounded by 1 to 2 layers of zygotene stage spermatocytes (data not shown). In contrast, 4-week-old UBR2−/− tubules nearly lacked round spermatids and contained abnormal spermatocytes, some of them apparently in pachytene, with ∼1 layer of type B spermatogonia at the periphery (data not shown). Thus, spermatocytes in UBR2−/− tubules were arrested at the prophase of meiosis I, between leptotene/zygotene and pachytene, followed by apoptosis. While most spermatocytes in the 129/B6 UBR2−/− tubules were arrested at leptotene/zygotene, a small number of spermatocytes proceeded through a pachytene-like stage and formed, by 5 to 6 weeks, what appeared to be round spermatids. However, most of these cells still died by 8 weeks, without forming spermatozoa (Fig. 5F). Taken together, these results (Fig. 5 and data not shown) indicated that most, though not all, germ line cells in UBR2−/− tubules were arrested at the prophase of meiosis I, between leptotene/zygotene and pachytene, followed by apoptosis.
UBR2 is required for homologous chromosome pairing during male meiosis.
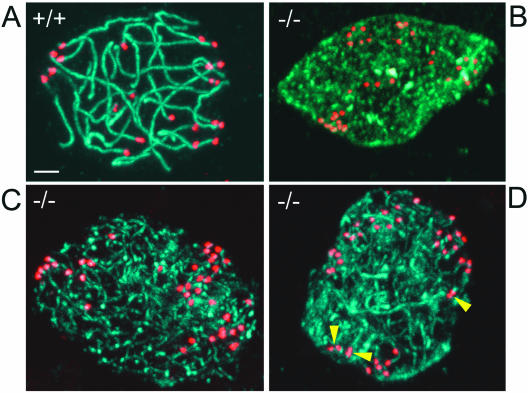
The completion of synapsis of homologous chromosomes marks the beginning of pachytene. We stained sections of 4-week-old 129/B6 +/+ and UBR2−/− testes with antibody to SCP3 (a gift from Peter Moens, York University), a major component of the lateral element of the SC (4, 33, 41, 56). Mostly pachytene stage spermatocytes, with their characteristic chromatin morphology, were present in +/+ tubules, in contrast to UBR2−/− tubules, where SCP3-specific staining of cell nuclei was much more diffuse (Fig. 6A and B). To characterize this perturbation further, we stained surface-spread +/+ and UBR2−/− spermatocytes from 4-week-old mice with both anti-SCP3 and anti-CREST antibodies, with the latter staining the centromeres. Observation of more than 1,000 stained spermatocytes from 10 pairs of +/+ and UBR2−/− testes of mice of different ages confirmed the disruption of SC formation and chromosome pairing. Examples are shown in Fig. 7. In contrast to +/+ pachytene spermatocytes, in which the homologous chromosomes, including their centromeres, were paired throughout their lengths, at most, short patches of SC were observed in UBR2−/− spermatocytes. In addition, the centromeres of individual homologous chromosomes were largely (but not entirely) unpaired in UBR2−/− spermatocytes, resulting in approximately twice as many (∼40) centromere-specific anti-CREST foci in UBR2−/− cells as in +/+ cells (Fig. 7, compare panel A with panels B to D). We conclude that UBR2 is required for the homologous chromosome pairing during meiosis I, at least in the 129/B6 genetic background.
FIG. 7.
Failure to assemble SC in UBR2−/− spermatocytes. Spermatocytes prepared from +/+ (A) and UBR2−/− (B to D) mice were analyzed by confocal microscopy, with antibody to SCP3 used to label the SC (green) and CREST antibody used to label centromeres (red). Yellow arrowheads, incompletely paired centromeres. Scale bar, 5 μm.
In situ hybridization was used to examine the expression, in testes, of m-UBR1, of the lacZ-marked m-UBR2− allele [UBR2−(lacZ)] and also of m-ATE1, which encodes Arg-tRNA-protein transferases (R-transferases) of the N-end rule pathway (Fig. 1A and 8A). The m-UBR2 expression, detected through lacZ mRNA expressed from the PUBR2 promoter, was high mainly in UBR2+/− primary spermatocytes (Fig. 8Aa). In contrast to UBR2+/− tubules, where the expression of m-UBR2 (lacZ) was prominent only in the 2 to 3 outer cell layers (containing primary meiotic spermatocytes), lacZ was expressed in UBR2−/− testes throughout the tubules (Fig. 8Ab, compare with Aa), suggesting that the abnormal pachytene stage UBR2−/− spermatocytes (Fig. 5 and 7) moved to the lumen without further differentiation. m-UBR1, encoding the other (E3α) Ub ligase of the mouse N-end rule pathway (Fig. 1A), was prominently expressed in spermatogonia but not in meiotic spermatocytes, in contrast to m-UBR2 (Fig. 8Ad and data not shown). m-ATE1, encoding R-transferases (Fig. 1A) (24), was expressed largely in primary spermatocytes of UBR2+/− tubules, in a pattern essentially indistinguishable from that of m-UBR2 expression (Fig. 8Ac, compare with Aa). The ramifications of these results are considered in Discussion.
Disrupted spermiogenesis in UBR2−/− mice.
tH2B is incorporated into the chromatin of both meiotic spermatocytes and round spermatids until the stage of elongating spermatids (7). Staining of testis sections from 4-week-old +/+ mice with anti-tH2B antibody (a gift from W. Baarends, Erasmus University) showed large numbers of tH2B-positive round spermatids (Fig. 6C). In contrast, most (though not all) UBR2−/− tubules of the same age nearly lacked tH2B-positive spermatids (Fig. 6D). The spermatocyte-derived round spermatids become elongated spermatids, whose nuclei continue to condense (43), with the concomitant replacement of tH2B and other histones by transition proteins 1 and 2 (TP1 and TP2) and later still by protamines 1 and 2 (7). Staining of testis sections from 4-week-old +/+ mice with anti-TP2 antibody (a gift from W. Baarends) showed highly organized arrangements of the TP2-positive elongated and condensing spermatids (Fig. 6E). In contrast, UBR2−/− tubules almost completely lacked TP2-containing cells (Fig. 6F). Thus, although UBR2−/− tubules contained occasional tH2B-positive round spermatids (Fig. 6D), only a small fraction of them proceeded to become TP2-positive spermatids, a pattern consistent with the possibility that m-UBR2, in addition to its function in meiosis (Fig. 5 and 7), may play an independent role in spermiogenesis as well.
Northern analysis of UBR2−/−, UBR2+/−, and +/+ testes.
We used Northern hybridization to compare the expression of genes in the testes of UBR2−/−, UBR2+/−, and +/+ mice by using total testis RNA from 6-week-old males (Fig. 8B to H). The TAF2Q, FTHL17, TKTL1, NXF2, and RBMY genes, which are expressed preferentially (or exclusively) in spermatogonia, were considerably up-regulated in UBR2−/− testes in comparison to those from UBR2+/− and +/+ testes (Fig. 8B). Among the genes that encode components of the SC and other chromosome-associated proteins, some of them (e.g., SA3) were significantly down-regulated in UBR2−/− testes, whereas little change was observed with several other genes of this set, for example, SCP3 and SA1 (Fig. 8C). In contrast, a strong down-regulation in UBR2−/− testes was observed for the genes encoding testis-specific transcription factors or kinases such as ALF, CREM, FHL4, CAMK4, ACT, and CALSPERMIN (Fig. 8E); postmeiotic proteins of various functions, such as HSP70T, FSC1, GAPD-S, and MCS (Fig. 8F); and the components of sperm precursors at late stages of differentiation such as PROTAMINE 1 and PROTAMINE 2, TP1 and i, and PROACROSIN (Fig. 8G). The findings of strong up- or down-regulation of specific genes in UBR2−/− testes (Fig. 8B to H) will facilitate the dissection of circuits regulated by m-UBR2.
DISCUSSION
The N-end rule pathway is defined as a set of molecular components that is necessary and sufficient, in the context of a specific compartment (the nucleus and the cytosol), for the recognition and degradation of proteins bearing N-degrons (53). This hardware-centric definition of the pathway bypasses semantic problems that would arise if, for example, a cognate Ub ligase recognizes not only N-degrons but a structurally unrelated degron as well. As described in the introduction, a Ub ligase of the N-end rule pathway does recognize structurally unrelated degrons through its distinct substrate-binding sites. In contrast to S. cerevisiae, where the N-end rule pathway is mediated by a single E3 (sc-UBR1), more than one E3 underlies this pathway in the mouse (see Introduction). The present work describes biochemical and genetic analyses of the 200-kDa m-UBR2, a close homolog of m-UBR1, the previously characterized E3 (E3α) of the mouse N-end rule pathway (26). We report the following main results. (i) Similarly to m-UBR1, m-UBR2 was found to interact with m-HR6B, an ortholog of the S. cerevisiae RAD6 E2 enzyme that mediates the yeast N-end rule pathway in a complex with sc-UBR1 (Fig. 1K). Further, m-UBR2, in a binding pattern indistinguishable from that of m-UBR1, specifically recognized test substrates bearing destabilizing N-terminal residues (Fig. 1C to J), indicating that m-UBR2 is the second Ub ligase of the N-end rule pathway. (ii) Most UBR2−/− female mice of the mixed (129/B6) background died as embryos. In contrast, UBR2−/− males were born at the normal frequency and were apparently normal at birth but exhibited testis degeneration by 2 months of age, owing to massive apoptosis of the UBR2−/− spermatocytes (Fig. 3 to 5). In the inbred (129/129) background, the UBR2−/− genotype was lethal for most embryos of either gender (Fig. 3B). (iii) UBR2−/− males were infertile while UBR2+/− males had a significantly reduced fertility. While the gross architecture of UBR2−/− testes was normal, and spermatogonia were intact as well, the UBR2−/− spermatocytes were arrested between leptotene/zygotene and pachytene and died through apoptosis (Fig. 5). A conspicuous defect of UBR2−/− spermatocytes was the absence of intact SCs (Fig. 7). Spermiogenesis was also disrupted in spermatids that resulted from meiotic divisions of those rare UBR2−/− spermatocytes that did not undergo apoptosis (Fig. 6). (iv) Northern analyses of RNA from UBR2−/−, UBR2+/−, and +/+ testes identified several genes that were strongly up- or down-regulated in the absence of m-UBR2 (Fig. 8B to H). (v) In situ hybridization indicated that m-UBR2 was expressed in the testes, primarily in spermatocytes, whereas the expression of m-UBR1 was prominent only in spermatogonia (Fig. 8A).
Thus, despite the high similarity of m-UBR1 and m-UBR2 (Fig. 1), their expression patterns in the testes appear to be either nonoverlapping or at most partially overlapping. If so, the N-end rule pathway may be either absent or down-regulated in UBR2−/− spermatocytes. In this model, the concentration of a (currently unknown) regulator of meiosis that is normally degraded by the N-end rule pathway would be abnormally high in UBR2−/− spermatocytes, resulting in the observed perturbations of SC assembly. Such perturbations would be expected to activate checkpoint circuits (39), leading to the observed pachytene stage apoptosis of UBR2−/− spermatocytes. Given the complexity of meiosis, a similar phenotype would be expected to result from mutations in other genes as well. For example, deletions of SCP3 (56), SPO11 (4, 41), SIAH1a (10), and HR6B (the latter is an E2 enzyme that interacts with m-UBR2) (Fig. 1K) (3, 40) have been shown to cause perturbations of meiosis in mouse spermatocytes. The SCs of HR6B−/− males (3) were damaged to a considerably lesser extent than those of UBR2−/− males (Fig. 7), presumably because of partial complementation, in the former case, by m-HR6A, a close homolog of m-HR6B. Interestingly, mutations in several genes that are involved in the meiotic prophase I events of chromosome synapsis and recombination also display sex-specific effects, in that these mutations cause disruption of spermatogenesis but are still compatible with oogenesis (20), a pattern reminiscent of the fact that the rare surviving UBR2−/− females were fertile, in contrast to invariably infertile UBR2−/− males (see Results).
To account for the fact that the UBR2−/− genotype confers embryonic lethality exclusively on females in one genetic background but on both genders in another background (Fig. 3), it would suffice to assume that in the mixed (129/B6) background, the differential expression patterns of m-UBR1 versus m-UBR2 would decrease the N-end rule pathway's activity below a critical threshold in UBR2−/− cell types that are essential for female but not male development. By contrast, in the inbred (129/129) background, the activity of the N-end rule pathway would become too low in a UBR2−/− cell type essential for the development of both genders, owing to the absent (or diminished) m-UBR1 expression in that cell type in that genetic background.
Whereas single UBR1−/− and UBR2−/− mutants were either viable as adults (UBR1−/− mice) or viable in a background- and gender-dependent manner (UBR2−/− mice), the recently constructed UBR1−/− UBR2−/− mouse strains were found to be completely penetrant E12.5-lethal mutants (Y. T. Kwon and A. Varshavsky, unpublished data). This result was consistent with the structural and substrate-binding similarities of m-UBR1 and m-UBR2 (Fig. 1). Strikingly, however, the EFs rescued from arrested double-mutant UBR1−/− UBR2−/− embryos were found to possess the N-end rule pathway, albeit diminished in activity, in comparison to +/+ EFs (Y. T. Kwon, I. V. Davydov, and A. Varshavsky, unpublished data). The identity of a third mouse E3 that mediates the N-end rule pathway in UBR1−/− UBR2−/− cells is unknown. The plant (Arabidopsis thaliana) RING domain protein PRT1, which is not a member of the UBR family (35), can specifically bind to a subset of type 2 (bulky hydrophobic) N-terminal residues, but not to type 1 residues, and has been shown to rescue a subset of the N-end rule pathway in ubr1Δ S. cerevisiae (A. Bachmair, personal communication). Thus, other E3s of the mouse N-end rule pathway might be PRT1-like, in that they may be specific for subsets of destabilizing N-terminal residues recognized by m-UBR1 and m-UBR2.
The phenotypes imposed by the UBR2−/− genotype were of high but incomplete penetrance. For example, the rare surviving UBR2−/− females (Fig. 3), while growth retarded, were largely normal otherwise and, in addition, fertile. Analogously, a small fraction of UBR2−/− spermatocytes in degenerating UBR2−/− testes did succeed in assembling a nearly complete set of SCs (data not shown). Thus, the absence of m-UBR2, while disrupting the critical UBR2-dependent steps, did not reduce the probability of these steps to zero, perhaps as a result of partial complementation by m-UBR1 and/or another E3 of the N-end rule pathway. The UBR2−/− phenotype of female-specific embryonic lethality and male-specific sterility is without precedent, to our knowledge. The few known genotypes that confer female-specific embryonic lethality include p53−/− (designated Trp53−/−) and MSH2−/− Trp53−/− (6, 44). The frequency of viable females of these genotypes strongly depended on the strain's genetic background, a pattern analogous to the phenotype of UBR2−/− mice (Fig. 3). Could the striking differences in the female-versus-male viability of UBR2−/− mice in the mixed (129/B6) background (Fig. 3A) be caused by a perturbation of the X chromosome inactivation (28), a process unique to females? Studies to address this possibility are under way.
What might be physiological substrates of the N-end rule pathway that underlie the observed effects of the UBR2−/− genotype on male meiosis? Previous work (37) has shown that S. cerevisiae SCC1, a subunit of the cohesin complex that is cleaved by the ESP1 separase at the metaphase-anaphase transition, yields a fragment (bearing N-terminal Arg) that is rapidly degraded by the N-end rule pathway. The failure, in ubr1Δ cells, to degrade the SCC1 fragment results in chromosome instability (37). A fraction of the orthologous SCC1 (RAD21) subunit of cohesin in somatic mammalian cells is also cleaved by separase (16), and the resulting C-terminal fragment of SCC1, similarly to its yeast counterpart, is a short-lived substrate of the mammalian N-end rule pathway (J. Zhou, Y. T. Kwon, and A. Varshavsky, unpublished data). In both yeast and mammalian meiotic cells, the SCC1 subunit of cohesin is replaced by its homolog REC8 (13, 21). In S. cerevisiae, the separase-mediated cleavage of REC8 yields an N-end rule substrate (5). It remains to be determined whether sporulation abnormalities of ubr1Δ S. cerevisiae, which lacks the N-end rule pathway's E3, are caused at least in part by the failure to degrade the REC8 fragment. The mammalian REC8 appears early in meiotic spermatocytes, where it forms axial element-like structures that eventually coalesce to form a part of SC. A fraction of REC8 persists along the chromosome arms until late in meiosis (13). Thus, a separase-produced fragment of mammalian REC8 is a potential N-end rule substrate whose stabilization in UBR2−/− spermatocytes may contribute to their arrest (Fig. 5 and 7). Previous work demonstrated a specific function of the N-end rule pathway in meiosis of the fission yeast Schizosaccharomyces pombe: MEI2, the inducer of meiosis, is a conditionally short-lived protein destroyed by the N-end rule pathway in vegetative cells (22). At present, it appears that a function of the N-end rule pathway that is perturbed in mouse UBR2−/− spermatocytes (Fig. 5 to 7) is distinct from the MEI2-linked function of this pathway in S. pombe.
Mouse c-MOS, a serine/threonine kinase, is yet another protein that may be relevant to the meiotic arrest of UBR2−/− spermatocytes. c-MOS is expressed largely in male and female germ line cells and is a key regulator of oocyte maturation (14). The function of c-MOS in male meiosis is unknown. In oocytes, c-MOS is targeted for degradation through its N terminus-proximal, apparently oocyte-specific degron that is distinct from the N-degron (45). Overexpression of MOS in the testes, through the expression, in transgenic mice, of a virus-derived, long-lived variant of c-MOS called v-MOS, resulted in the arrest of spermatocytes around the pachytene stage of meiosis (42), similar to the phenotype of UBR2−/− spermatocytes. Moreover, m-UBR2, when coexpressed with c-MOS in doubly transfected NIH 3T3 cells, can be specifically coimmunoprecipitated with c-MOS, suggesting at least an indirect (third protein-mediated) interaction between m-UBR2 and c-MOS (we could not detect, thus far, a direct interaction) (J. Sheng, Y. T. Kwon, and A. Varshavsky, unpublished data). If the meiotic catastrophe of UBR2−/− spermatocytes is caused by abnormally high levels of (metabolically stabilized) c-MOS in these cells, one might expect a suppression of this phenotype in UBR2−/− MOS−/− mice.
The m-ATE1 gene, encoding R-transferases that mediate the arginylation branch of the N-end rule pathway (Fig. 1A), is prominently expressed in spermatocytes of UBR2+/− tubules, in a pattern essentially indistinguishable from that of m-UBR2 expression (Fig. 8A). This finding raises an interesting possibility that the essential function of the N-end rule pathway in male meiosis might be carried out by the pathway's arginylation branch. ATE1−/− mouse strains, which lack N-terminal arginylation, die as embryos by E15, with heart defects and perturbation of angiogenic remodeling (24). While neither UBR1−/− mice (26) nor UBR2−/− mice (this work) exhibited significant cardiovascular defects (data not shown), UBR1−/− UBR2−/− double-mutant mice died as embryos by E12.5, and their terminal phenotypes included malformations of the neural tube and perturbed cardiovascular development (Kwon and Varshavsky, unpublished), with the latter defect being similar to that of ATE1−/− embryos. Given the lethality of the nonconditional ATE1−/− genotype (24), cell type-specific ablations of m-ATE1 are required to examine the role of arginylation in meiosis. Construction of such ATE1−/− mutants is under way.
Acknowledgments
Z. Xia and J. Y. An contributed equally to this work.
We are grateful to members of the Caltech Transgenic and Knockout Core Facility, especially S. Pease, B. Kennedy, and L. Sandoval, for care of mice and expert technical help. We thank F. Du (Yale University, New Haven, Conn.) for a precursor of SSC1-encoding plasmids of the present study; W. Baarends (Erasmus University) for anti-tH2B, anti-TP2, and advice; C. Höög (Karolinska Institute) for anti-SCP3 and the CREST serum; P. Moens (York University) for anti-SCP3; and H. P. Roest (Erasmus University) for anti-HR6B.
This work was supported by the NIH grants GM31530 and DK39520 and by a grant from the Kirsch Foundation to A.V.
REFERENCES
- 1.Alfonso, P. J., and W. S. Kistler. 1993. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol. Reprod. 48:522-529. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.). 2002. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
- 3.Baarends, W. M., E. Wassenaar, J. W. Hoogerbrugge, G. van Cappelen, H. P. Roest, J. Vreeburg, M. Ooms, J. H. J. Hoeijmakers, and J. A. Grootegoed. 2003. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiosis prophase. Mol. Cell. Biol. 23:1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 5.Buonomo, S. B. C., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann, and K. Nasmyth. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103:387-398. [DOI] [PubMed] [Google Scholar]
- 6.Cranston, A., and R. Fishel. 1999. Female embryonic lethality in Msh2-Trp53 nullizygous mice is strain-dependent. Mamm. Genome 10:1020-1022. [DOI] [PubMed] [Google Scholar]
- 7.Dadoune, J. P. 2003. Expression of mammalian spermatozoal nucleoproteins. Microsc. Res. Tech. 61:56-75. [DOI] [PubMed] [Google Scholar]
- 8.Davydov, I. V., and A. Varshavsky. 2000. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem. 275:22931-22941. [DOI] [PubMed] [Google Scholar]
- 9.deGroot, R. J., T. Rümenapf, R. J. Kuhn, and J. H. Strauss. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 88:8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickins, R. A., I. J. Frew, C. M. House, M. K. O'Brian, A. J. Holloway, I. Haviv, N. Traficante, D. M. de Kretser, and D. D. L. Botwell. 2002. The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol. Cell. Biol. 22:2294-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ditzel, M., R. Wilson, T. Tenev, A. Zachariou, A. Paul, E. Deas, and P. Meier. 2003. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 5:467-473. [DOI] [PubMed] [Google Scholar]
- 12.Du, F., F. Navarro-Garcia, Z. Xia, T. Tasaki, and A. Varshavsky. 2002. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA 99:14110-14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eijpe, M., H. Offenberg, R. Jessberger, E. Revenkova, and C. Heyting. 2003. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1-beta and SMC3. J. Cell Biol. 160:657-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrell, J. E. 1999. Xenopus oocyte maturation: new lessons from a good egg. BioEssays 21:833-842. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton, M. H., L. A. Cook, T. R. McRackan, K. L. Schey, and J. D. Hildebrandt. 2003. Gamma-2 subunit of G protein heterotrimer is an N-end rule ubiquitylation substrate. Proc. Natl. Acad. Sci. USA 100:5081-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauf, S., I. C. Waizenegger, and J.-M. Peters. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293:1320-1323. [DOI] [PubMed] [Google Scholar]
- 17.Heng, H. H. O., J. Suire, and L.-C. Tsui. 1992. High-resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc. Natl. Acad. Sci. USA 89:9509-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko, A., A. Ciechanover, and A. Varshavsky. 2000. The ubiquitin system. Nat. Med. 10:1073-1081. [DOI] [PubMed] [Google Scholar]
- 19.Heyting, C., and A. J. Dietrich. 1991. Meiotic chromosome preparation and protein labeling. Methods Cell Biol. 35:177-202. [DOI] [PubMed] [Google Scholar]
- 20.Hunt, P. A., and T. J. Hassold. 2002. Sex matters in meiosis. Science 296:2181-2183. [DOI] [PubMed] [Google Scholar]
- 21.Jessberger, R. 2002. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 3:767-778. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura, K., S. Katayama, S. Dhut, M. Sato, Y. Watanabe, M. Yamamoto, and T. Toda. 2001. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell 1:389-399. [DOI] [PubMed] [Google Scholar]
- 23.Kwon, Y. T., S. A. Balogh, I. V. Davydov, A. S. Kashina, J. K. Yoon, Y. Xie, A. Gaur, L. Hyde, V. H. Denenberg, and A. Varshavsky. 2000. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1 amidase and the asparagine branch of the N-end rule pathway. Mol. Cell. Biol. 20:4135-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon, Y. T., A. S. Kashina, I. V. Davydov, R.-G. Hu, J. Y. An, J. W. Seo, F. Du, and A. Varshavsky. 2002. An essential role of N-terminal arginylation in cardiovascular development. Science 297:96-99. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, Y. T., Y. Reiss, V. A. Fried, A. Hershko, J. K. Yoon, D. K. Gonda, P. Sangan, N. G. Copeland, N. A. Jenkins, and A. Varshavsky. 1998. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 95:7898-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon, Y. T., Z. Xia, I. V. Davydov, S. H. Lecker, and A. Varshavsky. 2001. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol. Cell. Biol. 21:8007-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson, T. G., D. L. Gronros, P. E. Evans, M. C. Bastien, K. M. Michalewich, J. K. Clark, J. H. Edmonds, K. H. Graber, J. A. Werner, B. A. Lurvey, and J. M. Cate. 1999. Identification and characterization of a protein destruction signal in the encephalomyocarditis virus 3C protease. J. Biol. Chem. 274:9871-9880. [PubMed] [Google Scholar]
- 28.Marahrens, Y. 1999. X-inactivation by chromosomal pairing events. Genes Dev. 13:2624-2632. [DOI] [PubMed] [Google Scholar]
- 29.Moens, P. B., R. Freire, M. Tarsounas, B. Spyropoulos, and S. P. Jackson. 2000. Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J. Cell Sci. 113:663-672. [DOI] [PubMed] [Google Scholar]
- 30.Mulder, L. C. F., and M. A. Muesing. 2000. Degradation of HIV-1 integrase by the N-end rule pathway. J. Biol. Chem. 275:29749-29753. [DOI] [PubMed] [Google Scholar]
- 31.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome-system controls transcription. Nat. Rev. Mol. Cell Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 32.Obin, M., E. Mesco, X. Gong, A. L. Haas, J. Joseph, and A. Taylor. 1999. Neurite outgrowth in PC12 cells. Distinguishing the roles of ubiquitylation and ubiquitin-dependent proteolysis. J. Biol. Chem. 274:11789-11795. [DOI] [PubMed] [Google Scholar]
- 33.Pelttari, J., M. R. Hoja, L. Yuan, J. G. Liu, E. Brundell, P. Moens, S. Santucci-Darmanin, R. Jessberger, J. L. Barbero, C. Heyting, and C. Höög. 2001. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell. Biol. 21:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickart, C. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 35.Potuschak, T., S. Stary, P. Schlogelhofer, F. Becker, V. Nejinskaia, and A. Bachmair. 1998. PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc. Natl. Acad. Sci. USA 95:7904-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugh, C. W., and P. J. Ratcliffe. 2003. The von Hippel-Lindau tumor suppressor, hypoxia-inducible factor (HIF-1) degradation, and cancer pathogenesis. Semin. Cancer Biol. 13:83-89. [DOI] [PubMed] [Google Scholar]
- 37.Rao, H., F. Uhlmann, K. Nasmyth, and A. Varshavsky. 2001. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410:955-960. [DOI] [PubMed] [Google Scholar]
- 38.Rechsteiner, M. 1998. The 26S proteasome, p. 147-189. In J. M. Peters, J. R. Harris, and D. Finley (ed.), Ubiquitin and the biology of the cell. Plenum Press, New York, N.Y.
- 39.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 40.Roest, H. P., J. van Klaveren, J. de Wit, C. G. van Gurp, M. H. Koken, M. Vermey, J. H. van Roijen, J. W. Hoogerbrugge, J. T. Vreeburg, W. M. Baarends, D. Bootsma, J. A. Grootegoed, and J. H. Hoeijmakers. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86:799-810. [DOI] [PubMed] [Google Scholar]
- 41.Romanienko, P. J., and R. D. Camerini-Otero. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6:975-987. [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg, M. P., C. R. Aversa, R. Wallace, and F. Propst. 1995. Expression of the v-Mos oncogene in male meiotic germ cells of transgenic mice results in metaphase arrest. Cell Growth Differ. 6:325-336. [PubMed] [Google Scholar]
- 43.Russel, L. D., R. A. Ettlin, A. P. S. Hakim, and E. D. Clegg. 1990. Histological and histopathological evaluation of the testis. Cache River Press, Clearwater, Fla.
- 44.Sah, V. P., L. D. Attardi, G. J. Mulligan, B. O. Williams, R. T. Bronson, and T. Jacks. 1995. A subset of p53-deficient embryos exhibit exencephaly. Nat. Genet. 10:175-180. [DOI] [PubMed] [Google Scholar]
- 45.Sheng, J., A. Kumagai, W. G. Dunphy, and A. Varshavsky. 2002. Dissection of c-MOS degron. EMBO J. 21:6061-6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sijts, A. J., I. Pilip, and E. G. Pamer. 1997. The Listeria monocytogenes-secreted p60 protein is an N-end rule substrate in the cytosol of infected cells. Implications for major histocompatibility complex class I antigen processing of bacterial proteins. J. Biol. Chem. 272:19261-19268. [DOI] [PubMed] [Google Scholar]
- 47.Solomon, V., S. H. Lecker, and A. L. Goldberg. 1998. The N-end rule pathway catalyzes a major fraction of the protein degradation in skeletal muscle. J. Biol. Chem. 273:25216-25222. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki, T., and A. Varshavsky. 1999. Degradation signals in the lysine-asparagine sequence space. EMBO J. 18:6017-6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taban, C. H., H. Hondermarck, R. A. Bradshaw, and B. Boilly. 1996. Effect of a dipeptide inhibiting ubiquitin-mediated protein degradation on nerve-dependent limb regeneration in the newt. Experientia 52:865-870. [DOI] [PubMed] [Google Scholar]
- 50.Turner, G. C., F. Du, and A. Varshavsky. 2000. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405:579-583. [DOI] [PubMed] [Google Scholar]
- 51.Unni, E., A. Mayerhofer, Y. Zhang, Y. M. Bhatnagar, L. D. Russell, and M. L. Meistrich. 1995. Increased accessibility of the N-terminus of testis-specific histone TH2B to antibodies in elongating spermatids. Mol. Reprod. Dev. 42:210-219. [DOI] [PubMed] [Google Scholar]
- 52.Varshavsky, A. 2003. The N-end rule and regulation of apoptosis. Nat. Cell Biol. 5:373-376. [DOI] [PubMed] [Google Scholar]
- 53.Varshavsky, A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93:12142-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varshavsky, A. 2000. Ubiquitin fusion technique and its descendants. Methods Enzymol. 327:578-593. [DOI] [PubMed] [Google Scholar]
- 55.Xie, Y., and A. Varshavsky. 1999. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 18:6832-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan, L., J. G. Liu, M. R. Hoja, J. Wilbertz, K. Nordqvist, and C. Höög. 2002. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296:1115-1118. [DOI] [PubMed] [Google Scholar]