Abstract
Fission yeast Mrc1 (mediator of replication checkpoint 1) is an adaptor checkpoint protein required for Rad3-dependent activation of the checkpoint kinase Cds1 in response to arrest of replication forks. Here we report studies on the regulation of Mrc1 by phosphorylation. Replication arrest induced by hydroxyurea (HU) induces Mrc1 phosphorylation that is detected by a change in Mrc1 electrophoretic mobility. Phosphorylation is maintained in cds1Δ, rad3Δ, and tel1Δ single mutants but eliminated in a rad3Δ tel1Δ double mutant. Mrc1 has two clusters of S/TQ motifs that are potential Rad3/Tel1 phosphorylation sites. Mutation of six S/TQ motifs in these two clusters strongly impairs Mrc1 phosphorylation. Two motifs located at S604 and T645 are vital for HU resistance. The T645A mutation strongly impairs a Cds1-Mrc1 yeast two-hybrid interaction that is dependent on a functional forkhead-associated (FHA) domain in Cds1, indicating that phosphorylation of T645 mediates Mrc1's association with Cds1. Consistent with this model, the T645 region of Mrc1 effectively substitutes for the T11 region of Cds1 that is thought to be phosphorylated by Rad3 and to mediate FHA-dependent oligomerization of Cds1. The S/TQ cluster that includes S604 is needed for Mrc1's increased association with chromatin in replication-arrested cells. These data indicate that Rad3 and Tel1 regulate Mrc1 through differential phosphorylation to control Cds1.
The fission yeast Schizosaccharomyces pombe has been a valuable model system for studies of genome surveillance and checkpoint mechanisms that control cell cycle progression and regulate DNA repair. Two distinct DNA structure checkpoints have been defined in fission yeast. One, known as the DNA replication checkpoint, is operative only during S phase and is activated in response to arrest of replication forks. The other system, known as the DNA damage checkpoint, is functional during G2 phase and is activated by various forms of DNA damage. Rad3, functioning in a complex with Rad26, acts as a sensor protein in both checkpoint mechanisms (5). Rad3 and the related kinase Tel1, which is involved in telomere maintenance (22), belong to a subgroup of a phosphatidylinositol 3-kinase (PI3K)-like family that includes the human homologs ATM (ataxia-telangiectasia mutated; similar to Tel1) and ATR (ATM and Rad3 related) (5). These kinases are thought to recognize certain abnormal DNA structures and act together with other checkpoint proteins to phosphorylate downstream effector protein kinases. Upon inhibition of DNA replication, Rad3 appears to directly phosphorylate the effector kinase Cds1 at threonine-11 (28). Activated Cds1 enforces the S-M checkpoint by regulating Cdc25 and Mik1 (24, 31). When DNA is damaged during G2 phase, Rad3 appears to directly phosphorylate the effector kinase Chk1 at serine-345 (7, 21). Regulation of Cdc25 and Mik1 by activated Chk1 controls the inhibitory phosphorylation of the cyclin-dependent kinase Cdc2 (3, 12, 13, 31).
Signal transduction from PI3K-like kinases to effector kinases is mediated by adaptor proteins such as Mrc1 (mediator of replication checkpoint 1). In both fission yeast and budding yeast, Mrc1 was identified as a vital component of the system that activates Cds1 (or its budding yeast homolog Rad53) in response to replication arrest (2, 29). Cds1 interacted with Mrc1 in the yeast two-hybrid system (29), indicating that a direct physical interaction between the two proteins may be necessary for Cds1 activation. Exactly how Mrc1 mediates Cds1/Rad53 activation is unknown. In budding yeast, Rad53 has a second adaptor protein known as Rad9. DNA damage induces Rad9 phosphorylation in a Mec1 (Rad3 homolog)/Tel1-dependent manner (11). Mec1 and Tel1 appear to phosphorylate Rad9 at multiple sites within S/TQ motifs that are preferred phosphorylation sites for PI3K-like kinases, and these phosphorylation events have been implicated in activation of Rad53 (25). Forkhead-associated (FHA) domain-mediated recognition of Rad9 phosphothreonine peptides is thought to couple Rad53 activation to the DNA damage checkpoint pathway. The FHA domain is a key domain of Cds1 and its homologs. The FHA domain was first identified in forkhead transcription factors and later shown to be present in many proteins with diverse functions (4). The FHA domain mediates protein-protein interactions (27). The FHA domain optimal binding sequence is TXXD, and phosphothreonine has much higher affinity than phosphoserine (9, 10).
Xenopus and human claspin proteins appear to be structurally and functionally related to yeast Mrc1 proteins. In both organisms, Chk1 is phosphorylated in an ATR and claspin-dependent manner in response to interference with replication (8, 16). In Xenopus, claspin becomes phosphorylated at S864 and S895, and phosphoclaspin binds to Chk1 (17). ATR is required for claspin phosphorylation, which probably occurs through an indirect mechanism because ATR cannot directly phosphorylate claspin at S864 and S895 in vitro. Claspin associates with chromatin during S phase in a manner that depends on the prereplication complex and Cdc45, but not on ATR or replication protein A (18). Claspin's chromatin association increases in response to replication arrest.
Cds1 activation appears to require Rad3-dependent phosphorylation of T11, but exactly how Rad3 targets Cds1 and how T11 phosphorylation leads to Cds1 activation are unknown. In budding yeast, it is thought that phosphorylation of Rad9 by Mec1/Tel1 induces Rad9 dimerization (26) and consequent recruitment of Rad53, bringing Rad53 proteins in close association and leading to intermolecular autophosphorylation of Rad53 (27). It has not been reported whether Rad53 activation requires direct phosphorylation by Mec1/Tel1. In human cells, studies suggest that phosphorylation of Chk2 at T68 allows binding of another Chk2 molecule through an FHA domain interaction (1, 30). This dimerization leads to Chk2 intermolecular autophosphorylation in the activation loop of the kinase domain.
In this report, we show that fission yeast Mrc1 is phosphorylated in response to replication arrest in a Rad3/Tel1-dependent manner. Two S/TQ clusters are required for the electrophoretic mobility shift induced by this phosphorylation. Threonine-645, a site in the first S/TQ cluster, is required for the interaction with the FHA domain of Cds1. Serine-604, located in the second S/TQ cluster, appears to be involved in Mrc1's association with chromatin. These findings identify two mechanisms controlling Mrc1's function in the replication checkpoint response.
MATERIALS AND METHODS
Fission yeast strains, growth medium, and genetic and molecular methods.
All of the strains listed in Table 1 were leu1-32 ura4-D18. Standard growth media and general biochemical and genetic methods were used. Yeast cultures were grown at 32°C in YES medium (0.5% yeast extract, 3% glucose, supplements) or Edinburgh minimal medium unless indicated otherwise. Hydroxyurea (Sigma) was used at the indicated concentration. To mutate phosphorylation sites at the mrc1 genomic locus, a site-directed mutagenesis kit (Stratagene) was used to mutate the indicated site(s) in plasmid pUC28-Mrc1. PCR was then used to amplify the 1357-to-2704 region of mrc1. Purified PCR products were transformed into an mrc1::ura4+ strain that had ura4+ inserted at nucleotide 1612. 5-Flouroorotic acid-resistant transformants were picked, and mutations were confirmed by sequencing. Other regions of mrc1 were sequenced to detect spurious mutations.
TABLE 1.
Yeast strains used in this study
| Straina | Genotype | Origin or reference |
|---|---|---|
| PR109 | h− | Our laboratory stock |
| HZ3345 (HZ100) | h− mrc1-S599A | |
| HZ3346 (HZ101) | h− mrc1-S604A | |
| HZ3347 (HZ102) | h− mrc1-S614A | |
| HZ3348 (HZ103) | h− mrc1-T634A | |
| HZ3349 (HZ104) | h− mrc1-S637A | |
| HZ3350 (HZ105) | h− mrc1-T645A | |
| HZ3351 (HZ106) | h− mrc1-S599AS604AS614A | |
| HZ3352 (HZ107) | h− mrc1-T634AS637AT645A | |
| HZ3353 (HZ108) | h− mrc1-S599AS604AS614AT634AS637AT645A | |
| KT2791 | h− mrc1-13myc::kanMX6 | 25 |
| HZ3354 (HZ109) | h− mrc1-S599AS604AS614A-13myc::kanMX6 | |
| HZ3355 (HZ110) | rad3::ura4+tel1::kan+mrc1-13myc::kanMX6 | |
| HZ3356 (HZ111) | h− mrc1-S604A chk1::ura4+ | |
| HZ3357 (HZ112) | h− mrc1-S599AS604AS614A chk1::ura4+ | |
| HZ3358 (HZ113) | h− mrc1-S599AS604AS614AT634AS637AT645A chk1::ura4+ | |
| BM2681 | h+ cds1-13myc::kanMX6 | Our laboratory stock |
| HZ3359 (HZ114) | h+ cds1::ura4+-13myc::kanMX6 | |
| HZ3360 (HZ115) | h+ cds1-S2T-13myc::kanMX6 | |
| HZ3361 (HZ116) | h+ cds1-T634AS637AT645A-13myc::kanMX6 | |
| HZ3362 (HZ117) | h+ cds1-T645A-13myc::kanMX6 | |
| KT2902 | rad3::leu2+ cds1::ura4+ | |
| KT2870 | h− mrc1-TAP::kanMX6 |
All are leu1-32 ura4-D18. The designations in parentheses are used in the text.
Cds1-S2T hybrid.
The ura4+ marker was inserted into cds1 between codons encoding glutamate-13 and alanine-14 to make a cds1::ura4+ null mutant. A PCR-based gene targeting method was used to delete the T8-to-S19 region by knocking in T629 to T650 of Mrc1. Plasmids pUC28-Mrc1, pUC28-Mrc1-T645A, and pUC28-S2TA were used as templates with which to amplify the Mrc1 T629-to-T650 region containing either the wild-type sequence, a T645A mutation, or a T634A S637A T645A triple mutation. PCR products were transformed into the cds1::ura4+ null mutant described above, and 5-flouroorotic acid-resistant colonies were selected for further confirmation and analyses.
ATM kinase assays.
PCR was performed to amplify cDNA expressing either the wild-type Mrc1 C terminus (amino acids 560 to 1019) or Mrc1 phosphorylation site mutant forms. PCR fragments were inserted into pGEX-KG (14). Glutathione S-transferase (GST)-Mrc1 fusion proteins were expressed in bacteria and purified with glutathione-Sepharose (Amersham Pharmacia). ATM was immunoprecipitated from HeLa cells and used in kinase assays as described previously (28).
Triton extraction and immunolocalization.
Asynchronous cells were treated with hydroxyurea (HU; 15 mM) for 2 to 3 h. Cells were harvested and subjected to Triton X-100 extraction to analyze chromatin association of Mrc1 as previously described (15). After extraction with 1% Triton X-100, cells were washed once and then fixed with formaldehyde and subjected to indirect immunofluorescence studies with 9E10 antibody as previously described (20). For DNase I treatment, cells were incubated with 5 U of DNase I (Sigma) together with Triton as previously described (15). Anti-myc (9E10 Covance) and Alexa Fluor 488 goat anti-mouse immunoglobulin G (IgG; Molecular Probes) were used as primary and secondary antibodies, respectively.
Antibody production and immunoblotting.
The Mrc1 N-terminal fragment including amino acids 1 to 317 was expressed as a GST-Mrc1 fusion protein in bacteria, purified, and used to make antisera in rabbits. Purified anti-Mrc1 antibody was verified by immunoblotting. To make anti-phospho-S604 antibody, the phosphorylated peptide CSQPSA-pS-QLT-amide was conjugated to keyhole limpet hemocyanin and injected into rabbits. Antisera were affinity purified with the phosphorylated peptide and depleted with the unphosphorylated form of the peptide. The purified antibodies were used for immunoblotting. For anti-Mrc1 immunoblotting, cells were harvested, washed, and then resuspended in loading buffer (50 mM Tris HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 10% β-mercaptoethanol, 0.1% bromophenol blue). Cells were boiled at 100°C for 5 min, vortexed with glass beads for 5 min, and boiled again for 5 min. Cell lysates were centrifuged for 10 min at 20,000 × g. Supernatants were subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Immunoblots were blotted with anti-Mrc1 antibody or anti-myc antibody (9E10; Covance). For anti-Cds1 immunoblotting, cells were grown for 2 h in selective medium supplemented with 15 mM HU. Cells were lysed by glass beads in a mixture of 50 mM Tris-HCl (pH 8), 150 mM NaCl, 0.2% NP-40, 5 mM EDTA, 10% glycerol supplemented with protease and phosphatase inhibitors. Total lysates were sonicated briefly and then immunoprecipitated with myc-agarose conjugate (Santa Cruz), and precipitates were used for immunoblotting with Cds1 antibody (gift from T. Wang). The secondary antibody used was horseradish peroxidase-conjugated anti-rabbit IgG (1:10,000) or anti-mouse IgG (1:4,000).
HU sensitivity and checkpoint studies.
Phosphorylation site mutants were grown in YES medium and then transferred to YES plates with HU at the indicated concentrations. Cells were grown for 2 days. To study the significance of DNA replication checkpoint phosphorylation mutations, a PCR-based gene-targeting method was used to delete chk1 in mrc1 phosphorylation site mutants. Log-phase cells grown in YES medium were treated with 15 mM HU for 6 h. Cells were fixed with glutaraldehyde and then stained with 4′,6′-diamidino-2-phenylindole (DAPI) before microscopic analysis.
Yeast two-hybrid assay.
To make pGAD424-S2T and its mutant form, two oligonucleotides (forward, 5′-AATTCACCAACACCTCATCTACACAGCCGAGTCAAGTAGATAGTCTAGTTCCTACTCAATTAGATTCCGG-3′; reverse,5′-GATCCCGGAATCTAATTGAGTAGGAACTAGACTATCTACTTGACTCGGCTGTGTAGATGAGGTGTTGGTG-3′) were denatured in 100°C water and annealed in 10 mM Tris Cl (pH 7.9)-50 mM NaCl-10 mM MgCl2-1 mM dithiothreitol. The annealing temperature was decreased gradually from 100 to 65°C. The annealed fragments with EcoRI and BamHI sticky ends were subcloned into pGAD424 (Clontech) to make pGAD424-S2T. This plasmid was then used as the template to make a T634A, S637A, or T645A mutation. The pGAD424-Mrc1 plasmid (29) was used to make phosphorylation mutant forms with full-length Mrc1. The Cds1 FHA mutant form was amplified from the pAS2-cds1 FHA mutant form (6) and subcloned into pBTM116 to make LexDB-cds1-fha. pGAD424 and pBTM116 derivatives were cotransformed into KT2906 cells (29), and transformants were picked to test their growth on minimum medium without histidine and supplemented with the indicated amount of 3-aminotriazole.
RESULTS
Rad3 and Tel1 control Mrc1 phosphorylation.
In response to HU treatment, fission yeast Mrc1 undergoes a posttranslational modification that retards its electrophoretic mobility (Fig. 1A). Treatment with lambda phosphatase showed that this modification was phosphorylation (Fig. 1A). To understand how Mrc1 phosphorylation is regulated, we monitored Mrc1 mobility in several checkpoint mutant backgrounds (Fig. 1B). In a cds1Δ strain, substantial Mrc1 phosphorylation was detected in the absence of any treatment (lane 3) and more phosphorylation occurred upon HU treatment (lane 4). These data suggested that the absence of Cds1 may lead to DNA replication abnormalities or DNA damage that result in Mrc1 phosphorylation. This pattern was markedly different from that seen in budding yeast (2), in which HU-induced phosphorylation of Mrc1 was substantially decreased in rad53Δ cells and no phosphorylation of Mrc1 was detected in the absence of treatment with HU or methyl methanesulfonate.
FIG. 1.
Rad3 and Tel1 are required for Mrc1 phosphorylation. The indicated strains were grown in YES medium supplemented with 12 mM HU for 4 h. Cells lysates were then subjected to Western blotting with anti-Mrc1 antibody or anti-tubulin antibody as a control. (A) Mrc1 is phosphorylated in response to HU treatment. Mrc1-TAP cells were treated with HU, and cell lysates were either mock treated or treated with phosphatase (λPPase) or sodium orthovanadate together with phosphatase before being subjected to SDS-PAGE. (B) Wild-type (WT) cells with the indicated genetic background were treated with HU and then lysed for SDS-PAGE. Anti-Mrc1 sera were used in this immunoblot assay.
Mrc1 phosphorylation was largely intact in rad3Δ and tel1Δ single mutants but fully abrogated in a rad3Δ tel1Δ double mutant (Fig. 1B). In contrast to the cds1Δ mutant, no Mrc1 phosphorylation was observed in a rad3Δ cds1Δ double mutant in the absence of HU (Fig. 1B). These findings suggest that Rad3 and Tel1 have overlapping roles in controlling Mrc1 phosphorylation in response to HU-induced replication arrest. However, in the absence of Cds1 and HU, only Rad3 is involved in the control of Mrc1 phosphorylation.
HU sensitivity of Mrc1 S/TQ cluster mutants.
Control of Mrc1 phosphorylation by Rad3 and Tel1 was interesting in view of the fact that Mrc1 has a preponderance of SQ and TQ motifs that are potential Rad3 and Tel1 phosphorylation sites. There are 14 S/TQ sequences in Mrc1, 7 of which are found in two clusters near the central region of the polypeptide. The 3S region contains SQ motifs at positions 599, 604, and 614. The S2T cluster includes S/TQ motifs at threonine-634, serine-637, and threonine-645 (Fig. 2A). (There is also a TQ motif at threonine-653.) Interestingly, the Saccharomyces cerevisiae Rad9 and Mrc1 proteins have similar S/TQ clusters. We performed a series of site-directed mutagenesis studies of these S/TQ clusters to evaluate if they are important for Mrc1 function. All mutations were made by conversion of the endogenous genomic locus. Mutations of the 3S cluster (3SA; S599A S604A S614A) caused severe HU sensitivity (Fig. 2B). Mutations of the S2T cluster (S2TA; T634A S637A T645A) also conferred sensitivity to HU, although to a slightly lesser extent than 3SA mutations. Mutations of both S/TQ domains showed an additive effect (Fig. 2B). The HU sensitivity of mrc1-6A cells is almost the same as that of an mrc1Δ mutant (data not shown).
FIG. 2.
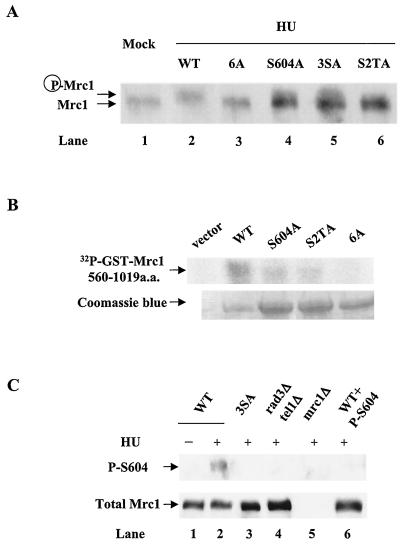
Phosphorylation of Mrc1 is required for cell survival in the presence of HU. (A) Schematic diagram of Mrc1. All of the potential Rad3/Tel1 phosphorylation sites are indicated. Two S/TQ clusters are designated 3S (for S599, S604, and S614) and S2T (for T634, S637, and T645). 6S/T indicates the combination of these two. a.a., amino acids. (B) Two S/TQ clusters are required for cell survival in the presence of HU. Serial dilutions of wild-type (WT) cells or cells with mutations of either one of the S/TQ clusters (3SA or S2TA) or both (6A) were plated on either YES medium or YES medium supplemented with the indicated amount of HU. (C) S604 and T645 are important residues for cell survival in the presence of HU. Cells with the indicated single mutations or 3SA or S2TA multiple mutations were subjected to spot assay on YES medium or YES medium supplemented with the indicated concentration of HU.
To further refine the analysis of the S/TQ clusters, cells with single mutations of S/TQ motifs in these regions were compared for sensitivity to HU (Fig. 2C). The S604A mutation caused severe HU sensitivity, similar to that caused by the 3SA triple mutation. The S599A and S614A mutations had no effect on HU survival (Fig. 2C), suggesting that S604 is the most important residue in the 3S cluster. Tests of single mutations in the S2T cluster showed that T645A caused HU sensitivity to a degree similar to that caused by the S2TA triple mutation. Mutants with S599A, S614A, T634A, and S637A single mutations showed the same HU resistance as wild-type cells (data not shown). These findings suggested that serine-604 and threonine-645, two putative Rad3/Tel1 phosphorylation sites, are critical for Mrc1 function in response to replication arrest.
Checkpoint defect of Mrc1 S/TQ cluster mutants.
In a chk1Δ background, mrc1Δ mutants fail to arrest division in HU and display a “cut” phenotype arising from division without prior replication (29). This defect is attributed to a failure to activate Cds1. To further assess the functional importance of the Mrc1 S/TQ clusters in the replication checkpoint response, we carried out microscopic observations of mutants treated with HU for 6 h at 30°C (Fig. 3). As expected, rad3Δ cells, which are unable to activate Cds1 and Chk1 in response to HU, displayed many cut cells, whereas the wild type and the chk1Δ, mrc1-3SA, and mrc1-6A mutants arrested division with very few cut cells. Double-mutant mrc1-S604A chk1Δ (25%) and mrc1-3SA chk1Δ (33%) cells showed a mixed phenotype of moderately elongated and many cut cells, while mrc1-6A chk1Δ (72%) cells displayed a phenotype very similar to that of rad3Δ cells. These findings demonstrated the importance of the Mrc1 S/TQ clusters in mediating the Cds1-dependent checkpoint arrest in response to HU.
FIG. 3.
Mrc1 phosphorylation is required for DNA replication checkpoint. Cells with the indicated mutations in a chk1+ or chk1Δ background were grown in YES medium at 30°C with 12 mM HU for 6 h. Cells were then fixed by the glutaraldehyde method and stained with DAPI. WT, wild type.
S/TQ cluster mutations impair Mrc1 phosphorylation in vivo and in vitro.
To evaluate if the S/TQ clusters are important for Mrc1 phosphorylation in vivo, cells with different phosphorylation site mutations were treated with HU and migration of endogenous Mrc1 protein on SDS-PAGE was monitored by immunoblotting with anti-Mrc1 antibody (Fig. 4A). In wild-type cells, HU-induced phosphorylation slowed Mrc1 migration on SDS-PAGE (Fig. 4A). The 6A mutation abolished the electrophoretic mobility change induced by HU, whereas the S604A, 3SA, and S2TA mutations had partial effects. These findings showed that S/TQ clusters are important for Mrc1 phosphorylation.
FIG. 4.
Phosphorylation of Mrc1 in vivo and in vitro. (A) S/TQ clusters are required for Mrc1 phosphorylation. Cells with either wild-type or mutant mrc1 were grown in YES medium supplemented with 15 mM HU for 2 h. Total lysates were subjected to SDS-PAGE and Western blotting with anti-Mrc1 antibody. (B) ATM phosphorylated Mrc1 at S/TQ clusters in vitro. ATM was immunoprecipitated from HeLa cells and used to phosphorylate substrates. The sequence including GST-Mrc1 amino acids 560 to 1019 and its phosphorylation mutant forms were expressed and purified from Escherichia coli and used as substrates for ATM kinase assays. Kinase assays were performed at 30°C for 30 min. The reaction mixtures were subjected to electrophoresis, and phosphoproteins were visualized by phosphorimager (Molecular Dynamics). The proteins were then detected by Coomassie blue staining. (C) Serine-604 is phosphorylated in vivo. Cells with the indicated genetic backgrounds were grown in YES medium or in YES medium supplemented with 15 mM HU for 2 h. Total lysates were subjected to Western blotting with phospho-S604 antibody (top). The blot was also stripped and reblotted with anti-Mrc1 antibody to visualize total Mrc1 protein levels (bottom). WT, wild type; a.a., amino acids.
To evaluate whether the S/TQ clusters are likely to be directly phosphorylated by Rad3/Tel1, in vitro kinase assays of the S/TQ clusters were performed with immunoprecipitated ATM from HeLa cells (Fig. 4B). ATM was used as a Rad3/Tel1 surrogate because the fission yeast kinases have very poor activity in vitro, whereas ATM is a much more robust kinase and its in vitro specificity is similar to that of Rad3 (21, 28). Mrc1 wild-type and mutant substrates were made by expressing the C-terminal region of Mrc1 (amino acids 560 to 1019) as a GST fusion protein in bacteria. ATM was able to phosphorylate wild-type GST-Mrc1(560-1019). The S604A single mutation and S2TA triple mutation substantially decreased 32P incorporation, while the 6A mutation appeared to abolish phosphorylation of GST-Mrc1(560-1019). These results indicate that serine-604 and one or more of the S/TQ motifs in the T634- to-T645 region are phosphorylated by ATM in vitro.
Serine-604 is phosphorylated in vivo upon HU treatment.
To directly analyze the phosphorylation status of Mrc1 at serine-604, affinity-purified phosphospecific S604 antibodies were made (see Materials and Methods) and used for immunoblotting (Fig. 4C). In wild-type cells treated with HU, these antibodies detected a protein that migrated at the position of Mrc1 (lane 2). This signal was not detected in untreated cells (lane 1), nor was it seen in mrc1-3SA (lane 3) or mrc1Δ (lane 5) mutants treated with HU. Preincubation of the phospho-S604 antibodies with the phospho-S604 peptide also eliminated the signal (lane 6). These findings strongly indicated that the antibodies specifically detected Mrc1 that was phosphorylated on serine-604. Importantly, the S604 phosphorylation signal was abolished in a rad3Δ tel1Δ double mutant (lane 4) treated with HU, suggesting that HU-induced phosphorylation at serine-604 is catalyzed by Rad3 and Tel1.
Threonine-645 mediates Mrc1 interaction with the Cds1 FHA domain.
Mrc1 interacts with Cds1 in the yeast two-hybrid system (29) (Fig. 5A). To determine if a functional FHA domain in Cds1 is required for this interaction, a cds1-fha mutant that has two highly conserved residues (serine-85 and histidine-88) mutated to alanine was tested in the two-hybrid assay. Previous studies showed that cds1-fha cells are highly sensitive to HU (6). We found that Cds1-FHA failed to interact with Mrc1, demonstrating the importance of the FHA domain in mediating the interaction with Mrc1 (Fig. 5A). To determine if the S/TQ clusters are important for this interaction, we mobilized several S/TQ cluster mutations into the yeast two-hybrid vector and assayed interactions with Cds1 (Fig. 5A). The 3SA mutation did not impair the interaction with Cds1. However, the S2TA mutation (T634A S637A T645A) greatly decreased the interaction. To further define the most important residue(s), the T634A, S637A, or T645A single mutation was mobilized into the two-hybrid vector and tested for interaction with Cds1. Mutation of either T634 or S637 did not affect the Mrc1-Cds1 interaction (data not shown); however, the T645A single mutation decreased Mrc1's interaction with Cds1 to the same extent as the S2TA triple mutation did. These finding correlate with the HU sensitivity of the S2TA triple mutant and the individual single mutants.
FIG. 5.
Cds1 FHA domain can interact with Mrc1 T645. (A) Mrc1 T645 is required for its interaction with Cds1. pGAD424-Mrc1 or its phosphorylation mutant forms and LexDB-Cds1 or its fha mutant form were cotransformed into strain L40 for a yeast two-hybrid assay. (B) The DNA fragment encoding amino acids 629 to 639 of Mrc1 was subcloned into pGAD424 to make pGAD424-S2T. pGAD424-S2T and LexDB-Cds1 or its fha mutant form were cotransformed into strain KT2906 for a yeast two-hybrid assay. AD, activation domain.
To determine if the S2T region of Mrc1 alone is capable of interacting with Cds1, the 21-amino-acid peptide containing the T634-to-T645 region of Mrc1 was fused to the Gal4 activation domain and used for the two-hybrid system (Fig. 5B). The short peptide interacted with Cds1, although not quite as well as full-length Mrc1. Consistent with the studies carried out with full-length Mrc1, the interaction of Cds1 with the short peptide containing the T634-to-T645 region of Mrc1 was dependent on an intact FHA domain (Fig. 5B).
T645 mediates direct binding with the Cds1 FHA domain.
In the course of these studies, we noticed that the spacing of the S/TQ motifs in the Mrc1 T634-to-T645 region (S/TQXS/TQX6S/TQ) was similar to that of the Cds1 T8-to-S19 region and the Rad53 T12-to-S24 region (Fig. 6A). We have previously reported evidence that phosphorylation of Cds1 threonine-11 by Rad3 is vital for Cds1 activation (28). Studies of mammalian Chk2 have provided evidence that phosphorylation of threonine-68 (equivalent to Cds1 threonine-11) mediates an interaction with the FHA domain of another Chk2 protein, promoting oligomerization that leads to intermolecular autophosphorylation and full kinase activation (1, 30). We reasoned that if both the Cds1 T8-to-S19 and Mrc1 T634-to-T645 regions mediate FHA domain interactions, it may be possible to replace the T8-to-S19 region of Cds1 with the T634-to-T645 region of Mrc1. To accomplish this substitution in the genomic copy of cds1+, we first made a cds1 disruption strain in which the ura4+ gene was inserted between the threonine-11 and serine-19 codons of cds1. A PCR-based gene-targeting method was used to replace the Cds1 T8-to-S19 region with the T634-to-T645 region of Mrc1. The wild type (S2T) and an S2TA mutant were used to create cds1-S2T (wild type) and cds1-S2TA (mutant) hybrids. Cells with cds1+, cds1Δ, cds1-S2T, or cds1-S2TA were compared for sensitivity to HU (Fig. 6B). As expected, wild-type cells were resistant to HU, whereas cds1Δ cells were acutely sensitive. Remarkably, the cds1-S2T strain was only slightly sensitive to HU, indicating that the T634-to-T645 region of Mrc1 effectively substituted for the T8-to-S19 region of Cds1 (Fig. 6B). In contrast, the cds1-S2TA strain, containing the triply mutated (T634A S637A T645A) T634-to-T645 region of Mrc1, was acutely sensitive to HU. To further determine if T645 is the most important residue, the T645A mutation was introduced into the cds1-S2T hybrid strain. Like cds1-S2TA cells, cds1-T645A hybrid cells were very sensitive to HU (Fig. 6B).
FIG. 6.
Mrc1 threonine-645 mediates direct interaction with Cds1 FHA domain. (A) Sequence homology between the Mrc1 T634-to-T645 and Cds1 T8-to-S19 regions. The numbers indicate the position of the sequence within the protein. Grey boxes indicate the conserved S/TQ sequences. (B) Mrc1 T634-to-T645 region can replace Cds1 T8-to-S19 region. BM2681 (Cds1-13myc), HZ114 (Δcds1::ura4+-13myc), HZ115 (Cds1-S2T-13myc), HZ116 (Cds1-S2TA-13myc), or HZ117 (Cds1-T645A-13myc) cells were spotted on either YES medium or YES medium supplemented with the indicated amount of HU to test their survival. (C) Mrc1 T645 is required for its interaction with Cds1. BM2681 (Cds1-13myc), HZ115 (Cds1-S2T-13myc), HZ116 (Cds1-S2TA-13myc), or HZ117 (Cds1-T645A-13myc) cells were transformed with a multicopy plasmid expressing either wild-type Cds1, its fha mutant form, or the T8AT11A phosphorylation mutant form. Cells were grown in selective minimal medium and treated with 15 mM HU for 2.5 h before being harvested. Lysates were immunoprecipitated with anti-myc antibody, and precipitates were subjected to Western blotting with anti-Cds1 antibody. Total lysates were also blotted with anti-Cds1 antibody as the control. IP, immunoprecipitate; OE, overexpression.
Having found that the Mrc1 T634-to-T645 region can effectively replace the T8-to-S19 region of Cds1, we explored whether the Mrc1 T634-to-T645 region of Cds1-S2T mediates an interaction with wild-type Cds1 in a manner akin to that proposed for oligomerization of human Chk2. Cds1 was overexpressed in a Cds1-S2T strain, cells were treated with HU, and the Cds1-S2T hybrid protein was immunoprecipitated from lysates. Precipitates were subjected to immunoblotting with anti-Cds1 antibody. Cds1-S2T coimmunoprecipitated with Cds1 (Fig. 6C). Mutation of T645 to alanine in Cds1-S2T, or overexpression of Cds1-T11AT8A instead of wild-type Cds1, greatly reduced but did not eliminate the interaction (Fig. 6C). The combination of these two mutant constructs completely abolished the interaction (Fig. 6C). These data strongly suggested that phosphorylation of T645 in the S2T cluster of Mrc1 mediates interaction with Cds1.
Chromatin association of Mrc1.
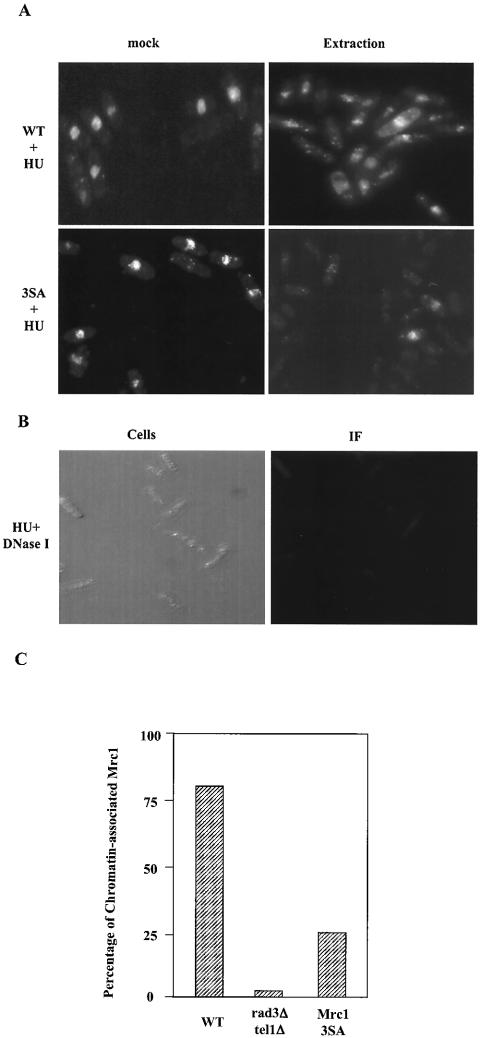
To better understand the role of Mrc1 in checkpoint signaling to Cds1, and in particular to characterize the role of the second S/TQ motif in Mrc1 function, we used an in situ chromatin binding assay to test the chromatin association activity of Mrc1. In this assay, Triton X-100 was used to extract proteins from the nucleus prior to immunofluorescence microscopy (15). Proteins that remained in the nucleus after extraction should have a close association with chromatin DNA or an insoluble component of the nuclear matrix. We first examined HU-arrested wild-type cells (Fig. 7A). In samples that were not treated with Triton X-100, about 90% of the cells displayed a visible Mrc1-13myc signal. In all of these cells, the signal was localized to the nucleus. A similar proportion (82%) of the cells retained the Mrc1-13myc signal after Triton X-100 extraction. The nuclear retention of Mrc1 was dependent on the presence of chromatin because DNase I digestion completely abolished the nuclear Mrc1 signals (Fig. 7B). In rad3Δ tel1Δ cells treated with HU, little Mrc1 signal was retained in the nucleus after Triton extraction, indicating that Rad3/Tel1-dependent phosphorylation mediates or stabilizes Mrc1's association with chromatin (Fig. 7C). To directly test if phosphorylation can affect Mrc1's chromatin binding activity, a 13myc-tagged form of mrc1-3SA was expressed from the endogenous locus and these cells were subjected to HU treatment and Triton extraction (Fig. 7A and C). In cells that were not treated with Triton, 80% of the cells had a nuclear Mrc1 signal, but this number was reduced to 27% upon Triton extraction. Because S604A is the critical mutation in the mrc1-3SA mutant, these data suggest that serine-604 is critical for high-affinity association of Mrc1 with chromatin.
FIG. 7.
Chromatin binding activity of Mrc1. (A) KT2791 cells (Mrc1-13myc) or HZ109 cells (Mrc1-3SA-13myc) were grown in minimal medium overnight and then either mock treated or treated with 15 mM HU for 3 h. Cells were harvested and extracted with 1% Triton X-100 for an in situ chromatin binding assay (see Materials and Methods). (B) Mrc1 chromatin association is sensitive to DNase I treatment. KT2791 cells (Mrc1-13myc) were grown in the presence of 15 mM HU and harvested for a chromatin binding assay. Cells were incubated with DNase I together with Triton X-100 and then subjected to an indirect immunofluorescence (IF) assay. The same cells are shown on the left side. (C) KT2791 cells (Mrc1-13myc), HZ109 cells (3SA-13myc), or HZ110 cells (rad3Δ tel1Δ Mrc1-13myc) were treated with 15 mM HU. Cells were subjected to Triton X-100 extraction for in situ chromatin binding assays. In each case, the number of cells with Mrc1 nuclear signal versus the total number of cells is represented on the y axis. At least 200 cells were counted from at least two different experiments, and the standard error is was 15% of the average. WT, wild type.
DISCUSSION
These studies have addressed the mechanism and functional significance of Mrc1 phosphorylation. We found that the phosphorylation-dependent mobility change of Mrc1 was not individually dependent on Cds1, Rad3, or Tel1, but it was abolished in a rad3Δ tel1Δ double mutant. Evidence of Tel1-dependent phosphorylation of Mrc1 in a rad3Δ background was surprising because Cds1 activation is completely dependent on Rad3 (19). Tel1 function is thought to be confined to telomere maintenance (22). The functional significance of Tel1-mediated phosphorylation of Mrc1 is unknown, but it raises the interesting possibilities that Mrc1 may have hitherto undiscovered functions in telomere maintenance or that Tel1 may have an unappreciated checkpoint activity.
Acting on the hypothesis that Mrc1 is a direct substrate of Rad3, and mindful of the fact that budding yeast Rad9 and fission yeast Mrc1 share clusters of S/TQ motifs in their central regions, we assayed the consequences of mutating two S/TQ clusters. These studies revealed that mutation of six S/TQ sites eliminated the phosphorylation-dependent mobility shift, and more detailed analyses indicated that sites in both S/TQ clusters were involved in phosphorylation. Further analyses identified two sites, S604 and T645, that are crucial for Mrc1 function. On the basis of the in vivo and in vitro data shown here, it is likely that Rad3 and Tel1 directly phosphorylate serine-604 and threonine-645. Use of phosphoserine-604-specific antiserum confirmed that serine-604 is phosphorylated in HU-treated cells by a mechanism that is abrogated in a rad3Δ tel1Δ double mutant.
In yeast two-hybrid analyses, Mrc1 and Cds1 interacted in a manner that required T645 in Mrc1 and an intact FHA domain in Cds1. These data strongly suggest that the T645 region of Mrc1 is directly involved in mediating the association between Cds1 and Mrc1. It was previously reported that the T68 region of human Chk2 associates with the FHA domain of Chk2 to promote intermolecular association and autophosphorylation (1). We therefore supposed that if the T645 region of Mrc1 associates with the FHA domain of Cds1, and if T645 is phosphorylated by Rad3, then the T645 region of Mrc1 might substitute for the T11 region of fission yeast Cds1, which corresponds to the T68 region of human Chk2. Remarkably, we found that the hybrid Cds1 containing the T645 region of Mrc1 conferred HU resistance on fission yeast. Even more remarkably, we found that this rescue was abolished by the T645A mutation. These phenotypes correlated with the ability of the hybrid Cds1 to associate with wild-type Cds1. Taken together, these data strongly suggest that the T645 region of Mrc1 is crucial because it is required for association with the FHA domain of Cds1. Our studies also suggest that S604 is required for stable association with chromatin. In Xenopus, claspin binds to chromatin even without activation of the replication checkpoint (18). In our assay, we could not detect Mrc1 chromatin binding activity in the absence of replication arrest, suggesting that fork stalling may increase the affinity of Mrc1 for chromatin.
Our data suggest the following model concerning the function and regulation of Mrc1. Mrc1 is probably a component of the replisome that is not required for replication but is essential for activation of the replication checkpoint response. Rad3 and Tel1 are recruited to stalled forks, where they phosphorylate Mrc1 on S604 and T645. Phosphorylation of S604 stabilizes Mrc1's interaction with the stalled replisome. Phosphorylation of T645 creates a binding site for the FHA domain of Cds1. The recruitment of Cds1 by Mrc1 allows Rad3 to phosphorylate Cds1 on T11, a critical step in Cds1 activation. Tel1 apparently cannot phosphorylate Cds1 efficiently on T11 because Cds1 activation is abolished in a rad3 mutant (28). It is interesting that Mrc1 is phosphorylated by the same upstream kinase(s) at two different sites that have distinct functions in the DNA replication checkpoint.
In budding yeast, HU-induced Mrc1 phosphorylation is reduced but not eliminated in mec1Δ cells (2). These data are consistent with our studies of Mrc1 in fission yeast and suggest that Tel1 may be required for the Mec1-independent phosphorylation of Mrc1 in budding yeast. It was recently shown that a budding yeast Mrc1 mutant that has all 17 S/TQ motifs mutated to AQ fails to undergo HU-induced phosphorylation; fails to support Rad53 phosphorylation, which is indicative of Rad53 activation; and fails to promote checkpoint arrest (23). Importantly, this 17AQ mutant did not exhibit the slow-replication phenotype seen in null mutant mrc1Δ mutant cells of budding yeast, showing that the mutant retained a basal function involved in DNA replication and was specifically defective in checkpoint signaling. This study also showed that Mrc1 associates with chromatin in concert with moving replication forks, suggesting that Mrc1 is a component of the replisome (23).
Acknowledgments
H.Z. and K.T. contributed equally to this work.
We thank all of the members of our laboratory and the Scripps Cell Cycle Groups for help and discussions. We especially appreciate T. Nakamura and B. Moser for providing yeast strains and T. Wang for providing anti-Cds1 antibody.
The work of K.T. was supported by the Naito Foundation, Kehara Memorial Foundation, Novartis Foundation (Japan), Yamanouchi Foundation for Research on Metabolic Disorders, and Radiation Effects Association. This work was supported by National Institutes of Health grant GM059447, which was awarded to P.R.
REFERENCES
- 1.Ahn, J. Y., X. Li, H. L. Davis, and C. E. Canman. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J. Biol. Chem. 277:19389-19395. [DOI] [PubMed] [Google Scholar]
- 2.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 3.Baber-Furnari, B. A., N. Rhind, M. N. Boddy, P. Shanahan, A. Lopez-Girona, and P. Russell. 2000. Regulation of mitotic inhibitor Mik1 helps to enforce the DNA damage checkpoint. Mol. Biol. Cell 11:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartek, J., J. Falck, and J. Lukas. 2001. CHK2 kinase—a busy messenger. Nat. Rev. Mol. Cell. Biol. 2:877-886. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, N. J., D. A. Holtzman, G. Flaggs, K. S. Keegan, A. DeMaggio, J. C. Ford, M. Hoekstra, and A. M. Carr. 1996. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 15:6641-6651. [PMC free article] [PubMed] [Google Scholar]
- 6.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capasso, H., C. Palermo, S. Wan, H. Rao, U. P. John, M. J. O'Connell, and N. C. Walworth. 2002. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J. Cell Sci. 115:4555-4564. [DOI] [PubMed] [Google Scholar]
- 8.Chini, C. C., and J. Chen. 2003. Human claspin is required for replication checkpoint control. J. Biol. Chem. 278:30057-30062. [DOI] [PubMed] [Google Scholar]
- 9.Durocher, D., J. Henckel, A. R. Fersht, and S. P. Jackson. 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4:387-394. [DOI] [PubMed] [Google Scholar]
- 10.Durocher, D., I. A. Taylor, D. Sarbassova, L. F. Haire, S. L. Westcott, S. P. Jackson, S. J. Smerdon, and M. B. Yaffe. 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6:1169-1182. [DOI] [PubMed] [Google Scholar]
- 11.Emili, A. 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2:183-189. [DOI] [PubMed] [Google Scholar]
- 12.Furnari, B., A. Blasina, M. N. Boddy, C. H. McGowan, and P. Russell. 1999. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell 10:833-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furnari, B., N. Rhind, and P. Russell. 1997. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science 277:1495-1497. [DOI] [PubMed] [Google Scholar]
- 14.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 15.Kearsey, S. E., S. Montgomery, K. Labib, and K. Lindner. 2000. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 19:1681-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumagai, A., and W. G. Dunphy. 2000. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 6:839-849. [DOI] [PubMed] [Google Scholar]
- 17.Kumagai, A., and W. G. Dunphy. 2003. Repeated phosphopeptide motifs in claspin mediate the regulated binding of Chk1. Nat. Cell Biol. 5:161-165. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J., A. Kumagai, and W. G. Dunphy. 2003. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell 11:329-340. [DOI] [PubMed] [Google Scholar]
- 19.Lindsay, H. D., D. J. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray, F. Osman, N. Walworth, and A. M. Carr. 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12:382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397:172-175. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Girona, A., K. Tanaka, X. B. Chen, B. A. Baber, C. H. McGowan, and P. Russell. 2001. Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc. Natl. Acad. Sci. USA 98:11289-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhind, N., and P. Russell. 2001. Roles of the mitotic inhibitors Wee1 and Mik1 in the G2 DNA damage and replication checkpoints. Mol. Cell. Biol. 21:1499-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 26.Soulier, J., and N. F. Lowndes. 1999. The BRCT domain of the S. cerevisiae checkpoint protein Rad9 mediates a Rad9-Rad9 interaction after DNA damage. Curr. Biol. 9:551-554. [DOI] [PubMed] [Google Scholar]
- 27.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka, K., M. N. Boddy, X.-B. Chen, C. H. McGowan, and P. Russell. 2001. Threonine-11, phosphorylated by Rad3 and ATM in vitro, is required for activation of fission yeast checkpoint kinase Cds1. Mol. Cell. Biol. 21:3398-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3:966-972. [DOI] [PubMed] [Google Scholar]
- 30.Xu, X., L. M. Tsvetkov, and D. F. Stern. 2002. Chk2 activation and phosphorylation-dependent oligomerization. Mol. Cell. Biol. 22:4419-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng, Y., K. C. Forbes, Z. Wu, S. Moreno, H. Piwnica-Worms, and T. Enoch. 1998. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature 395:507-510. [DOI] [PubMed] [Google Scholar]