Abstract
Idefix is a long terminal repeat (LTR)-retrotransposon present in Drosophila melanogaster which shares similarities with vertebrates retroviruses both in its genomic arrangement and in the mechanism of transposition. Like in retroviruses, its two LTRs flank a long 5′ untranslated region (5′UTR) and three open reading frames referred to as the gag, pol, and env genes. Here we report that its 5′UTR, located upstream of the gag gene, can fold into highly structured domains that are known to be incompatible with efficient translation by ribosome scanning. Using dicistronic plasmids analyzed by both (i) in vitro transcription and translation in rabbit reticulocyte or wheat germ lysates and (ii) in vivo expression in transgenic flies, we show that the 5′UTR of Idefix exhibits an internal ribosome entry site (IRES) activity that is able to promote translation of a downstream cistron in a cap-independent manner. The functional state of this novel IRES depends on eukaryotic factors that are independent of their host origin. However, in vivo, its function can be down-regulated by trans-acting factors specific to tissues or developmental stages of its host. We identify one of these trans-acting factors as the Gag protein encoded by Idefix itself. Our data support a model in which nascent Gag is able to block translation initiated from the viral mRNA and thus its own translation. These data highlight the fact that LTR-retrotransposons may autoregulate their replication cycle through their Gag production.
Long terminal repeat (LTR)-retrotransposons are mobile genetic elements present in the genomes of all eukaryotes that have been searched thoroughly for them. Their integrated form is composed of LTRs flanking a central region with one to three open reading frames (ORFs) (4). These ORFs encode predicted peptides resembling products of gag, pol, and env genes of retroviruses. In invertebrates, elements with three ORFs are presently classified as LTR-retrotransposons or endogenous retroviruses belonging to the gypsy family of elements. Another common feature of most LTR-retrotransposons and of related retroviruses is the presence of a long 5′ untranslated region (5′UTR) located between the 5′LTR and the gag gene.
These 5′UTR are multifunctional. They are involved in key steps of the replication cycle of retroelements, such as initiation of the proviral DNA synthesis, RNA dimerization, and encapsidation (8, 16). They have frequently been implicated in additional functions, such as the transcriptional enhancement of elements (7, 21, 31, 36). They can display the capacity to act as insulator elements able to isolate transcriptional units from the neighboring regulatory elements (26) and as a matrix attachment region (scaffold attachment region) involved in interactions with the nuclear matrix (22).
LTR-retrotransposons and vertebrate retroviruses all share a transposition mechanism which involves transcription of the integrated genomic DNA copy into RNA that contains all of the genetic information. Two functions have been attributed to this RNA. One is to be copied by reverse transcription into extrachromosomal DNA which becomes inserted into new chromosomal locations. The second is to be the template for both the proteic components of the virion core encoded by the gag gene and the enzymes involved in the replication process encoded by the pol gene. Thus, the full-length RNA serves as both mRNA and genomic RNA. It has been well established that the psi sequence present in the 5′UTR is recognized by the retroviral Gag through a combination of specific sequences and RNA structures in the RNA (1, 25). However, it remains unclear where and when this recognition assembly process occurs. Is there a single pool of full-length RNA within the cell that is alternatively translated and then encapsidated, or are there two independent pools of these RNAs, with one of them being the template for translation and the second being the template for encapsidation?
On another hand, the presence of long 5′UTRs raises an additional intriguing problem relative to gag and pol gene translation. The presence of several AUG and/or stable secondary structures encountered within 5′UTRs can possibly inhibit the procession of scanning ribosomes and result in a lack of translation of the downstream ORFs (18).
A decade ago, evidence was found for genetic elements termed internal ribosome entry sites (IRES) mediating the initiation of translation by direct internal ribosome entry. This mechanism, which allows ribosomes to avoid sequences upstream of a downstream gene and thus drives translation in a cap-independent manner, was first described for picornaviruses (15, 24) and has since been extended to cellular mRNAs (10, 37) and other viruses, such as several members of the retrovirus family (3, 5, 6, 11, 20, 23). These results prompted us to evaluate the capacity of the long 5′UTRs found in invertebrate LTR-retrotransposons to drive translation in a cap-independent manner.
We previously reported the identification of an LTR-retrotransposon, named Idefix, in Drosophila melanogaster (12). Like retroviruses, Idefix displays two LTRs flanking a long 5′UTR of 413 bp and three ORFs (gag, pol, and env). The present study shows for the first time that the 5′UTR of such an LTR-retrotransposon from the gypsy family is sufficient to drive internal initiation of translation. The Idefix IRES is functional in vitro in rabbit reticulocyte lysate (RRL) or wheat germ lysate (WGL) and in vivo in D. melanogaster; however, in the latter case, its activity is regulated by specific trans-acting factors that repress its function in some tissues or developmental stages. Through in vitro and in vivo experiments, we identified one of these factors that inhibits the cap-independent translation initiated from the 5′UTR as the Gag peptide encoded by Idefix itself.
MATERIALS AND METHODS
Plasmid constructs.
Standard procedures were used for restriction nuclease digestion and plasmid DNA construct (29). Escherichia coli strain SURE (RecA−) was used for the propagation of plasmids. The plasmid construct pXLJ0 has been described previously (2).
PCR was performed with sense primer 5′CGAATTCGGTTCGGTGTTCTTCT3′ and antisense primer 5′CGAATTCGTTGTGGGACTGCCATGATGTC3′, located at nucleotides (nt) 502 and 1024 of the Idefix sequence, respectively, to construct a translational fusion between the cyclin B2 gene of Xenopus laevis and NS′ of influenza virus. The amplified fragment, extending from position 502 to 1024, corresponds to an Idefix fragment starting 94 nt upstream from the end of its 5′LTR and ending 13 nt downstream from the AUG codon of its gag gene. This PCR product was first inserted into pGemTeasy (Promega). The EcoRI fragment of this clone, encompassing the full length of the 5′UTR, was subsequently cloned as an filled-in fragment into plasmid pXLJ0 which had been previously linearized with BamHI and had the cut ends filled in. This led to the pXL-Id clone. The orientation of the 5′UTR of Idefix was determined by using BamHI and EcoRI, which cleave two restriction sites located in the 5′part of Idefix UTR and in the vector, respectively. Nucleotide sequences throughout the entire Idefix insert were determined with Big Dye kits (Applied Biosystems).
The plasmid control was constructed with the IRES from encephalomyocarditis virus (EMCV) (Clontech) in pXLJ0. The pIRES plasmid was digested with XhoI and SalI, and the filled-in fragment was inserted into pXLJ0 which had been linearized with SalI. The orientation was determined by using EcoRI, which cleaves upstream of the EMCV IRES and within the vector.
Constructs used for transgenesis were designed as follows. A green fluorescent protein (GFP) EcoRI-BamHI fragment was inserted into the pUAST vector (20) which was previously linearized by EcoRI and BglII. This construct (pUAST-GFP) was digested with XhoI and XbaI. The 5′UTR region of Idefix was amplified by PCR with primers 5′GCGCAGTCGGTTAGGATCCAATA3′ and 5′GAGAGTTGTGGGAACTGCCATC3′, located at nt 596 and 1028 of the Idefix sequence, which correspond to the 5′ end of the 5′UTR of Idefix and the 5′end of its gag gene, respectively. The amplified fragment was then fused to LacZ in the pRCCMVneo-LacZ vector ((20) at the NheI site in both orientations (Id-LacZ and IdCi-LacZ). Id-LacZ and IdCi-LacZ fragments flanked by the XhoI and XbaI restriction sites were then cloned downstream of the GFP gene (provided by Alain Vincent), leading to the so-called UAS-GFP-Id-LacZ and UAS-GFP-IdCi-LacZ constructs, respectively.
The gag gene was amplified by PCR with primers 5′GGCAGTCCCACAACTCTCA3′ and 5′CATAGGGACTTGTATGTCCTT3′ at nt 1003 and 2028 of the Idefix sequence, respectively. The amplified fragment was cloned first in pGemTeasy and then in the pUAST vector at the EcoRI site. A vector named UAS-Gag was obtained.
Preparation of Gag fusion protein.
The Idefix Gag protein was expressed in bacteria as a fusion protein with glutathione S-transferase (GST). The region chosen for amplification spanned nt 1003 to 2028 of the Idefix sequence.
The oligonucleotides Idef1004 (5′-CGGATCCGACATCATGGCAGTCCCACAAC-3′) and Idef2028CI (5′-CGGATCCCATAGGGACTTGTATGTCCTT-3′) were designed as 5′ and 3′ primers, respectively. The additional BamHI restriction site used for cloning is underlined. The primer pair Idef1004-Idef2028CI was used to amplify the entire Gag protein sequence. The purified fragment was digested with BamHI and ligated to the expression plasmid pGEX-4T2 (Pharmacia), generating the pGEX-gag construct.
Expression of the GST-Gag fusion protein was induced in the E. coli BL21 by treatment of the log-phase bacterial culture with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 30°C. Bacteria were collected by centrifugation, and the pellet was resuspended in a lysis buffer containing an antiproteinase cocktail. Cells were broken briefly by sonication and cleared from insoluble material by centrifugation. Gag fusion protein was collected from the supernatant with 500 μl of glutathione agarose beads (Sigma) for 1 h at room temperature. The fusion protein was eluted from the beads by gentle shaking in 50 mM Tris-HCl (pH 9)-5 mM reduced glutathione for 10 min at 4°C. The fusion protein yield was estimated by Bradford spectrophotometric analysis, and the quality of each fraction was tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Transgenesis.
Drosophila lines with UAS-GFP-Id-LacZ, UAS-GFP-IdCi-LacZ, and UAS-Gag transgenes were obtained after P-element-mediated germ line transformation (28). Expression of these transgenes has been analyzed in a genetic background allowing Idefix expression and mobilization (13). The Gal4 drivers Actin-Gal4 (Mireille Galloni) and e22C-Gal4 (Bloomington Stock Center) were used. An actin promoter directs transcription of UAS transgenes at all stages of development. The e22C-Gal4 was used to specifically direct transcription in follicle cells of region 2A of the germarium, in which Idefix is expressed (14, 34).
In vitro RNA synthesis and translation.
Capped and uncapped RNAs were synthesized by using circulated DNA templates with T7 RNA polymerase (RNA transcription kit; Stratagene) according to the manufacturer's protocols. A total of 2 μg of plasmid DNA was used for RNA synthesis in 50-μl final reaction volumes. For capped RNAs, RNA synthesis was performed with a mix containing 2 μl of rGTP (2 mM) and 6.4 μl of 5′-terminal m7G cap (10 mM) (RNA cap structure analog; New England BioLabs). Capped and uncapped RNAs were synthesized for 1 h at 37°C. Capped RNAs were terminated by treatment with 6.4 μl of rGTP (10 mM) for 5 min at 37°C. Transcription was stopped by digestion of the template DNA with 40 U of DNase I for 15 min at 37°C, and RNA was precipitated with lithium chloride. RNA was resuspended in 20 μl of RNase-free water. Capped and uncapped RNAs were translated in nuclease-treated RRL (Promega) at a 50% concentration with 1 μg of RNA and 10 μCi of [35S]methionine (NEN Life Science Products).
Transcription-translation reactions in RRL and WGL were performed with the TnT Quick Coupled kit (Promega) under the conditions described by the manufacturer. Reactions were performed with 1 μg of DNA with incubation at 30°C for 90 min, and the products resulting from the translation of the dicistronic mRNAs were analyzed on 12% polyacrylamide gels. Autoradiography was performed for 16 h. Reaction products were quantified by incorporation of [35S]methionine (1,175 Ci/mmol; New England Nuclear) and phosphorimager analysis (Bio-Rad).
X-Gal staining.
For X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of the transgenic lines expressing β-galactosidase fusion proteins, third-instar larvae and adults were dissected in phosphate-buffered saline (PBS). For β-galactosidase activity detection, tissues were fixed in 0.5% glutaraldehyde in PBS for 4 min. After being washed in saline buffer, tissues were incubated at 37°C for 3 h and/or overnight by using standard methodology. Embryos were dechorionated by being dipped into 50% bleach for 2 min. After being washed in water, embryos were fixed in 0.35 ml of 0.75% glutaraldehyde in PBS and 0.7 ml of n-heptane for 20 min at room temperature. After being washed in PBS with 0.3% Triton X-100 (PBT), embryos were incubated at 37°C for 3 h and/or overnight by using standard methodology.
Immunostaining of adult tissues.
Ovaries were dissected in PBS and fixed in 4% formaldehyde for 15 min. After being washed in PBT, tissues were permeabilized and saturated for 4 h in PBT at room temperature. The anti-LacZ primary polyclonal antibody (Sigma) was diluted 1/2,000. The Cy3-conjugated anti-rabbit secondary antibody (Molecular Probes) was diluted 1/300. The anti-α-tubulin antibody (Sigma) was diluted 1/2,000. The Cy5-conjugated anti-mouse secondary antibody (Molecular Probes) was diluted 1/300.
Fluorescent staining and microscopy.
Light and fluorescence microscopies were performed with an Axiophot microscope (Zeiss), and confocal microscopy was performed with an Olympus confocal microscope.
RESULTS
The Idefix 5′UTR directs in vitro translation of a second cistron in a dicistronic RNA.
The Idefix structure bears a long 5′UTR of 413 bp. Since secondary structures observed in such a long 5′UTR may potentially cause a premature stop of the scanning ribosomes and thus inhibit translation of the downstream gag and pol genes, this region was tested for its ability to regulate translation by an internal ribosome entry process.
The 5′UTR of Idefix was inserted between two cistrons of a plasmid called pXLJ0. Plasmid pXLJ0 contains the cyclin B2 gene of X. laevis as an upstream cistron and the NS′ gene of influenza virus as a downstream cistron (2). The T7 promoter controls transcription of this dicistronic construct. The Idefix 5′UTR and the first 13 nt of its gag coding sequence were fused in frame with the NS′ coding sequence (pXL-Id). As a positive control for internal initiation of translation, we inserted the IRES of the EMCV (Clontech) between the two cistrons of pXLJ0. This led to the pXL-EMCV construct.
RNAs were synthesized and translated in vitro in an RRL or a WGL (by using the TnT kit from Promega) from plasmids pXLJ0, pXL-EMCV, and pXL-Id. Two species of proteins can potentially arise from such plasmids, with apparent molecular masses of 45.5 kDa for cyclin B2 and 26 kDa if the NS′ product is synthesized.
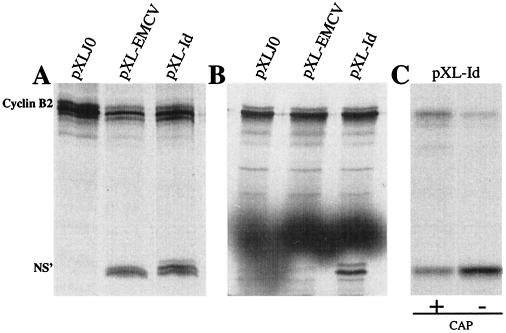
The results of transcription-translation experiments showed that a cyclin polypeptide was obtained for all three plasmids tested, i.e., pXLJ0, pXL-Id, and pXL-EMCV (Fig. 1A and B). When pXLJ0 was used in this experiment, the cyclin B2 polypeptide at 45.5 kDa was the only peptide detected on polyacrylamide gels. No translation of the second cistron occurred. In presence of either the 5′UTR of Idefix in plasmid pXL-Id or the IRES of EMCV in pXL-EMCV, an additional polypeptide with the predicted molecular mass of 26 kDa corresponding to the NS′ gene product was synthesized in RRL (Fig. 1A). The NS′ product was also detected for pXL-Id, but not for pXL-EMCV, when tested in WGL (Fig. 1B). This result is expected if the Idefix 5′UTR brings an IRES sufficient to drive the translation of a second cistron. This IRES is efficient in crude lysates from animals such as rabbit or plants such as wheat. This last property contrasts with that of the previously described IRES from EMCV, which is active only in animal lysates (results herein and from the manufacturer).
FIG. 1.
Dicistronic analysis of the Idefix 5′UTR sequence. (A) Transcription-translation in RRL of dicistronic mRNAs bearing the cyclin B2 and NS′ cistrons of pXLJ0 with either no insert between the two cistrons (pXLJ0), the IRES from EMCV (lane pXL-EMCV), or the 5′UTR of Idefix (lane pXL-Id). (B) Transcription-translation in WGL of dicistronic mRNAs from pXLJ0, pXL-EMCV, and pXL-Id. (C) Translation of capped (+) and uncapped (−) dicistronic RNAs from pXL-Id in RRL. The positions of cyclin B2 (45.5 kDa) and NS′ (26 kDa) are indicated on the left.
To ascertain whether the NS′ product was produced from a cap-independent translation of the pXL-Id construct, capped and uncapped dicistronic mRNAs were generated and used in in vitro translation assays. As shown in Fig. 1C, uncapped RNAs from pXL-Id were able to promote expression of NS′, thus independently from the first cistron, which is not expressed under these conditions or is expressed at a very low level due to an endogenous cap activity present in RRL and generating a small population of capped RNAs. In a parallel experiment performed with preliminary capped RNAs from pXL-Id, both products of the dicistronic transcript, i.e., cyclin B2 and NS′, were obtained. An increase in the amount of the second cistron product was clearly seen when translation of the first cistron was absent. This suggests that a competition between cap and IRES initiation for the recruitment of translational factors probably occurs. These data confirm that the NS′ protein is indeed translated by a cap-independent mechanism due to the presence of an IRES within the 5′UTR of Idefix.
From these results, we conclude that the 5′UTR of Idefix is sufficient to initiate translation within a dicistronic construct. Its efficiency is high in crude host-independent lysates, and it can operate in absence of any complementation with host specific proteins.
The Idefix 5′UTR functions as an IRES in transgenic Drosophila.
Since we have shown above that the activity of Idefix IRES depends on eukaryotic factors common to animals and plants, we could expect this IRES also to be active overall in vivo in its host organism, D. melanogaster. To verify that this is indeed the case, we studied the expression of a dicistronic plasmid by using Drosophila transgenesis and the UAS-Gal4 system (see Materials and Methods). A transgene, UAS-GFP-Id-LacZ (Fig. 2), with the GFP gene as a first cistron and LacZ as a second cistron placed downstream of the 5′UTR of Idefix was constructed and injected into flies. Two independent transgenic lines were recovered. In a first assay, we verified that neither of the two cistrons was expressed in these transgenic lines without the action of a Gal4 driver. Under these conditions, no GFP or LacZ expression was detected in any tissue or developmental stages (data not shown). This control brought two essential pieces of information: first, a Gal4 driver is needed to activate the dicistronic transgene, and second, no internal promoter that could eventually account for translation of the second cistron is present within the 5′UTR.
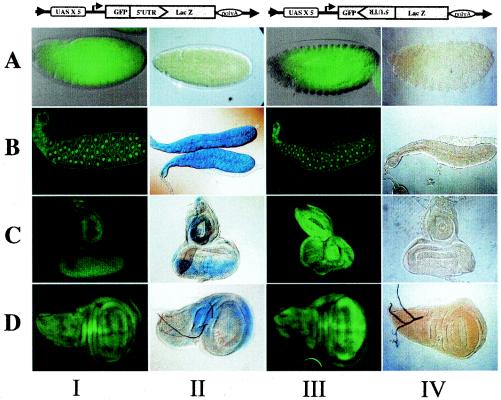
FIG. 2.
Analysis of dicistronic mRNA expression directed by an Actin-Gal4 driver in transgenic flies. The structures of the UAS-GFP-Id-LacZ and UAS-GFP-IdCi-LacZ transgenes are shown at the top. Green fluorescence reflects Gal4-activated transcription-translation of GFP from UAS-GFP-Id-LacZ and UAS-GFP-IdCi-LacZ transgenes (columns I and III, respectively). X-Gal staining reflects expression of the second cistron (LacZ) when placed downstream of the Idefix IRES in the sense orientation (plasmid UAS-GFP-Id-LacZ) (column II) or the reverse orientation (UAS-GFP-IdCi-LacZ) (column IV). Results for embryos (A), third-instar larva salivary glands (B), eye-antenna disks (C), and wing disks (D) are shown.
Thus, an Actin-Gal4 transgene was introduced by crossing to drive ubiquitous expression of the UAS-GFP-Id-LacZ transgene. As a negative control, similar experiments were performed with transgenic lines established from the injection of an additional construct bearing the Idefix 5′UTR in the opposite orientation (UAS-GFP-IdCi-LacZ [Fig. 2]).
Coexpression of first and second cistrons was then tested throughout the development of the fly, i.e., at the embryonic, larval, and adult stages. Translation of the first and second cistrons was examined by GFP fluorescence and LacZ staining, respectively.
The results showed that no LacZ was detected in lines bearing the UAS-GFP-IdCi-LacZ transgene, while GFP was detected. This was observed for all tissues and development stages, as exemplified Fig. 2, columns III and IV. This result indicates that the 5′UTR of Idefix in the opposite orientation yields no IRES activity and confirms results already observed in vitro in RRL or WGL (not shown).
When translation of the UAS-GFP-Id-LacZ transgene was analyzed, cell type-specific differences in IRES function were detected. In embryos, no LacZ staining was detected at any developmental stage, while the GFP was clearly translated (Fig. 2A, columns I and II). This indicates that this particular IRES is not functional at embryonic stages of Drosophila development.
By contrast, UAS-GFP-Id-LacZ expression under control of the Actin driver resulted in the synthesis of both GFP and LacZ products in differentiated larval tissues. Staining was observed in salivary glands and in all the imaginal disks of third-instar larvae, such as eye-antenna and wing disks, as shown in columns I and II of Fig. 2B, C, and D, respectively.
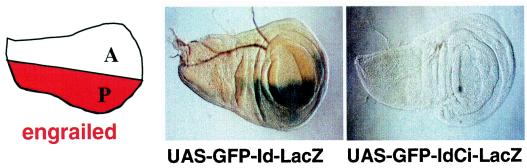
An en-Gal4 driver specifically directs expression of UAS transgenes to the posterior compartment of imaginal disks (14). Under these conditions, LacZ expressed from the dicistronic UAS-GFP-Id-LacZ transgene was detected in a portion of the disks corresponding to the posterior compartment, as illustrated by the wing disks (Fig. 3). No LacZ staining was observed in the anterior compartment, where en-Gal4 is not active. This result confirmed that no internal promoter that could account for translation of the second cistron is present within the 5′UTR.
FIG. 3.
Analysis of dicistronic mRNA expression directed by an en-Gal4 driver in transgenic flies. Engrailed is expressed in the posterior (P) compartmental part of wing disks (left panel). X-Gal staining reflects expression of the second cistron (LacZ) from the UAS-GFP-Id-LacZ and UAS-GFP-IdCi-LacZ transgenes driven by en-Gal4 (center and right panels, respectively).
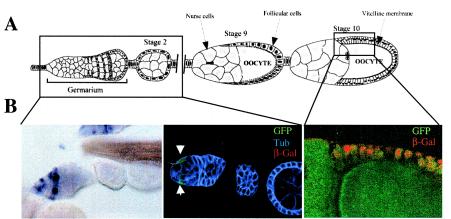
Since Idefix transcription occurs in somatic cells of adult ovaries in a structure called germarium (Fig. 4) (34), we were interested in searching for the IRES activity in these specific cells. Thus, transcription of the dicistronic transgene was specifically activated in this somatic lineage by using the selective driver e22C-Gal4 (14), and translation of the first and second cistrons was analyzed by immunostaining (see Materials and Methods). As expected, translation of the first cistron was detected by GFP staining (green) in the germarium; however, no LacZ staining (red), corresponding to translation of the second cistron initiated from the 5′UTR of Idefix, was ever detected, either after 3 h or after overnight coloration (Fig. 4B). The overall structure of the tissue is revealed by an α-tubulin staining (blue). When an Actin-Gal4 driver was used, both GFP and LacZ staining were detected in the follicle cells of later stages of oogenesis, where Idefix is not expressed (Fig. 4B, right panel).
FIG. 4.
Expression of the dicistronic transgene UAS-GFP-Id-LacZ in adult ovaries. (A) Schematic representation of an adult ovary. The ovariole is composed of the germarium (early stages of oogenesis) and later of two follicles in stages 9 and 10. (B) Left panel, in situ hybridization of Idefix RNAs in the germarium. Center panel, expression of GFP (green) and β-galactosidase (red) from the dicistronic transgene activated by the e22C-Gal4 driver. A strong immunostaining corresponding to GFP expression is observed in the germarium. This staining is detected in the somatic cells, where Idefix RNA is also detected (left panel). No staining corresponding to LacZ expression is detected in these cells. The overall structure of the ovariole is labeled for tubulin (blue). Right panel, merge of GFP (green) and β-galactosidase (red) staining observed at stage 10 of oogenesis, resulting from the activation of the dicistronic transgene by the Actin-Gal4 driver. LacZ expression is detected in the follicle cells, where Idefix is not expressed.
These results indicate that the IRES present within the 5′UTR sequence of Idefix is functional in vivo; however, it can be down-regulated in some tissues and developmental stages of D. melanogaster, certainly because of specific trans-acting factors that repress its function. One of them is specifically expressed within tissues where Idefix starts its replication cycle, which are the somatic cells of the germarium.
The Gag polypeptide encoded by Idefix inhibits its IRES-directed translation.
The fact that no LacZ staining was observed in the somatic cells where the retroelement Idefix is expressed could easily be explained if a factor encoded by Idefix itself and thus present when its replication cycle is occurring is able to suppress translation initiated from the cognate 5′UTR present within the dicistronic transgene. Since such a regulation has already been described for hepatitis C virus (HCV) regulation, which is down-regulated by its core-coding sequence that is able to affect translation initiated by the HCV IRES (17, 38), we tested the influence of the Gag polypeptide encoded by Idefix on its IRES-dependent translation.
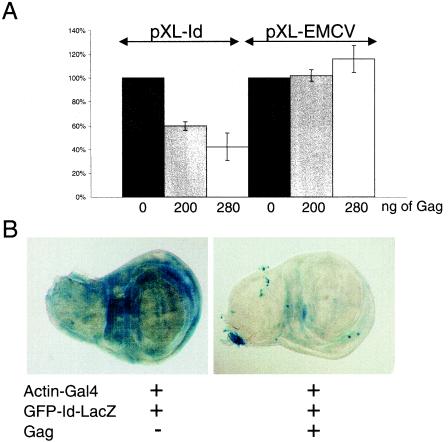
A recombinant GST-Gag product was expressed in E. coli and used in a TnT experiment performed on plasmids pXL-Id and pXL-EMCV. The NS′ translation in lysates was measured and quantified, and the results are shown in Fig. 5A. NS′ translation from pXL-Id or pXL-EMCV without any Gag addition was arbitrarily taken as 100%. The addition of GST-Gag to the reaction mixtures substantially inhibited Idefix IRES-dependent translation. Addition of 200 ng decreased NS′ translation to 60%; addition of 280 ng decreased it to 42%. No effect of Gag on EMCV IRES-dependent translation was observed (Fig. 5A), and Idefix or EMCV IRES-directed translation was not affected by addition of GST alone (data not shown).
FIG. 5.
Influence of Idefix Gag on IRES-dependent translation. (A) pXL-Id and pXL-EMCV were transcribed and translated in vitro with RRL in the absence or presence of 200 or 280 ng of Gag. NS′ translation from pXL-Id and pXL-EMCV without Gag addition was arbitrarily taken as 100%, and the NS′ activities of other products were normalized to this. Means and standard deviations from three independent triplicate TnT experiments are shown. (B) Wing imaginal disks from a transgenic fly with two transgenes (Actin-Gal4 and UAS-GFP-Id-LacZ) (left) and a transgenic fly with three transgenes (UAS-Gag, Actin-Gal4, and UAS-GFP-Id-LacZ) (right). IRES activity leading to LacZ expression is high when Gag is absent, as in transgenic lines with two transgenes (left), and is very weak or null in flies expressing UAS-Gag, as in transgenic lines with three transgenes (right).
From these experiments, we conclude that the Gag polypeptide encoded by Idefix acts as a trans-regulating factor that is able to repress the function of the Idefix IRES in vitro, while it seems to have no effect on the IRES function of EMCV.
Since the above data had been obtained in vitro, we performed experiments to verify that Gag also acts as a negative trans-regulating factor in vivo. We reasoned as follows: if Gag is ectopically expressed in tissues where the IRES is known to be active, such as in imaginal disks of third-instar larvae, its expression should result in the IRES inactivation. To test this, we constructed a P-transformation vector, pUAS-Gag, bearing the gag gene of Idefix fused to the UAS promoter. In such a construct, the expression of Gag may be placed under control of any Gal4 driver. The construct was introduced into flies by microinjection, and four transgenic lines were isolated.
In a line with three transgenes (UAS-Gag, Actin-Gal4, and UAS-GFP-Id-LacZ) in its genome, the Actin-Gal4 driver ubiquitously activates transcription of both transgenes UAS-Gag and UAS-GFP-Id-LacZ and allows the analysis of IRES function through LacZ expression in the presence of the Gag protein. On the other hand, in a line lacking UAS-Gag but bearing the two other transgenes (Actin-Gal4 and UAS-GFP-Id-LacZ), the IRES function can be analyzed in the absence of Gag (Fig. 2).
When analyzing LacZ expression in imaginal disks of these two types of lines, we found that expression from the dicistronic construct was dependent on the presence or absence of Gag (Fig. 5B). As exemplified by the wing disk, the expression was very weak or undetectable in imaginal disks of flies with three transgenes and thus in flies in which Gag and the dicistronic construct are coexpressed in these tissues. LacZ is highly expressed in lines in which Gag is not (Fig. 5B).
These results confirm the data obtained in vitro and extend them to in vivo. The polypeptide Gag encoded by Idefix acts as a trans-regulating factor that is able to repress translation initiated from the 5′UTR of Idefix.
DISCUSSION
One important feature of numerous LTR-retrotransposons is a long UTR located between the end of their 5′LTR and their gag gene. This region is essential in the life of the element, since it includes many control domains required for replication as the primer target site and the encapsidation signal. The present work provides evidence that the 5′UTR and the Gag product of an endogenous retrovirus from an insect may act together to initiate a switch between a translated state of the genomic mRNA and an untranslated state of this RNA, which will ultimately be encapsidated.
The 5′UTR of Idefix is sufficient for efficient cap-independent translation depending on eukaryotic translational factors independent of their host origin.
Once a retroviral mRNA is transcribed from its integrated proviral DNA copy, its level of expression is dependent upon the efficiency of a number of interconnected cellular processes. These include polyadenylation and splicing, mRNA export from the nucleus, degradation, and localization in the cytoplasm. The expression of this mRNA also depends upon the translational machinery and the presence of sequence elements that either activate or inhibit its translation. In that context, IRES, which are detected in numerous viral 5′UTRs, are essential for the viral translational control. The presence of an IRES allows initiation of translation and may imprint cell-specific translational control and modify virulence.
In the present study, we showed that the replication cycle of an LTR-retrotransposon from an insect may be subjected to such a translational control exerted by an IRES located within its 5′UTR. The complete and highly structured 5′UTR of Idefix from D. melanogaster shows the capacity to promote cap-independent translation, as shown by the following: (i) insertion of this 5′UTR into the intercistronic spacer region of a dicistronic mRNA is sufficient to mediate translation of the second cistron, (ii) this translation occurs in vitro and in vivo, and (iii) it occurs independently of the 5′ cap of the mRNA. Interestingly, we found that this IRES is functional in crude lysates as different as RRL and WGL, with no need for specific Drosophila factors. Thus, cellular factors not specific for a defined host organism but common to the eukaryotic translational machinery are sufficient to recognize and allow this IRES activity.
Cap-independent translation of Idefix may be silenced in vivo in specific tissues.
From the above-described results indicating that crude lysates alone are sufficient in vitro to allow translation from this domain, one might expect this IRES to be active in every cell of an organism. Unexpectedly, transgenic assays with dicistronic constructs indicated that, in vivo, the Idefix IRES exhibits clear on-off controls of its activity depending on tissue or developmental stages of D. melanogaster. When the dicistronic transgene is placed under the control of an ubiquitous driver and thus its transcription is driven in the whole organism, this IRES is not functional in all of the tissues. As examples, it is inactive in the embryonic stages of the fly development and is active in third-instar larval tissues. These results indicate that beside general eukaryotic translational factors, additional regulatory elements associated with the 5′UTR may integrate a regulatory input into the IRES function. These observations may be related to those from other studies performed with transgenic flies and transgenic mice, indicating that the IRES contained in the 5′UTRs of Antennapedia and Ultrabithorax (37) and of bFGF and c-Myc (9, 10) exhibit a high degree of spatial and developmental regulation, which suggests that they are recognized by trans-acting factors modulating their activity. The Idefix IRES has specific silencing in tissues of its host organism, D. melanogaster.
The major outstanding question regarding the Idefix IRES is thus whether its presence acts as a novel point of control in the replication cycle of Idefix that is able to allow or inhibit Idefix translation depending on the tissues or stages where it is transcribed.
We have previously reported that Idefix transcripts are detected in tissue of adult female ovaries corresponding to a somatic lineage of cells present in a structure called the germarium (34). The Gag product has also been detected in this tissue through Western blot analyses (unpublished data). Surprisingly, when dicistronic transgenes were specifically transcribed in these cells, where the Idefix replication cycle was occurring, no translation of the second cistron was ever observed. On the other hand, when the Actin-Gal4 driver was used, coexpression of both GFP and β-galactosidase was detected to later stages of oogenesis. This result highly supports the possibility that Idefix translation might be tightly regulated by a trans-acting factor present in these somatic cells of the germarium and acting on the IRES function. A paradoxical question thus arises: how is Idefix able to start a new round of replication if its translation is suppressed by a cellular factor expressed in the tissue where it is transcribed? One hypothesis reconciling this paradox is that Idefix by itself encodes the specific trans-acting factor that is able inhibit translation of its cognate RNA. Once transcribed and translated, it is then able to autoregulate its own replication cycle by controlling further translation initiated from the 5′UTR and responsible for its first and second ORF products.
The Gag product of Idefix blocks IRES function.
In RNA viruses, specific or preferential interactions between the nucleocapsid protein and its viral sense RNA have been demonstrated (19, 25, 33, 35). On another hand, binding of these core proteins to RNA structure in the proximity of an initiation codon has been documented as a mechanism of translational repression. For example, the core protein encoded by HCV has been shown to bind to its cognate RNA and then suppress translation (30, 38). Additionally, it has been reported that a competition between ribosomes and the Gag protein encoded by Rous sarcoma virus determines the fate of nascent retroviral RNA (32). These results support the hypothesis that Rous sarcoma virus Gag proteins autogenously regulate their synthesis and encapsidation of retroviral RNA. Similarly, the Tya protein of the yeast Ty1 retrotransposon, which is equivalent to the retroviral Gag, has been found to interact in vitro with the Ty1 RNA and exert a regulatory function during transposition (27).
On the basis of these studies, we tested the involvement of the gag gene product of Idefix and found that it is able to down-regulate translation initiated from the Idefix IRES. First, we demonstrated that Gag inhibits the IRES function in vitro, since a Gag polypeptide expressed as a recombinant protein in bacteria and used in a TnT experiment inhibits cap-independent translation initiated from the 5′UTR of Idefix. Second, in D. melanogaster, a Gag polypeptide ectopically expressed in imaginal disks of third-instar larvae down-regulates the IRES-dependent translation of a cistron that was otherwise expressed in the absence of Gag.
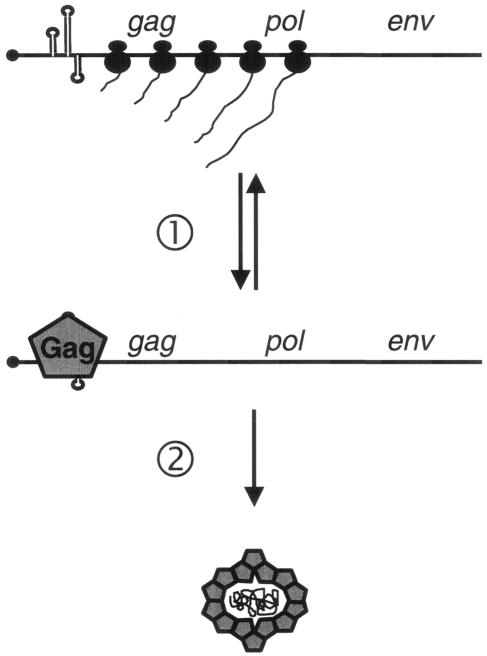
It has been well described that binding of Gag to the 5′UTR of its cognate RNA is an essential step in the replication cycle of retroviruses. The data reported here indicate that Idefix Gag may coordinate two essential processes during the replication cycle of this invertebrate retrovirus: first, its binding to the 5′UTR induces a down-regulation of the gag and pol gene translation; second, while bound to this domain, it initiates encapsidation (Fig. 6). Overall, the following regulated pathway of Idefix replication can be proposed: The initial step is to be transcribed in the germarium. The resulting full-length transcript is then translated and gives rise to Gag and Pol products from a cap-independent translation promoted by the IRES of the 5′UTR. At that time, the full-length transcript is used as a template for translation. Once an adequate concentration of Gag is reached, it binds to the 5′UTR, suppresses the IRES function, and shuts down Idefix translation. Then, as already described for retroviruses, this association of Gag with the 5′UTR initiates encapsidation. In this second step of the Idefix life cycle, the full-length transcript that was previously used as a template for translation is now used as a genomic transcript that will be reverse transcribed into extrachromosomal DNA and ultimately inserted within the genome.
FIG. 6.
Proposed model for Gag-mediated control of the Idefix replication cycle. When synthesized, the full-length Idefix RNA is first translated, giving rise to Gag and Pol products. Once an adequate concentration of these peptides is produced, Gag binds to the 5′UTR (step 1), shuts down translation, and drives the RNA to encapsidation (step 2).
It is still a matter of debate as to whether one or two pools of full-length viral RNA exist within the cell. If a unique pool exists, then it serves both as a template for translation and as genomic RNA that is packaged into virion. Alternatively, full-length RNA could be separated in two pools with different functions, one functioning as an mRNA that produces viral proteins and another serving as genomic RNA awaiting encapsidation by Gag. In the scenario proposed for Idefix, Gag may successively coordinate two main processes of the life cycle from a single pool of RNA. Its binding to the mRNA generates the switch of the full-length RNA from a translated to an untranslated state that can then be packaged in viral particles. These two successive functions favor the model predicting a single pool of full-length RNA.
Analysis of Idefix Gag properties is currently under way to further dissect its function in the repression of translation and the process of encapsidation. However, it will also be interesting to investigate how many IRES there are likely to be within other retrotransposons described for insects genomes and how they are implicated in the regulation of their mobilization.
Acknowledgments
We are grateful to A.C. Prats for comments on a draft of the manuscript, J. L. Darlix for general discussions, A. Vincent for providing clones, and M. Galloni and the Bloomington Stock Center for flies.
This work was supported by grants from the Association de Recherche contre le Cancer (ARC 5585), CNRS (GDR 2157), INSERM (U384), and Université Franco-Italienne (UFI) through the “Vinci 2002” program. C.M. received a graduate grant from MESR and FRM, and F.A. received a graduate grant from MESR.
REFERENCES
- 1.Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491-511. [DOI] [PubMed] [Google Scholar]
- 2.Bailly, J. L., A. M. Borman, H. Peigue-Lafeuille, and K. M. Kean. 1996. Natural isolates of ECHO virus type 25 with extensive variations in IRES sequences and different translational efficiencies. Virology 215:83-96. [DOI] [PubMed] [Google Scholar]
- 3.Berlioz, C., and J. L. Darlix. 1995. An internal ribosomal entry mechanism promotes translation of murine leukemia virus Gag polyprotein precursors. J. Virol. 69:2214-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke, J. D., T. Eickbush, S. B Sandmeyer, and D. F. Voytas. 1999. Virus taxonomy, ICTV VIIth report. Springer-Verlag, New York, N.Y.
- 5.Brasey, A., M. Lopez-Lastra, T. Ohlmann, N. Beerens, B. Berkhout, J. L. Darlix, and N. Sonenberg. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 77:3939-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck, C. B., X. Shen, M. A. Egan, T. C. Pierson, C. M. Walker, and R. F. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75:181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavarec, L., S. Jensen, J. F. Casella, S. A. Cristescu, and T. Heidmann. 1997. Molecular cloning and characterization of a transcription factor for the copia retrotransposon with homology to the BTB-containing lola neurogenic factor. Mol. Cell. Biol. 17:482-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. M., S. H Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 9.Creancier, L., P. Mercier, A. C. Prats, and D. Morello. 2001. c-myc internal ribosome entry site activity is developmentally controlled and subjected to a strong translational repression in adult transgenic mice. Mol. Cell. Biol. 21:1833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creancier, L., D. Morello, P. Mercier, and A. C. Prats. 2000. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol. 150:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deffaud, C., and J. L. Darlix. 2000. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 74:846-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desset, S., C. Conte, P. Dimitri, V. Calco, B. Dastugue, and C. Vaury. 1999. Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol. 16:54-66. [DOI] [PubMed] [Google Scholar]
- 13.Desset, S., C. Meignin, B. Dastugue, and C. Vaury. 2003. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164:501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy, J. B., D. A. Harrison, and N. Perrimon. 1998. Identifying loci required for follicular patterning using directed mosaics. Development 125:2263-2271. [DOI] [PubMed] [Google Scholar]
- 15.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansen, L. K., and C. D. Morrow. 2000. The RNA encompassing the internal ribosome entry site in the poliovirus 5′ nontranslated region enhances the encapsidation of genomic RNA. Virology 273:391-399. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y. K., S. H. Lee, C. S. Kim, S. K. Seol, and S. K. Jang. 2003. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 83:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Z., D. Yang, Z. Qiu, K. T. Lim, P. Chong, and S. Gillam. 1996. Identification of domains in rubella virus genomic RNA and capsid protein necessary for specific interaction. J. Virol. 70:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Lastra, M., C. Gabus, and J. L. Darlix. 1997. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum. Gene Ther. 8:1855-1865. [DOI] [PubMed] [Google Scholar]
- 21.Matyunina, L. V., I. K. Jordan, and J. F. McDonald. 1996. Naturally occurring variation in copia expression is due to both element (cis) and host (trans) regulatory variation. Proc. Natl. Acad. Sci. USA 93:7097-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nabirochkin, S., M. Ossokina, and T. Heidmann. 1998. A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 273:2473-2479. [DOI] [PubMed] [Google Scholar]
- 23.Ohlmann, T., M. Lopez-Lastra, and J. L. Darlix. 2000. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 275:11899-11906. [DOI] [PubMed] [Google Scholar]
- 24.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 25.Poon, D. T., E. N. Chertova, and D. E. Ott. 2002. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): functional linkage between translation and RNA packaging. Virology 293:368-378. [DOI] [PubMed] [Google Scholar]
- 26.Roseman, R. R., V. Pirrotta, and P. K. Geyer. 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth, J. F., S. M. Kingsman, A. J. Kingsman, and E. Martin-Rendon. 2000. Possible regulatory function of the Saccharomyces cerevisiae Ty1 retrotransposon core protein. Yeast 16:921-932. [DOI] [PubMed] [Google Scholar]
- 28.Rubin, G. M., and A. C. Spradling. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218:348-353. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, P. A., and V. G. Corces. 1995. The suppressor of Hairy-wing protein regulates the tissue-specific expression of the Drosophila gypsy retrotransposon. Genetics 139:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonstegard, T. S., and P. B. Hackett. 1996. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 70:6642-6652. [PMC free article] [PubMed] [Google Scholar]
- 33.Stohlman, S. A., R. S. Baric, G. N. Nelson, L. H. Soe, L. M. Welter, and R. J. Deans. 1988. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol. 62:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tcheressiz, S., V. Calco, F. Arnaud, L. Arthaud, B. Dastugue, and C. Vaury. 2002. Expression of the Idefix retrotransposon in early follicle cells in the germarium of Drosophila melanogaster is determined by its LTR sequences and a specific genomic context. Mol. Genet. Genomics 267:133-141. [DOI] [PubMed] [Google Scholar]
- 35.Weiss, B., U. Geigenmuller-Gnirke, and S. Schlesinger. 1994. Interactions between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 22:780-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, S., L. V. Matyunina, and J. F. McDonald. 1998. An enhancer region within the copia untranslated leader contains binding sites for Drosophila regulatory proteins. Gene 209:239-246. [DOI] [PubMed] [Google Scholar]
- 37.Ye, X., P. Fong, N. Iizuka, D. Choate, and D. R. Cavener. 1997. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol. 17:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, J., O. Yamada, H. Yoshida, T. Iwai, and H. Araki. 2002. Autogenous translational inhibition of core protein: implication for switch from translation to RNA replication in hepatitis C virus. Virology 293:141-150. [DOI] [PubMed] [Google Scholar]