Abstract
A common feature of diverse chemopreventive agents is the ability to activate expression of a genetic program that protects cells from reactive chemical species that, if left unchecked, would cause mutagenic DNA damage. The bZIP transcription factor Nrf2 has emerged as a key regulator of this cancer-preventive genetic program. Nrf2 is normally sequestered in the cytoplasm by a protein known as Keap1. Chemopreventive agents allow Nrf2 to escape from Keap1-mediated repression, although the molecular mechanism(s) responsible for activation of Nrf2 is not understood. In this report, we demonstrate that Keap1 does not passively sequester Nrf2 in the cytoplasm but actively targets Nrf2 for ubiquitination and degradation by the proteosome under basal culture conditions. We have identified two critical cysteine residues in Keap1, C273 and C288, that are required for Keap1-dependent ubiquitination of Nrf2. Both sulforaphane, a chemopreventive isothiocyanate, and oxidative stress enable Nrf2 to escape Keap1-dependent degradation, leading to stabilization of Nrf2, increased nuclear localization of Nrf2, and activation of Nrf2-dependent cancer-protective genes. We have identified a third cysteine residue in Keap1, C151, that is uniquely required for inhibition of Keap1-dependent degradation of Nrf2 by sulforaphane and oxidative stress. This cysteine residue is also required for a novel posttranslational modification to Keap1 that is induced by oxidative stress. We propose that Keap1 is a component of a novel E3 ubiquitin ligase complex that is specifically targeted for inhibition by both chemopreventive agents and oxidative stress.
There is compelling evidence from epidemiological studies and from laboratory-based experiments that the incidence of cancer can be reduced by dietary consumption of various plant-derived chemicals (2, 16, 17, 38, 42). Indeed, a structurally diverse array of plant-derived phytochemicals, including isothiocyanates, coumarins, indoles, and lactones, have been shown to be efficacious in the prevention or reduction of cancer in both humans and laboratory animals (42). Despite their structural diversity, these phytochemicals, along with synthetic chemopreventive agents such as the 1,3-dithiolethiones, share chemical and biological features that are thought to be responsible for their cancer-preventive properties. On the chemical level, these anticarcinogens are all electrophilic compounds that have a striking propensity to react with sulfhydryls, leading to the suggestion that the cellular target(s) of these chemicals might be cysteine residues in one or more proteins that control key regulatory pathways leading to cancer (8, 9). On the biological level, these anticarcinogens share the ability to activate expression of a battery of genes that are regulated, in part, by the presence of cis-acting DNA sequences referred to as the antioxidant response elements (AREs) (11, 17, 30, 38, 41). The ARE gene battery, which includes classical phase 2 genes such as glutathione S-transferase Ya and NAD(P)H oxidoreductase (NQO1), has recently been extended to include a number of other genes with diverse roles in cellular metabolism (21, 23, 31, 39). These ARE-dependent genes are also activated by induction of oxidative stress by agents such as tert-butyl hydroxyquinone (tBHQ) and diethylmalate (30). Induction of ARE-dependent genes represents a coordinated response to electrophilic reactive chemicals and to oxidative stress that increases the capacity of cells to detoxify reactive chemicals, to minimize oxidative DNA damage, and to restore cellular redox homeostasis.
The transcription factor Nrf2 has emerged as the critical regulator of ARE-dependent transcription. Nrf2 is a member of a small family of transcription factors that share a conserved bZIP dimerization/DNA-binding domain and the ability to bind ARE-like DNA sequence motifs (28, 30). The characterization of mice that lack Nrf2 has revealed an essential role for Nrf2 in both basal and inducible expression of classical phase 2 genes (4, 5, 12, 20, 24, 33). Nrf2-dependent transcription is critically required for cancer prevention by anticarcinogens, as Nrf2-deficient mice display increased sensitivity to carcinogenic chemicals and cancer incidence in these mice is not reduced by chemopreventive agents such as oltipraz, a synthetic 1,3-dithiolethione (33). Nrf2 may also participate in other physiological processes, as a recent report indicates that Nrf2-deficient mice have a lupus-like autoimmune disorder (43).
The ability of Nrf2 to activate transcription of its target genes is regulated, in large part, through association with a cytoplasmic protein termed Keap1 (7, 14). Keap1 was first identified in a two-hybrid screen that used the N-terminal regulatory domain of Nrf2 as bait. Keap1 is a member of a large family of proteins that contain an N-terminal Broad complex, Tramtrack, and Bric a brac (BTB) domain and a C-terminal Kelch repeat domain (1). Previous reports have demonstrated that Keap1 retains Nrf2 in the cytoplasm via a direct protein-protein interaction between the C-terminal Kelch repeat domain of Keap1 and the N-terminal Neh2 regulatory domain of Nrf2 (14, 15). The role of Keap1 as a physiological regulator of Nrf2 is further supported by the observation that macrophages from mice that lack Keap1 have constitutive nuclear accumulation of Nrf2 (15). However, the molecular mechanisms responsible for cytoplasmic retention of either Keap1 or the Keap1-Nrf2 complex are poorly understood. In this report, we demonstrate that the linker region located between the BTB and Kelch domains is required for cytoplasmic localization of both the Keap1 protein and the Keap1-Nrf2 complex.
The ability of Keap1 to sequester Nrf2 in the cytoplasm provides a plausible mechanism for repression of Nrf2-dependent transcription. However, as Nrf2-dependent transcription is increased as a protective response to electrophilic chemicals and oxidative stress, Nrf2 must be able to escape Keap1-mediated repression, translocate to the nucleus, and activate expression of its target genes. Several mechanisms have been proposed for the ability of Nrf2 to escape Keap1-mediated repression. One attractive mechanism is based on the fact that chemicals which enable Nrf2 to escape Keap1-mediated repression share a common propensity to react with protein thiols (9). Hence, it has been postulated that one or more of the 27 cysteine residues in Keap1 may be the molecular target(s) of thiol-reactive chemical inducers (9). All 27 cysteine residues in Keap1 can be quantitatively labeled by thiol-specific reagents in vitro, although four cysteine residues located in the linker domain between the BTB and Kelch domains (C257, C273, C288, and C297) are the preferred sites of labeling (8). Furthermore, formation of the Keap1-Neh2 complex is dependent upon the presence of strong reducing agents in vitro, and the electrophoretic mobility of the Keap1-Neh2 complex can be altered by thiol-reactive reactive compounds, including the naturally occurring isothiocyanate, sulforaphane (8). However, the role of specific cysteine residues in Keap1 for induced release of Nrf2 in vivo has not been established.
The phosphorylation of Nrf2 has also been proposed as a mechanism for escaping Keap1-mediated repression. Several different protein kinase-dependent signal transduction pathways have been implicated in the release of Nrf2 from Keap1-mediated repression, including the ERK and JNK mitogen-activated protein kinase pathways, protein kinase C, and the phosphatidylinositol 3-kinase-dependent pathway (13, 18, 22, 30, 44). However, the only molecular target of these pathways on the Keap1-Nrf2 complex identified to date is a single serine residue in Nrf2, S40. This serine residue can be phosphorylated by protein kinase C in vitro, and replacement of this residue with alanine impairs the ability of protein kinase C to induce dissociation of Keap1 from Nrf2 in vitro (13). However, phosphorylation of this residue has not been demonstrated in vivo, and the S40A mutant of Nrf2 behaves identically to the wild-type Nrf2 protein in terms of both Keap1-mediated repression and escape from Keap1-mediated repression by chemical inducers (unpublished data).
Several recent reports have implicated proteosome-mediated degradation of Nrf2 as a potential regulatory mechanism. Steady-state levels of Nrf2 are increased in cells treated with known inducers of Nrf2-dependent transcription, including oltipraz, tBHQ, sulforaphane, and cadmium (15, 19, 25, 29, 37). Inhibitors of the 26S proteosome increase steady-state levels of Nrf2, and ubiquitination of Nrf2 has been demonstrated (25, 36, 37). However, the relationship between Keap1-mediated repression of Nrf2, escape by Nrf2 from repression by Keap1, and proteosome-dependent degradation of Nrf2 is not clear. Ectopic expression of Keap1 with Nrf2 can decrease the stability of Nrf2, and mice that lack Keap1 have elevated levels of nuclear Nrf2 (15, 25). However, another report has suggested that Keap1 may stabilize Nrf2 (36).
In this report, we provide evidence that Keap1 actively targets Nrf2 for ubiquitination and proteosome-mediated degradation. The N-terminal Neh2 domain of Nrf2 is targeted for Keap1-dependent ubiquitination. Two cysteine residues located in the linker domain of Keap1, C273 and C288, are critically required for both Keap1-dependent ubiquitination and for Keap1-mediated repression of Nrf2-dependent transcription under basal conditions. Two well-characterized chemical inducers of Nrf2-dependent transcription, tBHQ and sulforaphane, disrupt the ability of Keap1 to target Nrf2 for proteosome-mediated degradation and markedly increase the stability of Nrf2, leading to increased nuclear accumulation of Nrf2. A third cysteine residue located in the BTB domain of Keap1, C151, is required for stabilization of Nrf2 and for activation of Nrf2-dependent transcription by tBHQ and sulforaphane. We have identified a novel posttranslational modification on Keap1 that is induced by chemical inducers of Nrf2 in a dose-dependent manner. This posttranslational modification to Keap1 is uniquely dependent upon the integrity of C151 and is stable under reducing electrophoretic conditions. Thus, C151 provides a genetic link between the ability of chemical inducers to block Keap1-mediated repression of Nrf2 and to induce posttranslational modification(s) of Keap1. We propose that Keap1 is a component of an E3 ubiquitin ligase complex that is regulated by oxidative stress and chemopreventive agents.
MATERIALS AND METHODS
Construction of recombinant DNA molecules.
The Keap1 cDNA used in this study was obtained from Kazusu DNA Research Institute (Kisarazu, Japan) as KIAA 0132. The Keap1 expression vector was constructed by cloning a PCR-generated fragment containing the entire open reading frame of Keap1 into the BamHI/XhoI sites of pcDNA3 (Invitrogen). A series of deletion mutants of Keap1 in pcDNA3 were constructed by PCR and standard recombinant DNA techniques (34). The Keap1-ΔN construct lacks the first 66 codons of the Keap1 open reading frame, the Keap1-ΔBTB construct lacks the BTB domain (codons 67 to 178), the Keap1-ΔL construct lacks the linker domain between the BTB and Kelch domains (codons 179 to 321), the Keap1-ΔK construct lacks the Kelch domain (codons 322 to 608), and the Keap1-ΔC construct lacks the C-terminal 15 codons (609 to 624). Mutant Keap1 cDNAs containing serine codons in place of specific cysteine codons were generated by oligo-directed mutagenesis with standard techniques. The Nrf2 cDNA was a gift from Y. W. Kan (University of California, San Francisco) (27). The N-terminal 16 codons missing from this cDNA were added back by a PCR-based approach, and a PCR-generated fragment containing the complete Nrf2 open reading frame was subcloned into SmaI/NotI sites of pCI (Promega) tagged with hemagglutinin (HA)-epitope. Deletion mutants within the Nrf2 open reading frame were constructed by PCR and fused to the Gal4 DNA-binding domain in a pcDNA3-derived vector by standard recombinant DNA techniques. Two Gal4-Nrf2 expression vectors were used in this study, containing either codons 1 to 97 of Nrf2 or codons 1 to 454 of Nrf2 fused to the open reading frame of the Gal4 DNA-binding domain. All of the genes used in this study were sequenced in the context of the expression vectors used for the experiments.
Cell culture and transfections.
COS1, NIH 3T3, and MDA-MB-231 cells were purchased from the American Type Culture Collection. Cell cultures were maintained in either Dulbecco's modified Eagle's medium or Eagle's minimal essential medium in the presence of 10% fetal bovine serum (FBS). Transfections were performed with Lipofectamine Plus (Gibco BRL) according to the manufacturer's instructions. Chemical inducers of Nrf2-dependent transcription were added to medium for 16 h prior to cell lysis for analysis of reporter gene activity or other biochemical assays.
Reporter gene assays.
The ARE TATA-Inr luciferase reporter plasmid, pARE-Luc, was a gift from Bill Fahl (University of Wisconsin—Madison) and contains the 41-bp ARE sequence from the mouse glutathione S-transferase Ya subunit gene cloned behind a minimal TATA-Inr promoter in pGL2-basic (Promega) (41). The Gal4-dependent luciferase reporter plasmid, pFR-Luc, contains 5 copies of the Gal4 binding site upstream of a minimal promoter and the firefly luciferase gene (Stratagene). The Renilla luciferase expression plasmid, pRL-TK, contains the herpes simplex virus thymidine kinase promoter region upstream of the Renilla luciferase cDNA (Promega). The Gal-Nrf2 gene used for the Gal4-dependent luciferase reporter gene assays encodes a fusion protein of the Gal4 DNA-binding domain and amino acids 1 to 454 of Nrf2. NIH 3T3 cells grown on 24-well plates were transfected with 100 ng of pARE-Luc or pFR-Luc reporter plasmid, 10 ng of pRL-TK reporter plasmid, either 40 ng of the Nrf2 expression plasmid (with pARE-Luc) or 1 ng of the Gal-Nrf2 expression plasmid (with pFR-Luc), and either 100 ng (with pARE-Luc) or 10 ng (with pFR-Luc) of the wild-type or mutant Keap1 expression plasmid. The total amount of DNA was maintained at 400 ng with pcDNA3. Both firefly and Renilla luciferase activities were measured 48 h after transfection with the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity to control for sample-to-sample variations in transfection efficiency. All reporter gene assays were repeated in at least three independent transfections.
Immunofluorescence assays.
NIH 3T3 cells were grown on glass coverslips on 35-mm-diameter plates. Cells were transfected with 0.5 μg (each) of the HA-Nrf2 expression plasmids and expression plasmids for either wild-type or mutant Keap1 proteins. Cells were fixed with 100% methanol at −20°C for 10 min. Fixed cells were incubated for 40 min with affinity-purified rabbit anti-Keap antibodies and mouse anti-HA antibodies (Covance) at a 1:100 dilution in phosphate-buffered saline (10 mM sodium phosphate [pH 8.0] and 150 mM NaCl) containing 10% (vol/vol) FBS. Coverslips were washed and incubated with fluorescein isothiocyanate-conjugated anti-mouse and Texas red isothiocyanate-conjugated anti-rabbit antibodies at a 1:100 dilution for another 40 min. Both secondary antibodies were obtained from Jackson Laboratories. Coverslips were washed and mounted on glass slides. At least 100 positive cells were scored for localization of Nrf2 and Keap1 proteins under a microscope. Images were obtained with a Bio-Rad Radiance 2000 confocal system coupled to an Olympus IX70 inverted microscope and a digital camera. The images were captured with ImagePro, processed with Metamorph to obtain merged images, and transferred to Adobe Photoshop for construction of the figure. Minimal alterations were performed on the digital images with either Metamorph or Photoshop.
Antibodies, immunoprecipitation, and immunoblot analysis.
The full-length Keap1 protein was expressed in Escherichia coli as a hexahistidine-tagged protein and purified by metal-chelate chromatography. Care was taken to maintain reducing conditions during all stages of the purification and subsequent use of the purified Keap1 protein. Antibodies were generated in rabbits against the full-length Keap1 protein and anti-Keap1 immunoglobulin G was purified over a column containing immobilized His-Keap1 protein with reagents obtained from Pierce. Antibodies against Nrf2 (Santa Cruz) and the HA epitope (Covance) were purchased from commercial sources. Horseradish peroxidase-coupled secondary antibodies for enhanced chemiluminescence were obtained from Jackson Laboratories.
COS1 cells on 60-mm-diameter plates were transfected with 1 μg of the Nrf2 expression plasmid and 1 μg of the wild-type or mutant Keap1 expression plasmid. Cells were lysed in RIPA buffer (10 mM sodium phosphate [pH 8.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.2% sodium dodecyl sulfate [SDS]) containing 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Novagen) at 48 h posttransfection. For immunoprecipitation of Keap1 proteins, soluble cell lysates were incubated with 2 μg of affinity-purified anti-Keap1 antibodies for 1.5 h at 4°C and then incubated at 4°C with protein A-agarose beads for 1.5 h. For immunoprecipitation of HA-Nrf2, soluble cell lysates were incubated with 20 μl of HA-agarose beads (Covance) at 4°C for 1.5 h. Unbound proteins were removed by washing four times with RIPA buffer. The immunoprecipitated proteins were eluted in sample buffer (100 mM Tris [pH 6.8], 4% SDS, 20% glycerol, 200 mM DTT, and 0.2% bromophenol blue), boiled for 4 min, electrophoresed through SDS-7.5% polyacrylamide gels, transferred to nitrocellulose membranes, and subjected to immunoblot analysis.
To obtain nuclear and cytoplasmic subcellular fractions, transfected MDA-MB-231 cells were rinsed twice with ice-cold phosphate-buffered saline. The cell pellets were lysed with ice-cold hypotonic buffer (20 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM Na3VO4, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) supplemented with 0.2% Nonidet P-40. Lysates were centrifuged at 13,000 × g for 10 s to separate nuclear and soluble cytoplasmic components. Supernatants were collected as cytoplasmic extracts. Nuclear extracts were prepared by resuspension of the crude nuclei in high-salt butter (hypotonic buffer containing 20% glycerol and 420 mM NaCl) at 4°C for 30 min, and the supernatants were collected after centrifugation at 13,000 × g for 5 min.
For pulse-chase analyses, transfected cells in 35-mm-diameter dishes were labeled with Dulbecco's modified Eagle's medium containing 100 μCi of [35S]methionine and [35S]cysteine supplemented with 10% dialyzed FBS for 15 or 30 min. The labeled cells were either collected in RIPA buffer or the labeling medium was replaced with complete growth medium. Cell lysates were collected in RIPA buffer following the indicated chase periods and subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitated proteins were electrophoresed through an SDS-7.5% polyacrylamide gel and visualized by fluorography. The relative intensities of immunoprecipitated Nrf2 were quantified by phosphorimager analysis (Bio-Rad FXImager).
For detection of ubiquitinated Nrf2 proteins, cells were transfected with expression vectors for HA-ubiquitin (40), either nontagged full-length Nrf2 or Gal4-Neh2, and either wild-type or mutant Keap1 proteins. The transfected cells were exposed to dimethyl sulfoxide (DMSO) or MG132 (Boston Biochem) for 5 h. Cells were lysed in 2% SDS and 10 mM N-ethylmaleimide (NEM) to block ubiquitin hydrolase activity. Immunoprecipitation with either anti-Gal4 antibodies (Santa Cruz) or anti-Nrf2 antibodies (Santa Cruz) was performed under denatured conditions as described previously (3). Immunoprecipitates were analyzed by immunoblotting with antibodies directed against the HA epitope (Covance).
RESULTS
The central linker domain of Keap1 is required for cytoplasmic sequestration of Nrf2.
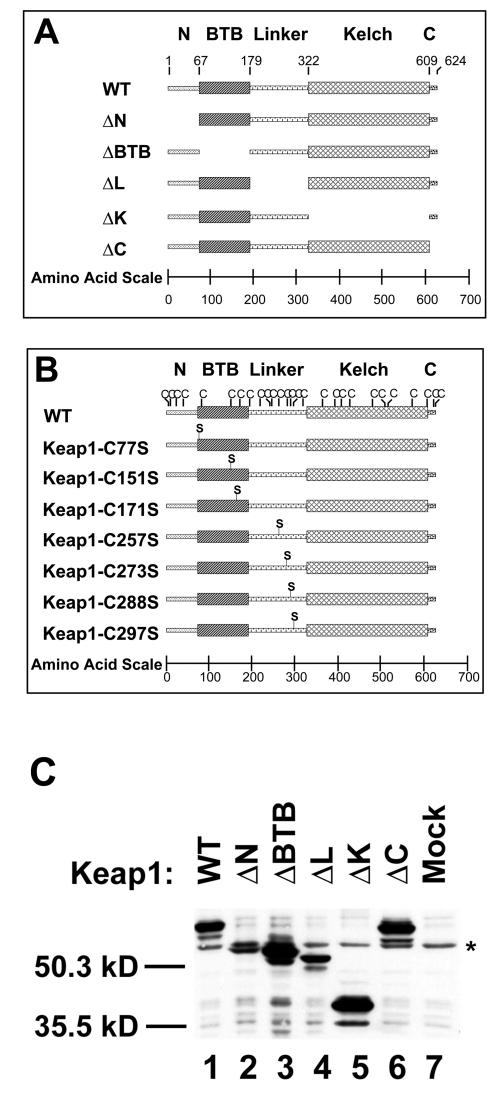
The Keap1 protein contains five discrete domains, including (i) the N-terminal 66 amino acids (termed N in this report); (ii) an N-terminal BTB domain, amino acids 67 to 178 (BTB in this report); (iii) a central linker domain, amino acids 179 to 321 (Linker, or L in this report); (iv) a C-terminal Kelch repeat domain, amino acids 322 to 608 (K in this report); and (v) a short C-terminal tail, amino acids 609 to 625 (C in this report). A series of mutant Keap1 proteins were constructed that contained precise deletions of each domain (Fig. 1A). Proteins of the expected sizes were readily detectable in lysates from COS1 cells transfected with expression vectors for the wild-type and mutant proteins (Fig. 1C).
FIG. 1.
Domain structure of the Keap1 protein. (A) Five discrete domains of the human Keap1 protein are designated as N (amino acids 1 to 66); BTB (amino acids 67 to 178), Linker (amino acids 179 to 321), Kelch (amino acids 322 to 608), and C (amino acids 609 to 624). Mutant Keap1 proteins lacking individual domains are illustrated. (B) The locations of all 27 cysteine residues in the human Keap1 proteins are shown. Mutant Keap1 proteins containing individual cysteine-to-serine mutations in the BTB and Linker domains utilized in this report are illustrated. (C) Expression of the wild-type (WT) Keap1 or deletion mutants. Lysates from mock-transfected COS1 cells or from COS1 cells transfected with expression vectors for either wild-type or mutant Keap1 proteins were subjected to immunoblot analysis with an antibody raised against the Keap1 protein. An endogenous protein that cross-reacts with the antibody is indicated by the asterisk. The locations of molecular mass markers are shown.
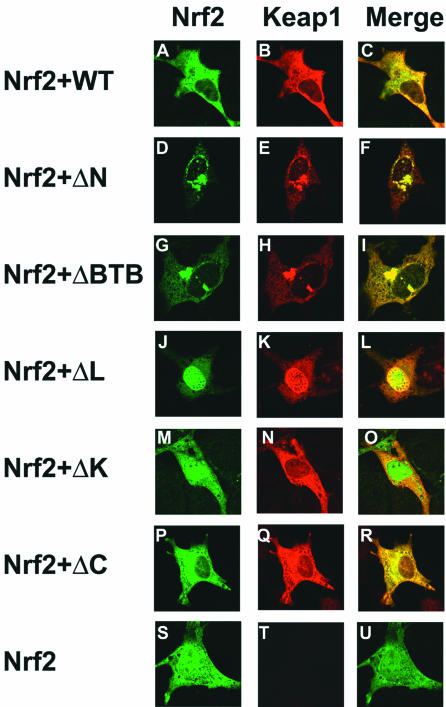
To determine the ability of the wild-type and mutant Keap1 proteins to sequester Nrf2 in the cytoplasm, NIH 3T3 cells were cotransfected with an expression vector for HA-tagged Nrf2 and expression vectors for either wild-type or mutant Keap1 proteins. NIH 3T3 cells were selected for these and subsequent reporter gene experiments, as NIH 3T3 cells contain very low levels of the endogenous Nrf2 and Keap1 proteins (data not shown). The subcellular distribution of the HA-Nrf2 and Keap1 proteins was visualized by confocal microscopy following double-label indirect immunofluorescence with anti-Keap1 and anti-HA antibodies (Fig. 2). The HA-Nrf2 protein was distributed in both the nucleus and the cytoplasm in singly transfected cells (Fig. 2S and Tables 1 and 2). The HA-Nrf2 protein was efficiently sequestered in the cytoplasm by coexpression of the wild-type Keap1 protein (Fig. 2A and Tables 1 and 2). Likewise, HA-Nrf2 was efficiently sequestered in the cytoplasm by the mutant Keap1-ΔN, Keap1-ΔBTB, and Keap1-ΔC proteins (Fig. 2D, G, and P and Tables 1 and 2). The Keap1-ΔN and Keap1-ΔBTB proteins displayed increased perinuclear localization relative to the wild-type Keap1 protein (Fig. 2, compare panels B, E, and H). The Keap1-ΔK protein, which lacked the C-terminal Kelch domain and is therefore unable to associate with Nrf2, was unable to retain HA-Nrf2 in the cytoplasm (Fig. 2M and Tables 1 and 2). In contrast, the Keap1-ΔL protein, which lacked the linker domain between the BTB and Kelch domains of Keap1, was mislocalized to the nucleus and was unable to retain HA-Nrf2 in the cytoplasm (Fig. 2J and K and Tables 1 and 2). These results indicate that the linker domain of the Keap1 protein is required to localize both Keap1 and Keap1-associated Nrf2 in the cytoplasm.
FIG. 2.
Cellular localization of the Nrf2 and Keap1 proteins. NIH 3T3 cells were cotransfected with an expression vector for HA-Nrf2 and expression vectors for either wild-type (WT) or mutant Keap1 proteins. The cellular localization of the Nrf2 and Keap1 proteins was determined by double-label indirect immunofluorescence with anti-HA (panels A, D, G, J, M, P, and S) or anti-Keap1 antibodies (panels B, E, H, K, N, Q, and T). Colocalization of the Nrf2 and Keap1 proteins is indicated by the presence of yellow in the merged images (panels C, F, I, L, O, R, and U).
TABLE 1.
Subcellular distribution of Keap1 proteins
| Keap1 proteina | % Subcellular distribution inb:
|
||
|---|---|---|---|
| N | N-C | C | |
| WT | 0 | 0 | 100 |
| ΔN | 0 | 0 | 100 |
| ΔBTB | 0 | 0 | 100 |
| ΔL | 85 | 15 | 0 |
| ΔK | 0 | 0 | 100 |
| ΔC | 0 | 0 | 100 |
The subcellular localization of the indicated Keap1 proteins in NIH 3T3 cells was determined by indirect immunofluoresence. Details of the Keap1 proteins are shown in Fig. 1. WT, wild type.
Percentages of cells that displayed predominantly nuclear (N), nuclear/cytoplasmic (N-C), and cytoplasmic (C) staining of Keap1 proteins are presented. A total of 100 cells that are positive for expression of Keap1 proteins were examined.
TABLE 2.
Subcellular distribution of HA-Nrf2a
| Keap1 proteinb | % Subcellular distribution inc:
|
||
|---|---|---|---|
| N | N-C | C | |
| Noned | 0 | 100 | 0 |
| WT | 0 | 0 | 100 |
| ΔN | 0 | 0 | 100 |
| ΔBTB | 0 | 0 | 100 |
| ΔL | 50 | 50 | 0 |
| ΔK | 0 | 100 | 0 |
| ΔC | 0 | 0 | 100 |
The subcellular localization of HA-Nrf2 in NIH 3T3 cells was determined by indirect immunofluoresence.
The indicated Keap1 proteins were coexpressed with Nrf2 in NIH 3T3 cells. WT, wild type.
Percentages of cells that displayed predominantly nuclear (N), nuclear/cytoplasmic (N-C), and cytoplasmic (C) staining of Nrf2 are presented. A total of 100 cells that are positive for expression of Nrf2 and the indicated Keap1 protein were examined.
HA-Nrf2 was expressed alone (without a Keap1 protein) in NIH 3T3 cells.
The N-terminal domains of Keap1 are required for efficient repression of Nrf2.
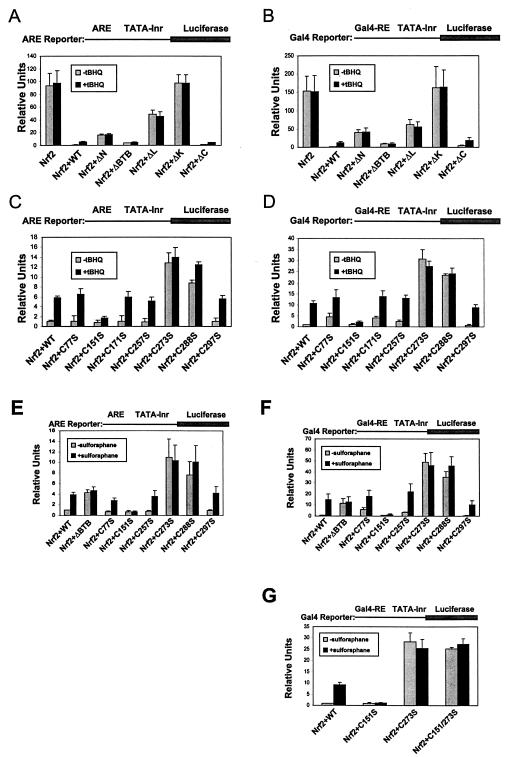
The wild-type and mutant Keap1 proteins were characterized for their ability to regulate Nrf2-dependent transcriptional activation in transient transfection experiments. An ARE-dependent firefly luciferase reporter gene was used to assess transcriptional activation by full-length Nrf2 (Fig. 3A). A parallel set of reporter gene assays was performed with a Gal4-dependent firefly luciferase reporter gene to assess transcriptional activation by a Gal4-Nrf2 fusion protein containing the N-terminal Neh2 domain, which contains residues required for binding to Keap1 (14), and the central transactivation domain of Nrf2 (Fig. 3B). Reporter gene assays were performed with NIH 3T3 cells grown under normal cell culture conditions to measure the ability of the Keap1 proteins to repress Nrf2-dependent transcriptional activity. Parallel reporter gene assays were performed with NIH 3T3 cells exposed to 25 μM tBHQ for 16 h prior to measurements of luciferase activity to assess the ability of Nrf2 to escape Keap1-mediated repression under conditions of oxidative stress.
FIG. 3.
Regulation of Nrf2-dependent transcriptional activity. (A) NIH 3T3 cells were cotransfected with plasmids containing an ARE-dependent firefly luciferase reporter gene and expression plasmids for Nrf2 and either wild-type (WT) or mutant Keap1 proteins containing deletions of individual domains. A plasmid encoding Renilla luciferase driven by the herpes simplex virus thymidine kinase promoter was included in all transfections to normalize for transfection efficiency. The transfected cells were exposed to 25 μM tBHQ for 16 h prior to analysis of firefly and Renilla luciferase activities in cell lysates. All samples were run in duplicate for each experiment, and the data shown represent the means of the results from three independent experiments. The error bars indicate the standard deviations from the means of the results from three experiments. (B) Transfections into NIH 3T3 cells and subsequent analyses of firefly and Renilla activities were performed as described for panel A, except that the firefly luciferase gene was driven by a Gal4-dependent promoter and a truncated Nrf2 protein (amino acids 1 to 454, containing the N-terminal Neh2 domain and the central transactivation domain) was expressed as a fusion protein with the Gal4 DNA-binding domain. (C) Transfections and reporter gene analyses were performed as described for panel A, except that the mutant Keap1 proteins contained singlecysteine-to-serine substitutions as illustrated in Fig. 1B. (D) Transfections and reporter gene analyses were performed as described for panel B, except that the mutant Keap1 proteins contained single cysteine-to-serine substitutions as illustrated in Fig. 1B. (E) Reporter gene assays were performed as described for panel A, except that transfected cells were treated with 4 μM sulforaphane for 16 h. (F) Reporter gene assays were performed as described for panel B, except that transfected cells were treated with 4 μM sulforaphane for 16 h. (G) Reporter gene assays were performed as described for panel F. The Keap1 C151/273S protein contains serine residues in place of both cysteine 151 and cysteine 273.
Ectopic expression of Nrf2 or the Gal4-Nrf2 fusion protein resulted in potent activation of the ARE-dependent or Gal4-dependent reporter genes under basal conditions. As expected, reporter gene activity was markedly reduced, to approximately 1%, by coexpression of wild-type Keap1 along with either full-length Nrf2 or the Gal4-Nrf2 fusion protein (Fig. 3A and B). Of all the mutants tested, only the Keap1-ΔC protein, which lacked the C-terminal 15 amino acids, was able to repress basal Nrf2-dependent gene expression as efficiently as the wild-type Keap1 protein. The Keap1-ΔK protein, which lacks the Kelch domain and is unable to bind Nrf2, was unable to repress Nrf2-dependent transcription. The other Keap1 proteins, Keap1-ΔN, Keap1-ΔBTB, and Keap1-ΔL, all of which contain an intact Kelch domain but lack specific N-terminal domains, were also impaired in the ability to repress Nrf2-dependent transcription. Repression of Nrf2-dependent transcription by these mutant Keap1 proteins varied widely. Deletion of the linker domain between the Kelch and BTB domains had the greatest impact on repression of Nrf2-dependent reporter gene activity. In contrast, the Keap1-ΔBTB protein, lacking just the N-terminal BTB domain, was nearly as effective as the wild-type Keap1 protein for repression of Nrf2-dependent transcriptional activity.
The ability of either Nrf2 or the Gal4-Nrf2 fusion protein to escape repression by wild-type and mutant Keap1 proteins in response to oxidative stress was assessed following treatment of transfected cells with 25 μM tBHQ (Fig. 3A and B). In cells singly transfected with either of the Nrf2 expression vectors, no further increase in Nrf2-dependent reporter gene expression was observed in tBHQ-treated cells, presumably a reflection of increased expression of the vector-encoded Nrf2 proteins relative to that of the endogenous Keap1 protein. However, tBHQ treatment of cells cotransfected with expression vectors for the wild-type Keap1 protein and expression vectors for either full-length Nrf2 or the Gal4-Nrf2 fusion protein resulted in a 6- to 10-fold increase in reporter gene activity. A similar tBHQ-induced increase in Nrf2-dependent reporter gene activity was observed in cells transfected with expression vectors for the Keap1-ΔC protein and either of the two Nrf2 proteins. In contrast, there was no tBHQ-induced increase in Nrf2-dependent reporter gene activity in the presence of the Keap1-ΔN, Keap1-ΔBTB, Keap1-ΔL, and Keap1-ΔK proteins. In the case of the Keap1-ΔK and Keap1-ΔL proteins, the absence of increased Nrf2-dependent reporter gene activity in tBHQ-treated cells presumably reflects the failure of these two mutant Keap1 proteins to sequester Nrf2 in the cytoplasm under basal conditions (Fig. 2). In contrast, as the Keap1-ΔN and Keap1-ΔBTB proteins are able to efficiently sequester Nrf2 in the cytoplasm under basal conditions, the absence of increased Nrf2-dependent transcription in tBHQ-treated cells in the presence of the Keap1-ΔN and Keap1-ΔBTB proteins suggests that these N-terminal domains of Keap1 are required for activation of Nrf2-dependent transcription in response to tBHQ-induced oxidative stress. These N-terminal domains may be required for direct sensing of tBHQ-induced oxidative stress by Keap1. Alternatively, mislocalization of the Keap1-Nrf2 complex to a predominantly perinuclear localization by these mutant Keap1 proteins may be responsible for the absence of tBHQ-induced Nrf2-dependent gene expression.
Basal repression and oxidative stress-induced activation of Nrf2-dependent transcription require distinct cysteine residues in Keap1.
The Keap1 protein can both repress Nrf2-dependent transcription under basal conditions and enable increased Nrf2-dependent transcription under conditions of oxidative stress (4). As formation of the Keap1-Nrf2 complex requires reducing conditions in vitro and Keap1 contains a number of cysteine residues that can react with thiol-reactive chemicals (8), an attractive hypothesis is that the oxidation status of one or more cysteine residues in Keap1 is a component of a molecular switch that is responsive to perturbations of the intracellular redox environment.
To test the involvement of cysteine residues in Keap1 in the activation of Nrf2 following exposure to oxidative stress, a number of mutant Keap1 genes containing serine codons substituted for specific cysteine codons were constructed. In the experiments reported in this report, we focused our attention on three cysteine residues within the BTB domain of Keap1 (C77, C151, and C171) and four cysteine residues within the central linker domain (C257, C273, C288, and C297). The three residues within the BTB domain were selected on the ability of the Keap1-ΔBTB protein to repress basal Nrf2-dependent transcriptional activity nearly as effectively as the wild-type Keap1 protein and to prevent increased Nrf2-dependent transcription in response to tBHQ-induced oxidative stress (Fig. 3A and B). The four cysteine residues in the linker domain of Keap1 were selected on the basis of a recent report that these cysteine residues preferentially react with thiol-specific chemicals in vitro (8).
Mutant Keap1 proteins containing single cysteine to serine substitutions were constructed (Fig. 1B). The mutant Keap1 proteins containing single cysteine-to-serine mutations were expressed at levels comparable to the wild-type Keap1 protein in transient transfection experiments and displayed no detectable differences in their stability or subcellular localization (data not shown).
The ability of mutant Keap1 proteins containing individual cysteine-to-serine mutations to both repress basal Nrf2-dependent gene expression and to allow increased Nrf2-dependent gene expression in response to oxidative stress was first evaluated in reporter gene assays. Mutant Keap1 proteins containing serine substitutions for cysteine residues C77, C171, C257, and C297 behaved identically to the wild-type Keap1 protein in terms of both repression of Nrf2-dependent reporter gene activity under basal culture conditions and increased Nrf2-dependent reporter gene activity following exposure of transfected cells to tBHQ (Fig. 3C and D).
Two of the mutant Keap1 proteins, Keap1-C273S and Keap1-C288S, were impaired in their ability to repress Nrf2-dependent transcriptional activation under basal culture conditions (Fig. 3C and D). Exposure of transfected cells to either tBHQ-induced oxidative stress or to sulforaphane did not result in a further increase in Nrf2-dependent transcriptional activation (Fig. 3C to F).
A single mutant Keap1 protein, Keap1-C151S, was a constitutive repressor of Nrf2-dependent reporter gene expression both under basal culture conditions and following exposure of cells to either tBHQ or sulforaphane (Fig. 3C to F). A Keap1 protein containing both the C151S and C273S mutations was unable to efficiently repress Nrf2-dependent transcriptional activity under basal conditions (Fig. 3G), indicating that the loss of repression phenotype of the Keap1-C273 mutant protein is dominant over the constitutive repressor phenotype of the Keap1-C151S mutant protein.
Taken together, these results indicate (i) that C273 and C288 are critically required for Keap1-mediated repression of Nrf2 under basal culture conditions and (ii) that C151 is specifically required for escape by Nrf2 from Keap1-mediated repression in response to tBHQ-induced oxidative stress and sulforaphane.
Cysteine 151 is required for a novel oxidative stress-induced modification to Keap1.
Cysteine-to-serine mutations in Keap1 can either perturb Keap1-mediated repression of Nrf2 under basal culture conditions (C273S or C288S) or perturb the ability of Nrf2 to escape Keap1-mediated repression (C151S). An attractive model is that these cysteine residues constitute a multicomponent redox-sensitive switch that determines the ability of Keap1 to repress Nrf2-dependent transcription.
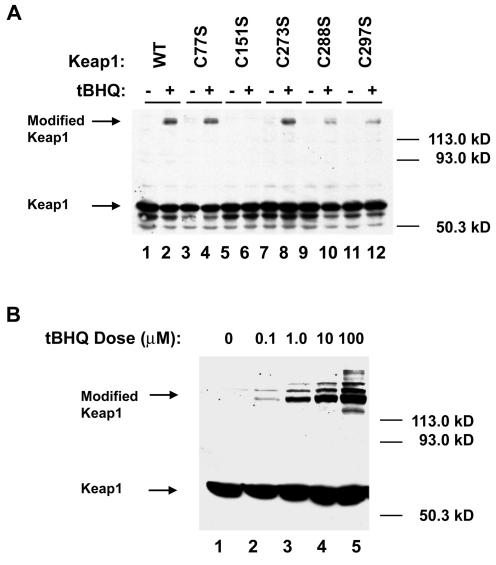
In other proteins in which cysteine residues function as redox-sensitive molecular switches, the critical cysteine residues often participate in reversible disulfide bonds that can alter the electrophoretic mobility of the protein under nonreducing conditions (6). The electrophoretic mobility of the wild-type Keap1 protein under both nonreducing and reducing conditions was carefully examined. For analysis under nonreducing conditions, COS1 cells were transfected with expression vectors for the wild-type Keap1 protein and lysed with trichloroacetic acid to trap cysteine residues in their native oxidation state (26). Cell lysates were treated with NEM, which will react with reduced cysteine thiols but not oxidized disulfide bonds, prior to electrophoresis through nonreducing SDS-polyacrylamide gels. No detectable alterations in the electrophoretic mobility of Keap1 in nonreducing SDS-polyacrylamide gels was observed between untreated and tBHQ-treated cells (data not shown).
However, under reducing conditions, while the majority of the Keap1 protein migrates with an apparent molecular mass of 68 kDa, a slow migrating form of Keap1 with an apparent molecular mass of greater than 120 kDa was observed in cell lysates from tBHQ-treated cells (Fig. 4A, lane 2). This slow migrating form of Keap1 was not present in untreated cells but was induced by tBHQ exposure in a dose-dependent manner (Fig. 4B). This slow migrating form of Keap1 was also observed in cells treated with other known inducers of ARE-dependent gene expression, including oltipraz, diethylmalate, and sulforaphane (data not shown).
FIG. 4.
A novel form of Keap1 with retarded mobility is induced by tBHQ in a dose-dependent manner and requires C151. (A) COS1 cells transfected with expression vectors for either wild-type (WT) (lanes 1 and 2) or the indicated mutant Keap1 proteins (lanes 3 to 12) were treated with DMSO (−) or 25 μM tBHQ (+) for 16 h. Cell lysates were collected, electrophoresed through an SDS-7.5% polyacrylamide gel and subjected to immunoblot analysis with an antibody against Keap1. The arrows on the left side of the figure indicate the location of Keap1 and of a tBHQ-induced form of Keap1 that migrates with retarded mobility through an SDS-polyacrylamide gel. The positions of molecular mass markers are indicated on the right side of the figure. (B) Transfections into COS1 cells and subsequent immunoblot analysis were performed as described for panel A, except that transfected cells were treated with the indicated doses of tBHQ.
A number of mutant Keap1 proteins were examined for the presence of this slow migrating form in tBHQ-treated cells (Fig. 4A, lanes 3 to 12; data not shown). The slow migrating form of Keap1 was observed in lysates from tBHQ-treated COS1 cells transfected with the mutant Keap1 proteins, with the notable exception of the Keap1-C151S protein. Thus, a single cysteine residue in Keap1 (C151) provides a genetic link between posttranslational modifications that occur on Keap1 during oxidative stress and escape by Nrf2 from Keap1-dependent repression.
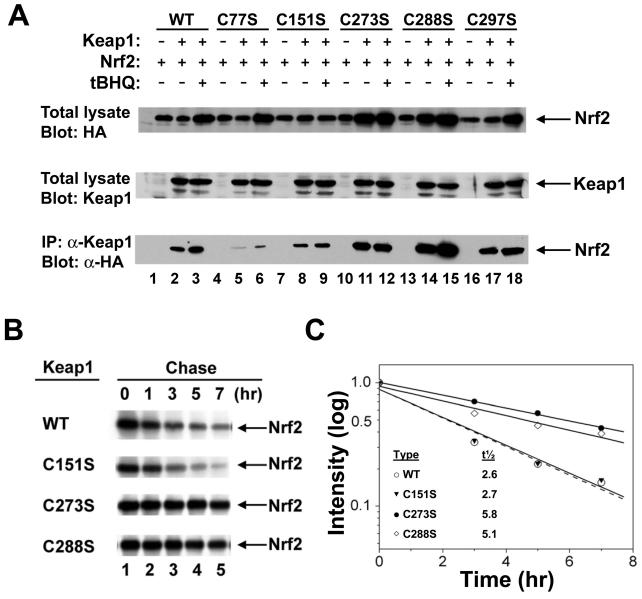
Cysteines 273 and 283 are required for Keap1-dependent degradation of Nrf2 under basal culture conditions.
In contrast to the Keap1-C151S protein,which functions as a constitutive repressor of Nrf2-dependent transcription, the Keap1-C273S and Keap1-C288S proteins are unable to efficiently repress Nrf2-dependent transcription. To determine whether the inability of the Keap1-C273S and Keap1-C288S proteins to efficiently repress Nrf2-dependent transcription is simply due to the failure of these mutant Keap1 proteins to associate with Nrf2, coimmunoprecipitation experiments were performed. COS1 cells were cotransfected with expression vectors for HA-Nrf2 and the wild-type or mutant Keap1 proteins. Cell lysates were immunoprecipitated with an affinity-purified anti-Keap1 antibody,and the presence of HA-Nrf2 in the anti-Keap1 immunoprecipitates was determined by immunoblot analysis. As expected, HA-Nrf2 was readily detectable in immunoprecipitates from cell lysates containing ectopically expressed wild-type Keap1 protein (Fig. 5A, lane 2). HA-Nrf2 was also readily detectable in anti-Keap1 immunoprecipitates from cell lysates containing the Keap1-C273S or the Keap1-C288S proteins (Fig. 5A, lower panel, lanes 11 and 14). The impaired ability of the Keap1-C273S and Keap1-C288S proteins to repress Nrf2-dependent transcription is not simply due to the inability of these mutant Keap1 proteins to associate with Nrf2.
FIG. 5.
Mutant Keap1 proteins increase the stability of Nrf2. (A) COS1 cells were cotransfected with an expression vector for HA-Nrf2 (all lanes) and expression vectors for either wild-type (WT) or mutant Keap1 proteins (lanes 2, 3, 5, 6, 8, 9, 11, 12, 14, 15, 17, and 18). The transfected cells were either untreated (lanes 1, 2, 4, 5, 7, 8, 10, 11, 13, 14, 16, and 17) or treated with 25 μM tBHQ for 16 h (lanes 3, 6, 9, 12, 15, and 18). Cell lysates were electrophoresed through an SDS-7.5% polyacrylamide gel and subjected to immunoblot analysis with anti-HA (α-HA) (top panel) or anti-Keap1 (α-Keap1) (middle panel) antibodies. Equivalent amounts of protein from each sample were also subjected to immunoprecipitation with an anti-Keap1 antibody. The immunoprecipitated proteins were electrophoresed through an SDS-7.5% polyacrylamide gel and subjected to immunoblot analysis with anti-HA (bottom panel). Only the relevant portions of the autoradiographs are shown. The Nrf2 and Keap1 proteins are indicated by arrows. +, present; −, absent. (B) COS1 cells were cotransfected with an expression vector for HA-Nrf2 and expression vectors for either wild-type Keap1 (top panel), Keap1-C151S (second panel from top), Keap1-C273S (third panel from top), or Keap1-C288S (bottom panel). Transfected cells were pulse labeled with media containing [35S]methionine and [35S]cysteine for 30 min and chased with media containing unlabeled methionine for 0, 1, 3, 5, and 7 h. Cell lysates were collected following the indicated chase periods and subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitated proteins were subjected to electrophoresis through an SDS-7.5% polyacrylamide gel and visualized by fluorography. The data shown are from one experiment. The pulse-chase analysis was repeated a second time with nearly identical results. (C) The relative intensities of immunoprecipitated Nrf2 were quantified by phosphorimager analysis and plotted on a semi-log graph. The amount of Nrf2 present at the end of the 30-min labeling period was set at 1. The value at each subsequent time point is the average of two individual experiments. The half-life of Nrf2 in the presence of the indicated Keap1 proteins is indicated in the inset.
Immunoblot analysis of total cell lysates revealed that Nrf2 levels were consistently elevated in COS1 cells cotransfected with expression vectors for HA-Nrf2 and either the Keap1-C273S or Keap1-C288S protein compared to cells cotransfected with expression vectors for HA-Nrf2 and the wild-type Keap1 protein (Fig. 5A, top panel, compare lanes 1 and 2 with lanes 10 and 11 and lanes 13 and 14). Pulse-chase labeling experiments were performed to determine whether the Keap1-C273S and Keap1-C288S proteins were able to alter the stability of the coexpressed Nrf2 protein (Fig. 5B). When coexpressed with the wild-type Keap1 protein in COS cells, the half-life of the HA-Nrf2 protein was found to be 2.6 h (Fig. 5C). Similarly, the half-life of the HA-Nrf2 protein in the presence of the Keap1-C151S protein was 2.7 h. However, the half-life of the HA-Nrf2 protein when coexpressed with the Keap1-C273S or Keap1-C288S protein was greater than 5 h (Fig. 5C).
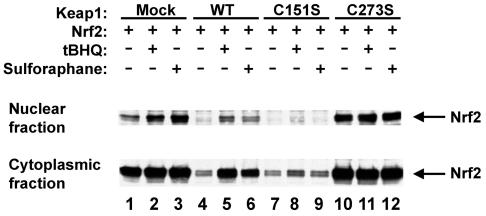
Elevated levels of HA-Nrf2 in the presence of coexpressed Keap1-C273S or Keap1-C288S proteins was observed in several different cell lines, most strikingly in the MDA-MB-231 breast cancer cell line (Fig. 6 and data not shown). The HA-Nrf2 protein is readily detectable in both cytoplasmic and nuclear fractions of singly transfected MDA-MB-231 cells (Fig. 6, lane 1). However, in cells transfected with expression vectors for both HA-Nrf2 and the wild-type Keap1 protein, the steady-state level of the cytoplasmic HA-Nrf2 protein was markedly decreased, and levels of nuclear HA-Nrf2 were below the level of detection by immunoblot analysis (Fig. 6, compare lanes 1 and 4). Coexpression of HA-Nrf2 with the Keap1-C151S protein also markedly reduced the steady-state levels of both nuclear and cytoplasmic HA-Nrf2 (Fig. 6, compare lanes 1 and 7). In contrast, coexpression of HA-Nrf2 with either the Keap1-C273S protein or the Keap1-C288S protein had no effect on steady-state levels of HA-Nrf2 in both the cytoplasmic and nuclear fractions of MDA-MB-231 cells (Fig. 6, compare lanes 1 and 10 and data not shown).
FIG. 6.
The Keap1-C151S protein blocks stabilization and subsequent nuclear translocation of Nrf2 by chemical inducers. Nuclear and cytoplasmic proteins were isolated from MDA-231 cells cotransfected with an expression vector for HA-Nrf2 (all lanes) and expression vectors for either wild-type (WT) Keap1 (lanes 4 to 6), Keap1-C151S (lanes 7 to 9), or Keap1-C273S (lanes 10 to 12). The transfected cells were either untreated (lanes 1, 4, 7, and 10), treated for 16 h with 25 μM tBHQ (lanes 2, 5, 8, and 11), or treated for 16 h with 4 μM sulforaphane (lanes 3, 6, 9, and 12) prior to isolation of nuclear and cytoplasmic proteins. Aliquots of nuclear and cytoplasmic proteins derived from equivalent numbers of cells were electrophoresed through an SDS-7.5% polyacrylamide gel and subjected to immunoblot analysis with anti-HA antibodies. +, present; −, absent.
The effects of the Keap1 proteins on protein stability of HA-Nrf2 in MDA-MB-231 cells paralleled the effects of the Keap1 proteins on steady-state levels of HA-Nrf2. The half-life of ectopically expressed HA-Nrf2 in the absence of Keap1 was more than 2 h, whereas the half-life of HA-Nrf2 in the presence of coexpressed wild-type Keap1 was approximately 40 min (Table 3). Coexpression of the Keap1-C151S protein with HA-Nrf2 resulted in a similar reduction in the half-life of HA-Nrf2. In contrast, the half-life of HA-Nrf2 in the presence of the Keap1-C273S protein was greater than 6 h (Table 3).
TABLE 3.
Half-life of HA-Nrf2a
| Keap1 proteinb | Treatmentc | t1/2 (h)d |
|---|---|---|
| None | − | 2.7 |
| None | + | 3.2 |
| WT | − | 0.6 |
| WT | + | 2.5 |
| C151S | − | 0.6 |
| C151S | + | 0.6 |
| C273S | − | 6.5 |
HA-Nrf2 was expressed in MDA-MB-231 cells in the absence or presence of Keap1 proteins and in the absence or presence of sulforaphane.
The indicated Keap1 proteins (Fig. 1B) were coexpressed in MDA-MB-231 cells with HA-Nrf2. WT, wild type.
Transfected MDA-MB-231 cells were treated with DMSO (−) or 4 μM sulforaphane (+) for 16 h prior to and during the pulse-chase experiment.
The half-life of HA-Nrf2 was determined by pulse-chase labeling.
Inducers of Nrf2-dependent transcription block Keap1-dependent degradation of Nrf2.
To further examine the relationship between Keap1-dependent degradation of Nrf2 and activation of Nrf2-dependent gene transcription by chemical inducers, MDA-MB-231 cells transfected with expression vectors for HA-Nrf2 and either the wild-type or mutant Keap1 proteins were exposed to either tBHQ or sulforaphane. Exposure of cells transfected with expression vectors for both HA-Nrf2 and wild-type Keap1 to either tBHQ or sulforaphane increased the abundance of HA-Nrf2 in both the nuclear and cytoplasmic fractions (Fig. 6, compare lane 4 with lanes 5 and 6). In contrast, steady-state levels of HA-Nrf2 in either the nucleus or the cytoplasm were not significantly altered by either tBHQ or sulforaphane in cells transfected with expression vectors for both HA-Nrf2 and the Keap1-C151S protein (Fig. 6, lanes 7 to 9). HA-Nrf2 levels in both the nucleus and the cytoplasm were elevated in untreated cells transfected with expression vectors for both HA-Nrf2 and the Keap1-C273S protein and were not further induced by tBHQ treatment (Fig. 6, lanes 10 to 12). Pulse-chase experiments were carried out to confirm that the sulforaphane-induced increase in steady-state levels of HA-Nrf2 resulted from a sulforaphane-dependent increase in the stability of HA-Nrf2. The half-life of HA-Nrf2 in the presence of the wild-type Keap1 protein was increased approximately fourfold in sulforaphane-treated cells, from approximately 40 min to more than 150 min (Table 3). In contrast, the half-life of HA-Nrf2 in the presence of the Keap1-C151S protein was approximately 40 min in both untreated and sulforaphane-treated cells. Taken together, these results indicate that exposure of cells to either tBHQ or sulforaphane blocks Keap1-dependent degradation of Nrf2, resulting in the stabilization of Nrf2 and increased nuclear translocation of Nrf2. In subsequent experiments, we have confirmed that both tBHQ and sulforaphane increase Nrf2-dependent reporter gene expression in MDA-MB-231 cells transfected with expression vectors for Nrf2 and the wild-type Keap1 protein (data not shown). In contrast, the Keap1-C151S protein functions as a constitutive repressor of Nrf2-dependent reporter gene expression in MDA-MB-231 cells (data not shown), in agreement with the reporter gene assays performed with NIH 3T3 cells (Fig. 3).
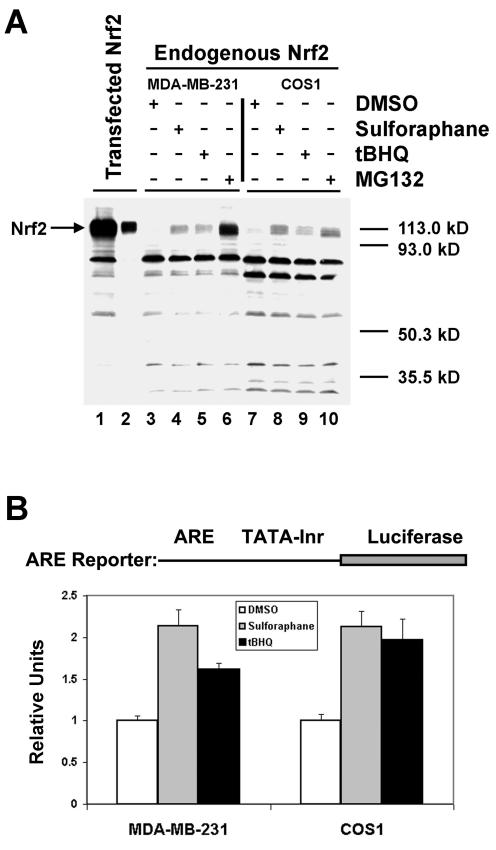
Several prior reports have demonstrated that endogenous Nrf2 is ubiquitinated in cells and is degraded in a proteosome-dependent manner (25, 36, 37). To confirm that the ability of the endogenous Nrf2 protein to activate ARE-dependent gene expression is regulated at the level of stability of the Nrf2 protein, the steady-state abundance of Nrf2 was examined by immunoblot analysis with both MDA-MB-231 cells and COS1 cells. Although ectopically expressed Nrf2 was readily detected in both MDA-MB-231 cells (Fig. 7A, lanes 1 and 2) and COS1 cells (data not shown), endogenous Nrf2 was not detectable in either cell type under basal culture conditions (Fig. 7A, lanes 3 and 7). In contrast, the endogenous Nrf2 protein was readily detectable following exposure of either MDA-MB-231 or COS1 cells to sulforaphane (Fig. 7A, lanes 4 and 8), tBHQ (Fig. 7A, lanes 5 and 9), or MG132 (Fig. 7A, lanes 6 and 10). Reporter gene assays performed in parallel cultures demonstrated that both sulforaphane and tBHQ were able to increase ARE-dependent gene expression in both cell types (Fig. 7B).
FIG. 7.
Stabilization of endogenous Nrf2 correlates with activation of ARE-dependent gene expression. (A) MDA-MB-231 and COS1 cells were either untreated (lanes 3 and 7), treated for 16 h with 10 μM sulforaphane (lanes 4 and 8), treated for 16 h with 25 μM tBHQ (lanes 5 and 9), or treated for 10 h with 10 μM MG132 (lanes 6 and 10). Whole-cell lysates were collected, and 50 μg of total cell protein from each lysate was electrophoresed through an SDS-7.5% polyacrylamide gel and subjected to immunoblot analysis with anti-Nrf2 antibodies. As a control, 50 μg (lane 1) or 5 μg (lane 2) of a whole-cell lysate derived from MDA-MB-231 cells transfected with an expression vector for Nrf2 was analyzed in parallel on the same immunoblot. (B) Transient transfection reporter gene assays were performed with MDA-MB-231 and COS1 cells essentially as described for Fig. 3. The cells were treated with 10 μM sulforaphane or 25 μM tBHQ for 16 h prior to analysis of reporter gene activity. +, present; −, absent.
Inhibition of Keap1-dependent degradation of Nrf2 does not result in quantitative release of Nrf2 from Keap1.
Keap1 is able to both sequester Nrf2 in the cytoplasm and target Nrf2 for proteosome-mediated degradation. The experimental results shown in Fig. 6 and 7 and in Table 3, together with several recently published reports (5, 19, 25, 29, 37), provide strong experimental support for the notion that inducers of Nrf2-dependent transcription inhibit Keap1-dependent targeting of Nrf2 for proteosome-mediated degradation. Inhibition of Keap1-dependent targeting of Nrf2 results in a marked increase in the steady-state levels of Nrf2 that is presumably responsible for increased transcriptional activation of ARE-dependent genes.
However, it is not clear how stabilization of Nrf2 in the cytoplasm leads to increased levels of nuclear Nrf2 and Nrf2-dependent gene expression. For example, the data shown in Fig. 6 indicate that only a small fraction of the total Nrf2 in tBHQ-treated cells transfected with expression vectors for both Nrf2 and Keap1 is present in the nuclear fraction while the bulk of the Nrf2 protein remains in the cytoplasm (Fig. 6, lanes 5 and 6, compare upper and lower panels). To determine whether cytoplasmic Nrf2 in tBHQ-treated cells was able to associate with Keap1, lysates from either untreated or tBHQ-treated COS1 cells cotransfected with expression vectors for both HA-Nrf2 and Keap1 were immunoprecipitated with anti-Keap1 antibodies. The immunoprecipitated proteins were analyzed for the presence of HA-Nrf2 by immunoblot analysis. Substantial HA-Nrf2 was recovered in anti-Keap1 immunoprecipitates from both untreated and tBHQ-treated COS1 cells (Fig. 5A, compare lanes 2 and 3). Consistent with this result, analysis of cells cotransfected with expression vectors for both Nrf2 and Keap1 by indirect immunofluorescence demonstrated that Nrf2 remains largely cytoplasmic in tBHQ-treated or sulforaphane-treated cells (data not shown). Thus, although inducers of Nrf2-dependent transcription inhibit Keap1-dependent degradation of Nrf2, these inducers do not cause quantitative release of Nrf2 from Keap1.
Cysteines 273 and 283 are required for Keap1-dependent ubiquitination of the N-terminal Neh2 domain of Nrf2.
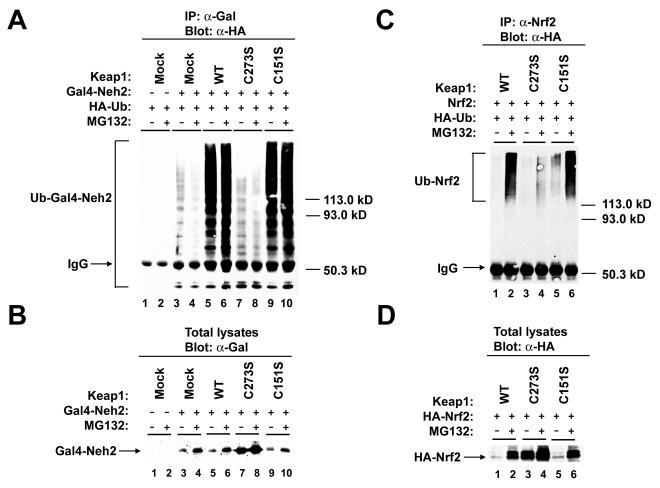
To determine if Keap1 targets Nrf2 for ubiquitin conjugation, MDA-MB-231 cells were cotransfected with an expression vector for HA-ubiquitin, an expression vector for a Gal4-Nrf2 fusion protein containing the N-terminal Neh2 domain (amino acids 1 to 97), and expression vectors for wild-type and mutant Keap1 proteins. The transfected cells were either left untreated or treated with MG132 and lysed in the presence of N-ethylmaleimide to block ubiquitin hydrolase activity. Cell lysates were immunoprecipitated with antibodies directed against the DNA-binding domain of Gal4 and the immunoprecipitated proteins were analyzed for the presence of HA-ubiquitin by immunoblot analysis. A ladder of anti-HA-immunoreactive polypeptides was detectable in anti-Gal4 immunoprecipitates derived from cells transfected with expression vectors for HA-ubiquitin and the Gal4-Neh2 fusion protein (Fig. 8A, lanes 3 and 4). Coexpression of either the wild-type Keap1 protein or the Keap1-C151S protein resulted in a marked increase in the levels of anti-HA immunoreactive polypeptides present in anti-Gal4 immunoprecipitates (Fig. 8A, lanes 5, 6, 9, and 10). In contrast, coexpression of the Keap1-C273S protein had no effect on the level of anti-HA immunoreactive polypeptides present in anti-Gal4 immunoprecipitates (Fig. 8A, lanes 7 and 8). Treatment of transfected cells with MG132 did not markedly increase detection of ubiquitinated Gal4-Nrf2 proteins, although the steady-state level of the Gal4-Nrf2 fusion protein was slightly increased by MG132 treatment (Fig. 8B). These results demonstrate that the wild-type Keap1 protein can increase ubiquitin conjugation onto the Gal4-Nrf2 protein. Furthermore, this Keap1-dependent increase in ubiquitination of the Gal4-Nrf2 fusion polypeptide is disrupted by the C273S substitution within Keap1.
FIG. 8.
Keap1-dependent ubiquitination of Nrf2 requires C273. (A) MDA-MB-231 cells were cotransfected with an expression vector for HA-ubiquitin (HA-Ub) (lanes 1 to 10), a Gal4-Neh2 fusion protein (lanes 3 to 10), or expression vectors for either wild-type (WT) Keap1 (lanes 5 and 6), Keap1-C273S (lanes 7 and 8), or Keap1-C151S (lanes 9 and 10). The transfected cells were exposed to DMSO (odd-numbered lanes) or MG132 (even-numbered lanes) for 5 h. Cells were lysed in 2% SDS and 10 mM NEM to block ubiquitin hydrolase activity. Anti-Gal4 (α-Gal) immunoprecipitates (IP) were analyzed by immunoblot with anti-HA (α-HA) antibodies. IgG, immunoglobulin G. (B) MDA-MB-231 cells were transfected with expression plasmids and treated with MG132 as described for panel A, except that no HA-ubiquitin plasmids were transfected. Total cell lysates were collected in sample buffer and subjected to immunoblot analysis with anti-Gal antibodies. (C) MDA-MB-231 cells were cotransfected with expression vectors for HA-ubiquitin and Nrf2 (lanes 1 to 6), and expression vectors for either wild-type Keap1 (lanes 1 and 2), Keap1-C273S (lanes 3 and 4), or Keap1-C151S (lanes 5 and 6). The transfected cells were exposed to DMSO (odd-numbered lanes) or MG132 (even-numbered lanes) for 5 h. Cells were lysed in 2% SDS and 10 mM NEM to block ubiquitin hydrolase activity. Anti-Nrf2 (α-Nrf2) immunoprecipitates were analyzed by immunoblot with anti-HA antibodies. (D) MDA-MB-231 cells were cotransfected with an expression vector for HA-Nrf2 (lanes 1 to 6) and for either wild-type Keap1 (lanes 1 and 2), Keap1-C273S (lanes 3 and 4), or Keap1-C151S (lanes 5 and 6). The transfected cells were exposed to DMSO (odd-numbered lanes) or MG132 (even-numbered lanes) for 5 h. Total cell lysates were collected in sample buffer and subjected to immunoblot analysis with anti-HA antibodies.
The ability of the Keap1-C273S protein to block ubiquitin conjugation to the full-length Nrf2 polypeptide was also determined. MDA-MB-231 cells were cotransfected with expression vectors for HA-ubiquitin, nontagged Nrf2, and the wild-type or mutant Keap1 proteins and either untreated or treated with MG132 prior to cell lysis. HA-ubiquitin was readily detected as a broad smear in anti-Nrf2 immunoprecipitates with an apparent molecular mass greater than 120 kDa from MG132-treated cells cotransfected with expression vectors for Nrf2 and the wild-type Keap1 protein (Fig. 8C, lane 2). MG132 treatment was necessary to detect ubiquitination of full-length Nrf2 in the presence of Keap1, presumably due to stabilization of Nrf2 (Fig. 8D). Abundant HA-ubiquitin conjugates were also present in anti-Nrf2 immunoprecipitates from MG132-treated cells that expressed the Keap1-C151S protein (Fig. 8C, lane 6). However, HA-ubiquitin conjugates in the anti-Nrf2 immunoprecipitates were markedly reduced in MG132-treated cells that were cotransfected with expression vectors for Nrf2 and the Keap1-C273S protein (Fig. 8C, lane 4).
DISCUSSION
Nrf2 is repressed by the Keap1 protein under basal conditions but can escape Keap1-mediated repression when cells are exposed to reactive chemicals, oxidative stress, or chemopreventive agents. In this report, we have identified three cysteine residues in Keap1 that have distinct roles in basal repression of Nrf2 and in the escape by Nrf2 from Keap1-mediated repression in response to chemical inducers. Two cysteine residues, C273 and C288, located in the central linker domain of Keap1, are required for basal repression of Nrf2 while a third residue located in the N-terminal BTB domain, C151, is required for induced escape by Nrf2 from Keap1-mediated repression. Furthermore, our results link these cysteine residues to the ability of Keap1 to target Nrf2 for ubiquitination and proteosome-dependent degradation, as both C273 and C288 are required for Keap1-dependent ubiquitination of Nrf2 while C151 is required for stabilization of Nrf2 in response to tBHQ-induced oxidative stress and sulforaphane. Our results support the hypothesis that these cysteine residues are components of a molecular switch that enables Keap1 to regulate steady-state levels of Nrf2 in response to perturbations in the intracellular redox environment.
A striking finding of our results is that Keap1 does not passively sequester Nrf2 in the cytoplasm but actively targets Nrf2 for ubiquitination and subsequent degradation by the proteosome. Our results are in agreement with several recent reports that steady-state levels of endogenous Nrf2 are increased upon treatment of cells with proteosome inhibitors (15, 25, 36, 37). Our results indicate that Keap1, by targeting the Neh2 domain of Nrf2 for ubiquitination, is a major regulator of Nrf2 stability. The N-terminal Neh2 domain of Nrf2 is sufficient for Keap1-dependent ubiquitination, suggesting that one or more of the seven lysine residues within the Neh2 domain are the acceptor site(s) of Keap1-targeted ubiquitination and are critical determinants of Nrf2 stability. Consistent with this notion, steady-state levels of the Gal4-Neh2 fusion protein are decreased by coexpression of Keap1 in a proteosome-dependent manner. However, it is likely that there are additional sequences within Nrf2 that contribute to the overall stability of Nrf2, as coexpression of Keap1 has a more pronounced effect on the steady-state level of full-length Nrf2 than on the Gal4-Neh2 fusion protein. Furthermore, it is possible that the stability of Nrf2 may also be regulated in a Keap1-independent manner (25), perhaps by additional ubiquitination sites outside the N-terminal Neh2 domain. Consistent with this notion, Keap1-independent ubiquitination of a Gal4 fusion protein containing the Neh2 domain and the central transactivation domain of Nrf2 is readily detectable in transfected cells (data not shown).
Our data indicate that the Keap1 protein targets Nrf2 for ubiquitination. It is most likely that Keap1-mediated ubiquitination of Nrf2 occurs in the context of an E3 ubiquitin ligase complex that includes, at a minimum, an E2 ubiquitin conjugating enzyme (32). Both single-subunit and multisubunit classes of E3 ubiquitin ligases have been described. The single-subunit E3 proteins are able to direct ubiquitination of a substrate with just an associated E2 ubiquitin conjugating enzyme while the multisubunit E3 ubiquitin ligases require additional protein components to accomplish ubiquitin transfer from the E2 protein to the substrate. It is not known at this point whether Keap1 is a single-subunit E3 ubiquitin ligase or whether it functions in the context of a larger complex to direct ubiquitination of Nrf2. If Keap1 functions in the context of a larger complex, it is possible that one or more of these Keap1-associated proteins that are required for Keap1-dependent ubiquitination of Nrf2 may be expressed in a cell type-specific manner. Cell type-specific differences in the expression of Keap1-associated proteins required for ubiquitination of Nrf2 may contribute to cell type-specific differences in the stability of Nrf2, including the marked difference in Nrf2 stability between COS1 (2.6 h) and MDA-MB-231 (40 min) cells. Furthermore, as ectopic expression of Keap1 in HepG2 cells can inhibit proteosome-mediated degradation of Nrf2 (36), HepG2 cells may lack one or more protein components required for Keap1-mediated degradation of Nrf2, such that ectopic expression of Keap1 sequesters Nrf2 in the cytoplasm away from the Keap1-independent degradation pathway(s).
Our results indicate that mutation of two critical cysteine residues in the linker domain abolishes ubiquitin transfer onto Nrf2 yet does not perturb the ability of Keap1 to associate with Nrf2. Two possible functions can be envisioned for these two critical cysteine residues. One possibility is that these two cysteine residues are simply required for binding of Keap1 to either an E2 ubiquitin conjugating enzyme or another component of a multisubunit E3 ubiquitin ligase complex. RING fingers, found in many proteins that are components of E3 ubiquitin protein ligase complexes, have a conserved arrangement of cysteine and histidine residues that bind a Zn ion and are thought to form a structural element required for assembly of a functional ubiquitin ligase complex. Although these residues in Keap1 do not have adjacent cysteine or histidine residues found in RING fingers (32), it is possible that C273 and C288 participate in binding a metal ion in the context of a homodimeric Keap1 protein. A second possibility is that C273 and C288 actively participate in ubiquitin transfer from the E2 protein to Nrf2. Although ubiquitin is brought into an E3 ubiquitin ligase complex as a thioester conjugate with an E2 ubiquitin conjugating enzyme, the E2 proteins require the participation of specific amino acids in E3 ubiquitin ligases to accomplish ubiquitin transfer to an acceptor lysine residue. For example, E6-AP, a HECT domain E3 ubiquitin ligase, has a specific cysteine residue that forms a thioester bond with ubiquitin during the transfer of ubiquitin from the associated E2 protein, Ubc H7, to the substrate, p53 (35). It will be of interest to determine whether either C273 or C288 forms thioester conjugates with ubiquitin.
The ability of chemical inducers of Nrf2-dependent transcription to stabilize Nrf2 is supported by both our experimental evidence and by several other recent reports (15, 19, 25, 29, 37). Our results demonstrate that Keap1 is the target of chemical inducers, as we have identified a mutant Keap1 protein that is a constitutive repressor of Nrf2-dependent transcription and blocks stabilization of Nrf2 by both tBHQ and sulforaphane. This mutant Keap1 protein contains a single cysteine-to-serine substitution at residue C151. At least two models can be envisioned that explain how Keap1-dependent ubiquitination of Nrf2 is disrupted by chemical inducers of Nrf2-dependent transcription. In one model, these inducers may directly modulate the oxidation status of either C273 or C288 and thereby inhibit Keap1-dependent ubiquitination of Nrf2. Both C273 and C288 display preferential reactivity with thiol-specific reagents in vitro (8), and our results demonstrate that serine substitutions at either of these residues disables Keap1-dependent ubiquitination of Nrf2. However, the Keap1-C151S protein, in which cysteine residues 273 and 288 are intact, functions as a constitutive repressor of Nrf2-dependent transcription. Furthermore, sulforaphane has no effect on the stability of Nrf2 in the presence of the Keap1-C151S protein. Thus, if modification of C273 or C288 by chemical inducers of Nrf2 is responsible for the inhibition of Keap1-dependent ubiquitination of Nrf2, our results indicate that a prior modification at C151 is required, perhaps to induce a conformational change that alters the accessibility of C273 and C288 to the cytoplasmic environment.
An alternative model is that C151 is the direct molecular target of chemical inducers of Nrf2-dependent transcription. We have identified a modified form of Keap1 that accumulates only upon exposure of cells to chemical inducers of Nrf2-dependent transcription. Importantly, formation of this modified form of Keap1 is abolished by the C151S substitution. Thus, the C151S mutation provides a genetic link between release of Nrf2 from Keap1-dependent repression and a posttranslational modification to Keap1. The molecular nature of the modification(s) responsible for the appearance of this modified form of Keap1 in cells is not known. The stability of the modified form of Keap1 under reducing conditions during electrophoresis through SDS-polyacrylamide gels indicates that the modified Keap1 protein is not simply a disulfide-linked dimeric complex. There is precedent for the participation of cysteine residues in the formation of amino acid cross-links that are stable under reducing conditions. For example, a cysteine residue located in the active site of galactose oxidase has been demonstrated to participate in a thioether linkage with a nearby tyrosine, thus forming an intramolecular amino acid cross-link (10). Given the marked decrease in the electrophoretic mobility of the modified form of Keap1, this modified form of Keap1 may result from an intermolecular protein ligation event. It should be noted that this modified form of Keap1 is present at substoichiometric levels in tBHQ-treated cells. In contrast, both tBHQ and sulforaphane increase the stability of Nrf2 in a nearly stoichiometric manner and stabilization of Nrf2 by these inducers is completely blocked by the Keap1-C151S protein. An attractive explanation is that the formation of the modified form of Keap1 simply reflects the ability of chemical inducers to modify the chemical reactivity of C151 in Keap1.
The results reported in this report, in conjunction with a number of recent reports that the Nrf2 protein is stabilized by chemical inducers of Nrf2-dependent transcription (15, 19, 25, 29, 37) have illuminated one important mechanism for Keap1-mediated repression and the ability of chemical inducers to enable Nrf2 to escape Keap1-mediated repression. It is likely that additional regulatory mechanisms cooperate with Keap1-dependent ubiquitination of Nrf2 to achieve precise regulation of Nrf2-dependent transcription. We note, for example, that although both tBHQ and sulforaphane treatment result in significant stabilization of Nrf2, only a small fraction of Nrf2 in transfected cells accumulates in the nucleus while the bulk of Nrf2 protein remains in the cytoplasm and can be immunoprecipitated with antibodies against Keap1. One possibility is that inhibition of Keap1-dependent degradation simply stabilizes Nrf2 and that additional posttranslational modifications to the Keap1-Nrf2 complex are required for dissociation of Nrf2 from Keap1. Phosphorylation of Nrf2 is an attractive candidate, as phosphorylation of Nrf2 on S40 by protein kinase C has been reported to reduce the affinity of Nrf2 for Keap1 (13). Another possibility is that accumulation of Nrf2 in the nucleus is also regulated by nuclear export. In this regard, we have recently identified an NES within Nrf2 that limits nuclear accumulation of Nrf2 following treatment of cells with chemical inducers of Nrf2 (unpublished data). It is likely that nuclear accumulation of Nrf2 is regulated at multiple levels, allowing for rapid yet precise regulation of Nrf2-dependent transcription in response to perturbations of the intracellular redox environment.
Acknowledgments
We gratefully acknowledge Bill Fahl, University of Wisconsin, for generous help and advice in the beginning stages of this project and for the gift of the pARE-luc reporter plasmid. We thank the Kazusa DNA Institute for the Keap1 cDNA clone, Y. Kan, University of California, San Francisco, for the gift of the Nrf2 cDNA clone, Shrikesh Sachdev for the expression vector for HA-ubiquitin, and Kent Gates and Alan Diehl for insightful advice. We thank Joyce Lo for sharing unpublished work with us.
This work was supported by the Molecular Biology Program and the Food for the 21st Century program at the University of Missouri, by research grants from the NIH (GM59213 and ES11721), and by the University of Missouri Research Board.
REFERENCES
- 1.Adams, J., R. Kelso, and L. Cooley. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10:17-24. [DOI] [PubMed] [Google Scholar]
- 2.Block, G., B. Patterson, and A. Subar. 1992. Fruit, vegetables, and cancer prevention: a review of the epidemiological evidence. Nutr. Cancer 18:1-29. [DOI] [PubMed] [Google Scholar]
- 3.Buschmann, T., S. Y. Fuchs, C. G. Lee, Z. Q. Pan, and Z. Ronai. 2000. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell 101:753-762. [DOI] [PubMed] [Google Scholar]
- 4.Chan, J. Y., and M. Kwong. 2000. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta 1517:19-26. [DOI] [PubMed] [Google Scholar]
- 5.Chanas, S. A., Q. Jiang, M. McMahon, G. K. McWalter, L. I. McLellan, C. R. Elcombe, C. J. Henderson, C. R. Wolf, G. J. Moffat, K. Itoh, M. Yamamoto, and J. D. Hayes. 2002. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 365:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaunay, A., D. Pflieger, M. B. Barrault, J. Vinh, and M. B. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471-481. [DOI] [PubMed] [Google Scholar]
- 7.Dhakshinamoorthy, S., and A. K. Jaiswal. 2001. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene 20:3906-3917. [DOI] [PubMed] [Google Scholar]
- 8.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. Wakabayashi, Y. Katoh, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99:11908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinkova-Kostova, A. T., M. A. Massiah, R. E. Bozak, R. J. Hicks, and P. Talalay. 2001. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc. Natl. Acad. Sci. USA 98:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firbank, S. J., M. S. Rogers, C. M. Wilmot, D. M. Dooley, M. A. Halcrow, P. F. Knowles, M. J. McPherson, and S. E. Phillips. 2001. Crystal structure of the precursor of galactose oxidase: an unusual self-processing enzyme. Proc. Natl. Acad. Sci. USA 98:12932-12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guyton, K. Z., and T. W. Kensler. 2000. Chemoprotection by phase 2 enzyme induction, p. 54-68. In G. M. Williams and O. I. Aruoma (ed.), Molecular drug metabolism in toxicology. OICA International Ltd., London, United Kindgom.
- 12.Hayes, J. D., S. A. Chanas, C. J. Henderson, M. McMahon, C. Sun, G. J. Moffat, C. R. Wolf, and M. Yamamoto. 2000. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem. Soc. Trans. 28:33-41. [DOI] [PubMed] [Google Scholar]
- 13.Huang, H. C., T. Nguyen, and C. B. Pickett.2002. Phosphorylation of Nrf2 at Ser40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 77:2769-274. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Connor, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 16.Kelloff, G. J., J. A. Crowell, V. E. Steele, R. A. Lubet, W. A. Malone, C. W. Boone, L. Kopelovich, E. T. Hawk, R. Lieberman, J. A. Lawrence, I. Ali, J. L. Viner, and C. C. Sigman. 2000. Progress in cancer chemoprevention: development of diet-derived chemopreventive agents. J. Nutr. 130:467S-471S. [DOI] [PubMed] [Google Scholar]
- 17.Kensler, T. W., T. J. Curphey, Y. Maxiutenko, and B. D. Roebuck. 2000. Chemoprotection by organosulfur inducers of phase 2 enzymes: dithiolethiones and dithiins. Drug Metab. Drug Interact. 17:3-22. [DOI] [PubMed] [Google Scholar]
- 18.Kong, A. N., E. Owuor, R. Yu, V. Hebbar, C. Chen, R. Hu, and S. Mandlekar. 2001. Induction of xenobiotic enzymes by the MAP kinase pathway and the antioxidant or electrophile response element (ARE/EpRE). Drug Metab. Rev. 33:255-271. [DOI] [PubMed] [Google Scholar]
- 19.Kwak, M. K., K. Itoh, M. Yamamoto, and T. W. Kensler. 2002. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 22:2883-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak, M. K., K. Itoh, M. Yamamoto, T. R. Sutter, and T. W. Kensler. 2001. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol. Med. 7:135-145. [PMC free article] [PubMed] [Google Scholar]
- 21.Kwak, M. K., N. Wakabayashi, K. Itoh, H. Motohashi, M. Yamamoto, and T. W. Kensler. 2003. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 278:8135-8145. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. M., J. M. Hanson, W. A. Chu, and J. A. Johnson. 2001. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J. Biol. Chem. 276:20011-20016. [DOI] [PubMed] [Google Scholar]
- 23.Li, J., J. M. Lee, and J. A. Johnson. 2002. Microarray analysis reveals an antioxidant responsive element-driven gene set involved in conferring protection from an oxidative stress-induced apoptosis in IMR-32 cells. J. Biol. Chem. 277:388-394. [DOI] [PubMed] [Google Scholar]
- 24.McMahon, M., K. Itoh, M. Yamamoto, S. A. Chanas, C. J. Henderson, L. I. McLellan, C. R. Wolf, C. Cavin, and J. D. Hayes. 2001. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 61:3299-3307. [PubMed] [Google Scholar]
- 25.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes.2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of ARE-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 26.Mezghrani, A., A. Fassio, A. Benham, T. Simmen, I. Braakman, and R. Sitia. 2001. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 20:6288-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moi, P., K. Chan, I. Asunis, A. Cao, and Y. W. Kan. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 91:9926-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motohashi, H., T. O'Connor, F. Katsuoka, J. Engel, and M. Yamamoto. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1-12. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen, T., P. J. Sherratt, H. C. Huang, C. S. Yang, and C. B. Pickett. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26S proteasome. J. Biol. Chem. 278:4536-4541. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43:233-260. [DOI] [PubMed] [Google Scholar]
- 31.Owuor, E., and A. Kong. 2002. Antioxidants and oxidants regulated signal transduction pathways. Biochem. Pharmacol. 64:765-770. [DOI] [PubMed] [Google Scholar]
- 32.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 33.Ramos-Gomez, M., M.-K. Kwak, P. M. Dolan, K. Itoh, M. Yamamoto, P. Talalay, and T. W. Kensler. 2001. Sensitivity to carcinogenesis is increased and chemopreventive efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA 98:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 35.Scheffner, M., U. Nuber, and J. M. Huibregtse. 1995. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373:81-83. [DOI] [PubMed] [Google Scholar]
- 36.Sekhar, K. R., X. X. Yan, and M. L. Freeman. 2002. Nrf2 degradation by the ubiquitin proteasome pathway is inhibited by KIAA0132, the human homolog to INrf2. Oncogene 21:6829-6834. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, D., E. Killeen, R. Naquin, S. Alam, and J. Alam. 2003. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 278:2396-2402. [DOI] [PubMed] [Google Scholar]
- 38.Talalay, P., and J. W. Fahey. 2001. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J. Nutr. 131:3027S-3033S. [DOI] [PubMed] [Google Scholar]
- 39.Thimmulappa, R. K., K. H. Mai, S. Srisuma, T. W. Kensler, M. Yamamoto, and S. Biswal. 2002. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 62:5196-5203. [PubMed] [Google Scholar]
- 40.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 41.Wasserman, W. W., and W. E. Fahl. 1997. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA 94:5361-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, C. S., J. M. Landau, M. T. Huang, and H. L. Newmark. 2001. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu. Rev. Nutr. 21:381-406. [DOI] [PubMed] [Google Scholar]
- 43.Yoh, K., K. Itoh, A. Enomoto, A. Hirayama, N. Yamaguchi, M. Kobayashi, N. Morito, A. Koyama, M. Yamamoto, and S. Takahashi. 2001. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 60:1343-1353. [DOI] [PubMed] [Google Scholar]
- 44.Yu, R., C. Chen, Y. Y. Mo, V. Hebbar, E. D. Owuor, T. H. Tan, and A. N. Kong. 2000. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J. Biol. Chem. 275:39907-39913. [DOI] [PubMed] [Google Scholar]