Abstract
The FACT complex facilitates transcription on chromatin templates in vitro, and it has been functionally linked to nucleosomes and putative RNA polymerase II (Pol II) elongation factors. In Saccharomyces cerevisiae cells, FACT specifically associates with active Pol II genes in a TFIIH-dependent manner and travels across the gene with elongating Pol II. Conditional inactivation of the FACT subunit Spt16 results in increased Pol II density, transcription, and TATA-binding protein (TBP) occupancy in the 3′ portion of certain coding regions, indicating that FACT suppresses inappropriate initiation from cryptic promoters within coding regions. Conversely, loss of Spt16 activity reduces the association of TBP, TFIIB, and Pol II with normal promoters. Thus, FACT is required for wild-type cells to restrict initiation to normal promoters, thereby ensuring that only appropriate mRNAs are synthesized. We suggest that FACT contributes to the fidelity of Pol II transcription by linking the processes of initiation and elongation.
Transcription of mRNA coding genes by RNA polymerase II (Pol II) is a complex process. Biochemical experiments have defined distinct steps of preinitiation complex formation, initiation of RNA synthesis, extension of abortive transcripts, promoter clearance, elongation, and termination (6, 35). The basic preinitiation complex at the promoter contains Pol II itself and general initiation factors. Upon assembly, TFIIH phosphorylates the C-terminal domain (CTD) of the largest Pol II subunit, and the phosphorylated form of Pol II mediates elongation. Biochemical and genetic approaches have identified putative elongation factors such as TFIIS, Spt4/5 (DSIF), Spt6, FACT, and the TREX and Paf complexes.
In Saccharomyces cerevisiae cells, the general initiation factors localize specifically to promoter regions (22-25, 37). Association of these initiation factors is mutually interdependent and is strongly correlated with transcriptional activity (23, 25). Thus, stable association of the preinitiation complex with promoters is the major, although not exclusive, limiting step for transcription in yeast cells. Unlike the initiation factors, putative elongation factors such as TFIIS, Spt4/5, Spt6, TREX, and the Paf complex associate with coding region of active genes (18, 37, 48). Kinetic analysis involving regulated expression of a long gene indicates that the Hpr1 and Tho2 subunits of the TREX complex travel with Pol II through the coding region (48). As TFIIS, Spt4/5, Spt6, and the Paf complex associate with Pol II (18, 45, 46, 51), Pol II-associated factors are exchanged during the transition between initiation and elongation.
The mammalian FACT complex was identified as a biochemical activity that acts subsequent to initiation to facilitate elongation on chromatin templates (34). Mammalian FACT is composed of two proteins that are homologous to yeast Spt16(Cdc68) and Pob3 (36). FACT specifically interacts with nucleosomes and H2A/H2B dimers, and FACT activity in vitro is blocked by cross-linking nucleosomal histones, suggesting that FACT might promote nucleosome disassembly upon transcription (36). In addition, FACT can affect elongation on naked DNA templates by acting in concert with P-TEFb CTD kinase to counteract the negative elongation activities of DSIF and NELF in vitro (50).
Yeast Spt16(Cdc68) was identified by its effects on transcription and cell cycle control (30, 41), whereas Pob3 was identified by copurification with DNA polymerase I (52). Spt16 and Pob3 form a heterodimer (4, 53) that interacts with the high-mobility group protein Nhp6 (3, 10), and both FACT subunits are essential for viability. Yeast FACT interacts with TFIIE (15) and with Sas3, the catalytic subunit of the NuA3 histone acetylase complex (14). Strains with spt16 or pob3 mutations show increased or decreased expression of various genes (3, 9, 28, 30, 41, 42), although the bases for these effects are unknown. Spt16 genetically interacts with TFIIS, Spt4/5, Spt6, and the Paf complex (7, 11, 27, 34, 47), proteins believed to be involved in elongation. Taken together, the results of these experiments indicate that FACT is an elongation factor whose activity is particularly relevant in the context of nucleosomal templates.
In accord with the idea that FACT is an elongation factor, we show that yeast FACT travels with elongating Pol II at transcriptionally active genes in vivo. Unexpectedly, FACT-deficient cells show decreased levels of preinitiation complexes at promoters and increased Pol II density and transcription in the 3′ portion of certain coding regions. Thus, in addition to associating with elongating Pol II, FACT contributes to transcriptional initiation at normal promoters and suppresses initiation within coding regions. We suggest that FACT contributes to transcriptional fidelity by linking the processes of initiation and elongation.
MATERIALS AND METHODS
Yeast strains and plasmids.
W303.1A yeast strains containing Spt16-(Myc)3, Pob3-(Myc)3, Tfg2-(Myc)3, and Hpr1-(Myc)13 were prepared as described previously (43), except for Hpr1-(Myc)13, for which the PCR product contained HIS3 and 13 Myc epitopes and HIS3 was not removed (48). The strain containing Spt16-(Myc)3 grows indistinguishably from the wild-type strain, indicating that Spt16-(Myc)3 fully complements the lethal phenotype due to loss of Spt16 function. The strain containing Pob3-(Myc)3 grows detectably slower than the wild-type strain, indicating that Pob3-(Myc)3 only partially complements the lethal phenotype. Strains expressing hemagglutinin (HA)-Rpb3 from the normal genomic locus were prepared by two-step gene replacement using a URA3 integrating plasmid containing a PCR fragment comprising the 700 bp surrounding the ATG codon of the HA-RPB3 locus of strain Z280 (kindly provided by Rick Young). GAL1-YLR454 strains were prepared by single-step integration of a linearized TRP3 plasmid containing the GAL1 promoter flanked downstream by the first 300 bp of the YLR454w coding region. The copper-inducible Spt16 depletion strain was prepared in strain ZMY117 (isogenic to ZMY60 except for leu2::PET56) as described previously (32). Isogenic FY56 (wild type) and L577 (spt16-197) strains (30) were kindly provided by Fred Winston; when necessary, they were converted to trp1 derivatives by using the marker swap plasmid pTU10 (8). Isogenic wild-type and kin28-ts16 strains (5) were analyzed as described previously (23). (HA)3-TBP strains were prepared by single-step integration of plasmid YIplac211-(HA)3-TBP (23). Strains were grown in synthetic complete medium containing 2% glucose as the carbon source unless indicated otherwise.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation in yeast cells was performed essentially as described previously (23, 48) using antibodies against the nonphosphorylated CTD of Pol II (8WG16; Covance), Rpb3 (NeoClone Biotechnology), HA epitope (12CA5), Myc epitope (06-549; Upstate Biotechnology), TATA-binding protein (TBP), TFIIB, Tfb3 (kindly provided by Michael Keogh and Steve Buratowski), Spt16, and Pob3 (the latter two were kindly provided by Tim Formosa). Comparable results for Pol II occupancy were obtained with the 8WG16, Rpb3, and 12CA5 (for strains containing HA-Rpb3) antibodies; presumably, this is due to the presence of nonphosphorylated heptad repeats even when the CTD is phosphorylated during the elongation process. For all experiments involving the YLR454 coding region, PCRs contained multiple primer pairs such that relative occupancy levels for the different regions were directly determined in the same reaction. Quantitated data are presented in arbitrary units that are directly related to the apparent immunoprecipitation efficiency (i.e., the amount of material immunoprecipitated relative to that of the input sample), as determined from PhosphorImager analysis.
Transcriptional analysis.
Total RNA was prepared by the hot phenol method (13), followed by treatment with DNase I (Promega) and removal of DNase I by phenol extraction. To determine RNA levels, RNA was randomly (for MDN1 and FLO8) or gene specifically (for YLR454) primed to generate cDNA that was subjected to PCR amplification and detection by ethidium bromide staining. For Northern blotting analysis, poly(A)-containing RNA was enriched from total RNA by using an Oligotex kit (Qiagen), electrophoretically separated on a 1% agarose gel containing formaldehyde, and transferred to a nylon membrane. RNA was detected by hybridization with random-primed probes comprising the 5′-most or 3′-most 800 bp of the YLR454 coding region.
RESULTS
FACT specifically associates with active Pol II genes.
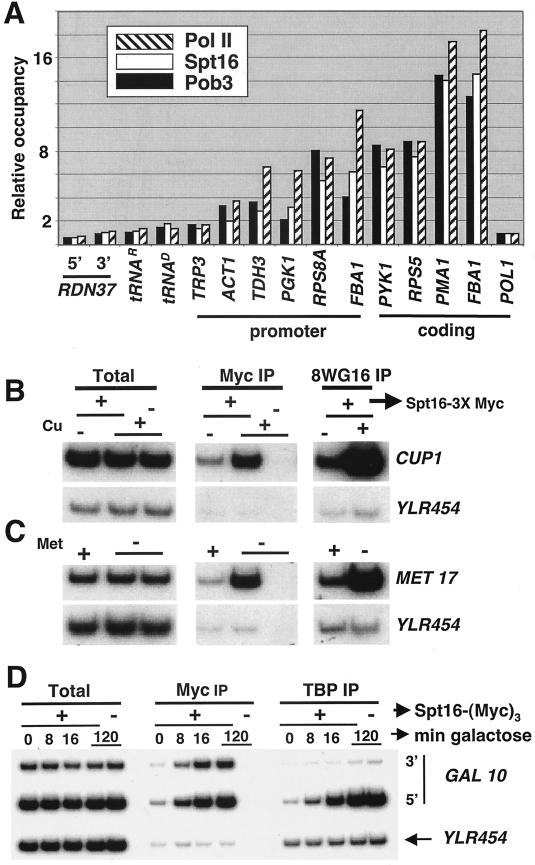
Spt16 and Pob3 associations with yeast genomic regions in vivo were analyzed by chromatin immunoprecipitation in strains expressing Myc-tagged derivatives from their natural promoters at their normal chromosomal loci. The levels of Spt16 and Pob3 association strongly correlated with each other and with the level of Pol II occupancy over a variety of Pol II-transcribed genes (Fig. 1A). In contrast, these FACT subunits did not associate with highly active sites of Pol I (RDN37) or Pol III (tRNAs glu and arg) transcription, indicating that FACT specifically associates with active Pol II genes. At certain promoters (FBA1, PGK1, and TDH3), the relative associations of Spt16 and Pob3 were about twofold below that of Pol II, suggesting that FACT association might occur just downstream of the preinitiation complex (see below). As Spt16 and Pob3 colocalize on active Pol II genes in vivo and form a stable heterodimer in vitro, we used Myc-tagged Spt16 to monitor FACT association in vivo in subsequent experiments.
FIG.1.
FACT associates with active Pol II genes. (A) Cells containing Spt16-(Myc)3 or Pob3-(Myc)3 were examined by chromatin immunoprecipitation with either Myc or 8WG16 antibodies followed by quantitative PCR using primers to the indicated regions. The relative level of Spt16 (white bars), Pob3 (black bars), and Pol II (8WG16) (striped bars) association (defined in arbitrary units as described in Materials and Methods) are shown. (B to D) For induction experiments, cells containing or lacking Spt16-(Myc)3 were induced by 10-min treatment with 1 mM CuSO4 (B), 3 h of growth in medium lacking methionine (C), or a shift from 2% raffinose to 2% galactose medium for the indicated times (D) and analyzed for the levels of Spt16, TBP, and Pol II (8WG16 antibody) at the indicated regions.
FACT association with active Pol II genes followed similar kinetics as the process of gene induction. FACT associated with CUP1 (Fig. 1B) and MET17 (Fig. 1C) upon the appropriate inducing treatment, and it accumulated at the GAL10 promoter at a rate similar to the rate of TBP recruitment upon galactose induction (Fig. 1D). Unlike TBP, FACT associated strongly with both the GAL10 promoter and the 3′ portion of the mRNA coding region.
FACT travels with elongating Pol II throughout the mRNA coding region.
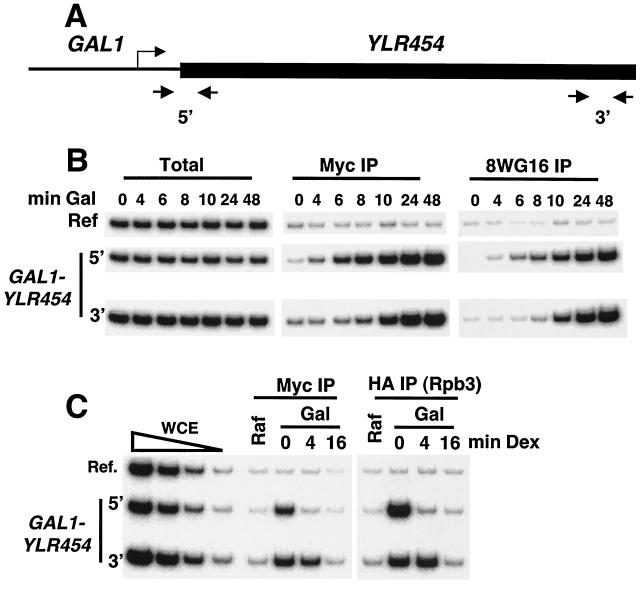
To examine whether FACT travels with elongating Pol II, we developed an approach that kinetically monitors association during the first or last “waves” of transcription under conditions where gene expression is rapidly induced or repressed. This approach relies on an allele in which the GAL1 promoter drives expression of the long (8 kb) YLR454 open reading frame at its natural locus (Fig. 2A) and has been used for analysis of the Hpr1 and Tho2 components of the TREX complex (48). To visualize the first wave of transcription, we analyzed FACT and Pol II occupancy at 2-min intervals after galactose addition (Fig. 2B). Pol II began to accumulate at the promoter by 4 min, whereas accumulation at the 3′ end began roughly 10 min after induction. The 6-min difference in Pol II occupancy at the 5′ and 3′ ends of this 8-kb gene indicates that the Pol II elongation rate is approximately 1.3 kb/min, in accord with previous results determined by other methods (13, 33). FACT association is similar to Pol II association, indicating that FACT enters the gene at or near the promoter and proceeds to the 3′ end of the gene during the first round of transcription.
FIG. 2.
FACT travels with Pol II throughout the mRNA coding region. (A) Schematic representation of the GAL1-YLR454 gene. (B) Spt16 and Pol II association with the indicated regions in cells containing Spt16-(Myc)3 that were grown in 2% raffinose (Raf) medium and shifted to 2% galactose (Gal) medium for the indicated times. (C) Spt16 and Pol II association with the indicated regions in cells containing Spt16-(Myc)3 and HA-Rpb3 that were induced with 2% galactose for 60 min and then repressed with 4% glucose (Dex) for the indicated times. WCE represents chromatin prior to immunoprecipitation.
To monitor the last wave of transcription, galactose-induced cells were treated with glucose to rapidly inhibit initiation. At 4 min after glucose addition, Pol II and FACT occupancies were drastically decreased in the vicinity of the promoter but were virtually unaffected near the 3′ end of the gene (Fig. 2C). However, Pol II and FACT were no longer associated with the 3′ end of the gene 16 min after transcriptional inactivation. This behavior is not an exclusive feature of the GAL1-YLR454 allele; similar results were obtained by monitoring YLR454 induction driven by the CUP1 promoter in response to copper or the GRE2 promoter in response to osmotic stress or glucose shutoff of GAL1-driven MDN1 (a 15-kb coding region; data not shown). Taken together, these observations demonstrate that FACT travels with elongating Pol II. However, our results do not address whether a single FACT molecule associates with Pol II and traverses the entire gene or whether different FACT molecules dynamically associate and disassociate with elongating Pol II.
FACT association appears to occur just downstream of the preinitiation complex.
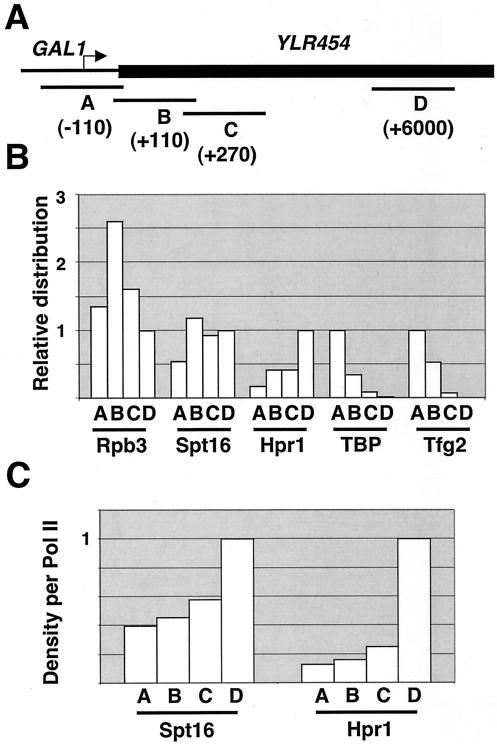
To address when FACT associates with the transcription complex, we compared its promoter-proximal distribution to Pol II and other transcription factors at the GAL1-YLR454 locus (Fig. 3A). For Rpb3, Spt16, and Hpr1 (a component of the TREX complex), we arbitrarily defined occupancy at a downstream location (region D) as 1.0 and directly measured the relative occupancy at three overlapping locations near the promoter (region A) and immediate 5′ portion of the coding sequence (regions B and C).
FIG. 3.
Spt16 association occurs just downstream of promoter. (A) Regions of the GAL1-YLR454 gene used to map protein occupancy at the promoter-proximal region. (B) Chromatin from galactose-induced cells containing either Hpr1-(Myc)13, HA-Rpb3, Spt16-(Myc)3, TBP, or Tfg2-(Myc)3 was immunoprecipitated with appropriate antibodies and analyzed by quantitative PCR. (C) Relative levels of Spt16 and Hpr1 with respect to Pol II at the indicated regions.
Two lines of evidence suggest that FACT association occurs after preinitiation complex formation. First, Spt16 occupancy at region A was reduced twofold in comparison to that in other regions, whereas occupancies of promoter-associated TBP and TFIIF (Tfg2 subunit) were two- to threefold higher in region A than in region B (Fig. 3B). Second, the Spt16-to-Pol II occupancy ratios at regions A to C were lower than at region D (Fig. 3C), even though Pol II occupancy at region A was comparable to (and perhaps slightly higher than) occupancy at region D. Interestingly, Pol II association at region B, which corresponds to the beginning of the mRNA coding region, was higher than at the other regions tested. Although the basis for this observation is unknown, we speculate that the increased Pol II density at this location might be related to “paused Pol II” (1, 2, 19, 39, 40) and/or to increased dwelling of Pol II during the step of promoter clearance. In accord with previous results suggesting that association of the TREX complex occurs after Pol II clears the promoter (56), the Hpr1-to-Pol II occupancy ratio was reduced at regions A to C in comparison to region D. At the promoter-proximal region, the Hpr1-to-Pol II occupancy ratio was lower in relative terms than the Spt16-to-Pol II occupancy ratio (Fig. 3B), suggesting that association of FACT precedes association of the TREX complex.
TFIIH is required for FACT association in vivo.
In yeast cells, TFIIH is localized at the promoter (37), where it plays a critical role in the transition between transcriptional initiation and elongation. The Kin28 subunit of TFIIH is required for phosphorylation of the Pol II CTD at serine 5 in the vicinity of the promoter (17, 44), although Kin28-mediated CTD phosphorylation per se is not essential for transcriptional activity (16, 17, 31). Thermal inactivation of Kin28 does not affect TBP association at promoters (23), although it significantly reduces association of Pol II (44).
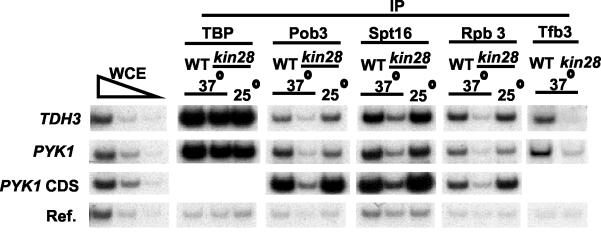
To address the mechanism of FACT recruitment, we examined Spt16 association in kin28-ts16 cells shifted to the restrictive temperature (Fig. 4). Under these conditions, TFIIH (assayed with antibodies against the Tfb3 subunit) association with the PYK1 and TDH3 promoters is drastically reduced. Interestingly, inactivation of Kin28 significantly reduces Spt16 and Pob3 association at the promoter and mRNA coding region in a manner similar to that of Pol II. In contrast, loss of Kin28 function does not affect the association of TBP at the promoter. Thus, TFIIH is required for FACT association in vivo, probably as a consequence of a functional connection (direct or indirect) between FACT and Pol II.
FIG. 4.
Spt16 association depends on TFIIH. Cross-linked chromatin from wild-type or kin28-ts16 cells grown at 25°C that were or were not shifted to 37°C for 1 h were immunoprecipitated with appropriate antibodies and analyzed by quantitative PCR with primers to the indicated regions.
FACT is important for association of TBP and TFIIB at the promoter.
Biochemically, a preinitiation complex is defined as the complete Pol II machinery stably assembled on a promoter that initiates transcription upon the addition of nucleotide precursors. In vivo, preinitiation complexes are likely to be transient, with initiation occurring rapidly after complex assembly, because nucleotide precursors are readily available. Furthermore, active promoters are likely to reinitiate transcription multiple times from an initial complex, such that Pol II itself is less stably associated with the promoter than other components of the Pol II machinery such as TBP, TFIIB, and TFIIA (49, 55). For this reason, measurements of Pol II occupancy at the promoter by chromatin immunoprecipitation cannot distinguish between preinitiation and early elongation complexes. The fact that inactivation of Kin28 reduces Pol II but not TBP association at the promoter suggests that most of the Pol II detected in wild-type strains is actually in elongation complexes just downstream from the promoter. However, the level of associations of TBP, TFIIA, and TFIIB at the promoter are strongly correlated with each other and with transcriptional activity (22-25), and TBP occupancy is strongly dependent on TFIIB and the Srb4 component of the mediator (23, 25). Thus, levels of TBP and TFIIB occupancy are good indicators of the amount of preinitiation complexes at promoters.
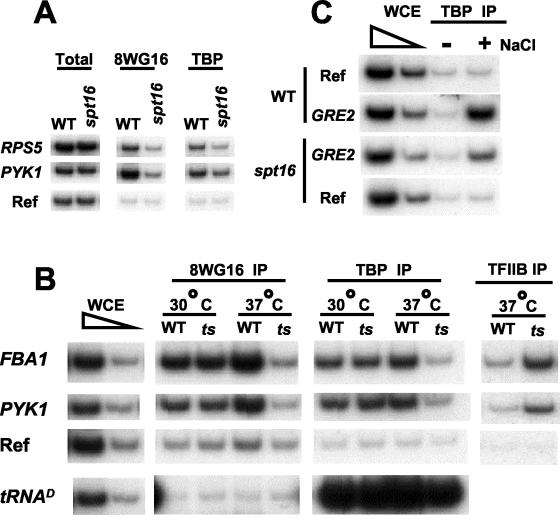
As FACT is essential for cell viability, we examined two conditional Spt16 alleles to assess the role of FACT in TBP occupancy in vivo. First, we generated a copper-inducible, double-shutoff allele (32) in which SPT16 transcription was repressed and Spt16 protein was rapidly degraded upon addition of copper (Fig. 5A). Second, we analyzed TBP occupancy in a strain containing spt16-197, a temperature-sensitive allele (30), that was shifted to the restrictive temperature (Fig. 5B). When Spt16 was inactivated by either of these two approaches, TBP occupancy at all three promoters tested was approximately threefold lower in the strain containing the conditional spt16 allele than in the isogenic strain containing the wild-type SPT16 allele. In addition, occupancy of TFIIB at these promoters was reduced to a comparable extent upon inactivation of Spt16. Thus, these results indicate that Spt16 contributes to TBP and TFIIB occupancy, and hence the level of preinitiation complexes, at normal promoters.
FIG. 5.
Spt16 depletion results in reduced TBP occupancy at promoters. (A) Strains lacking or containing a copper-inducible Spt16 depletion allele were treated with 0.75 mM copper sulfate for 3 h and analyzed for association of Pol II and TBP at the indicated promoters. (B) Pol II, TBP, and TFIIB association at the indicated promoters in wild-type or spt16-197 cells grown at 30°C and shifted to 37°C for 1 h. (C) TBP association at the GRE2 promoter in wild-type (WT) or copper-treated, Spt16-depleted cells that were or were not treated with 0.4 M NaCl for 5 min.
To provide additional evidence for a role of FACT in transcriptional initiation, we examined TBP and Pol II occupancy at the GRE2 promoter in cells subjected to osmotic stress for 5 min. As expected (38), wild-type cells showed a dramatic increase in TBP occupancy at the GRE2 promoter, indicating the formation of new preinitiation complexes (Fig. 5C). However, induction of TBP occupancy was reduced threefold upon osmotic stress in cells in which Spt16 was inactivated, indicating that FACT affects the de novo formation of preinitiation complexes in vivo.
Loss of FACT results in increased Pol II occupancy within the 3′ portions of certain mRNA coding regions.
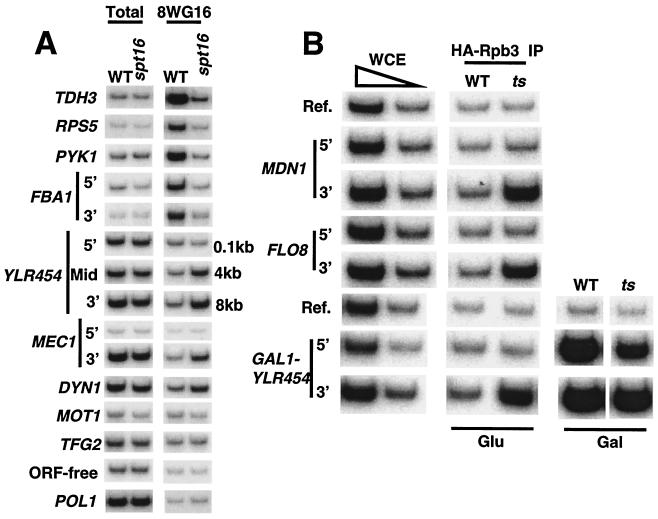
Given that FACT travels with elongating Pol II, we examined Pol II density in the conditional spt16 mutant strains. In accord with the decreased occupancy of TBP at promoters, copper-inducible depletion (Fig. 6A) or thermal inactivation (Fig. 6B) of Spt16 resulted in reduced Pol II density at both the proximal and distal regions of a variety of genes such as TDH3, RPS5, PYK1, and FBA1.
FIG. 6.
Inactivation of Spt16 causes increased Pol II occupancy within the 3′ regions of certain genes. (A) Strains lacking or containing a copper-inducible Spt16-depletion allele were treated with 0.75 mM copper sulfate for 3 h and analyzed for Pol II association (8WG16 antibody) at the indicated regions. (B) Pol II occupancy at the indicated regions in wild-type or spt16-197 cells containing HA-Rpb3 that were shifted to 37°C for 60 min.
Unexpectedly, Pol II density increased to abnormally high levels at the 3′ portions of several genes (YLR454, MEC1, and DYN1 [Fig. 6A]). Pol II density at the 5′ portions of these low-activity genes remained at the background level and hence cannot be measured. This effect was gene specific, because Pol II density at other genes such as MOT1 and TFG2 was not increased. Similarly, increased Pol II density was observed at the 3′, but not the 5′, portions of MDN1 and FLO8 in spt16-197 cells shifted to the restrictive temperature (Fig. 6B). In addition, when the YLR454 coding region was driven by the GAL1 promoter, Pol II occupancy increased at the 3′ end upon inactivation of Spt16 when cells were grown in glucose, conditions in which the GAL1 promoter is nearly inactive. The relative increase in Pol II density at the 3′ end of YLR454 with respect to the 5′ end was also observed when the gene was highly active (i.e., cells grown in galactose medium). The alterations in Pol II density at the 3′ portions of certain coding regions do not reflect changes in the phosphorylation status of the CTD (the 8WG16 antibody was made against the unphosphorylated CTD [Fig. 6A]), because comparable results were observed with HA antibodies using extracts from strains containing HA-Rpb3 (Fig. 6B).
FACT inhibits inappropriate transcriptional initiation from cryptic promoters within mRNA coding regions.
There are two possible explanations for how inactivation of FACT causes increased Pol II density at the 3′ portions of certain coding regions. One explanation is that loss of Spt16 affects the release of Pol II from the distal end of the affected genes to generate a “clogged pipe” in which Pol II molecules pile up behind the nonreleased Pol II molecule. We do not favor this explanation because increased Pol II density at the 3′ end of YLR454 is observed even when the GAL1 promoter driving the gene is nearly inactive, and because it is difficult to account for gene-specific effects. In addition, increased Pol II density occurred over at least a 4-kb region at the 3′ end of the YLR454 gene (Fig. 6A; see below), a result that differs from the relatively localized increase in Pol II distribution under conditions when Pol II elongation is arrested at a particular location in vivo (20). Alternatively, loss of Spt16 might result in inappropriate initiation at one or more sites within the mRNA coding region. This internal initiation model explains why increased Pol II density can occur over a relatively large region, and it can easily account for gene-specific effects.
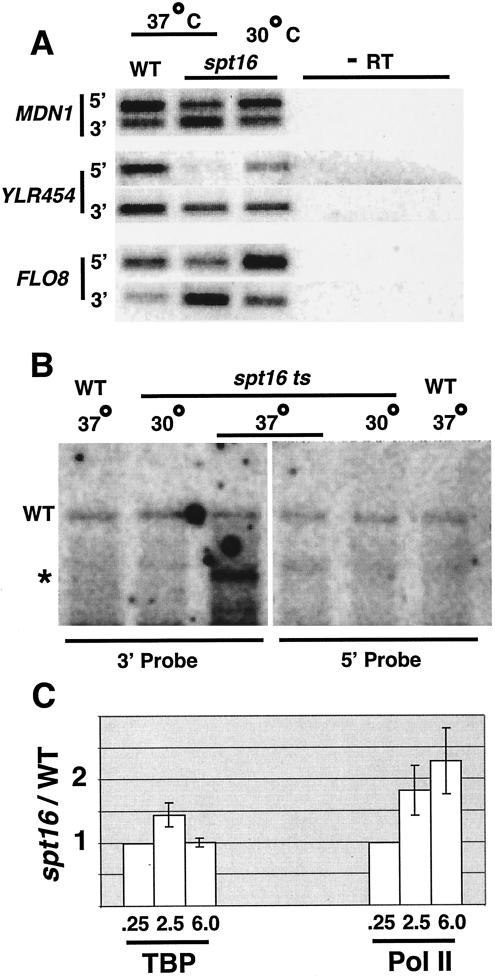
Three additional lines of evidence indicate that loss of Spt16 results in internal initiation within the normal mRNA coding region. First, for all three genes tested, inactivation of Spt16 caused increased RNA levels at the 3′ portions and decreased RNA levels at the 5′ portions (Fig. 7A). This observation is consistent with the internal initiation model but is not easily explained by the clogged-pipe model.
FIG. 7.
Inactivation of Spt16 causes transcription from a cryptic promoter within the YLR454 coding region. Wild-type and spt16-197 cells were shifted to 37°C for 60 min. (A) RNA levels (− RT indicates the absence of reverse transcriptase) at the indicated regions as determined by reverse transcriptase-PCR analysis. (B) Northern blot of mRNA hybridized to probes corresponding to the 5′ and 3′ regions of the YLR454 coding region. The wild-type 8-kb (WT) and internal 5.3-kb (*) transcripts are indicated. Molecular weights were calculated using the ribosomal RNAs as standards. The faint band that appears in all six lanes at a position just above that of the 5.3-kb transcript represents nonspecific hybridization that is unrelated to the YLR454 locus. The very faint band that occurs only under conditions of Spt16 inactivation at a position below that of the 5.3-kb transcript might represent another internal transcript, although we cannot exclude the possibility of nonspecific hybridization. (C) Ratio of TBP and Pol II association at the indicated YLR454 coding regions (defined in kilobases from the ATG initiation codon) in the spt16 strain with respect to the wild-type strain. The ratios for the 2.5 and 6.0 regions were normalized to that of the 0.25 regions, which was defined as 1.0.
Second, inactivation of Spt16 resulted in the appearance of a novel 5.3-kb, poly(A)-containing YLR454 RNA that was clearly distinct from the 8-kb RNA in wild-type strains (Fig. 7B). In accord with the results of the RNA analysis (Fig. 7A) and the Pol II occupancy experiments (Fig. 6B), the 5.3-kb RNA was four- to fivefold more abundant than the wild-type RNA. Importantly, this 5.3-kb RNA was detected with a hybridization probe for the 3′ end of the YLR454 gene but not with a probe for the 5′ end of the gene (Fig. 7B), and the position of the 3′ end of this 5.3-kb RNA was indistinguishable from that of the wild-type RNA (determined by 3′ rapid amplification of cDNA ends [data not shown]). The presence of this 5.3-kb RNA does not preclude the possibility of less abundant RNA species that arise from internal initiation. The presence of a discrete-sized, poly(A)-containing transcript essentially excludes the possibility that increased RNA levels at the 3′ portion of YLR454 arose from preferential stability of RNA at the 3′ end and/or preferential degradation of RNA at the 5′ end.
Third, the sequence TATAAAAT, a functionally optimal TATA element (54), lies within an AT-rich region (68% over 200 bp) just upstream of this 5.3-kb transcript. TBP occupancy (defined as the ratio in spt16-197 versus wild-type strains) in the vicinity of the TATAAAAT sequence was 1.4-fold higher than at other locations within the YLR454 coding region (Fig. 7C). Although this effect is quantitatively modest, it is significant (P = 0.0008 in four independent experiments in which relative TBP occupancies at the YLR454 regions were directly determined in an internally controlled manner). In addition, Pol II occupancy in the TATAAAAT region was about 1.8-fold higher than at the 5′ portion of the YLR454 coding region. The presence of TBP in the vicinity of an optimal TATA element just upstream of the novel 5.3-kb transcript demonstrates that loss of Spt16 results in internal initiation with the YLR454 coding region. Thus, FACT inhibits inappropriate initiation from cryptic promoters within mRNA coding regions.
DISCUSSION
FACT is important for fidelity of transcriptional initiation in vivo.
To express genetic information in an appropriate manner, it is essential that wild-type cells have mechanisms of transcriptional fidelity that restrict initiation to promoters, thereby ensuring that only appropriate mRNAs are synthesized. However, consensus sequences for TATA elements, initiator elements, and activator binding sites are short and occur within many mRNA coding regions. Thus, transcriptional fidelity is not achieved simply by restricting promoter elements to promoter regions. One mechanism that contributes to transcriptional fidelity is that proteins preferentially associate with promoter regions in comparison to protein-coding regions (21, 26, 29). Such preferential accessibility is due to general DNA sequence properties of promoter regions (29), and it may be related to nucleosome positioning.
FACT was characterized biochemically as a factor that acts subsequent to transcriptional initiation (34), and we show here that FACT travels with elongating Pol II in vivo. Unexpectedly, analysis of Spt16-defective cells indicated that FACT is also important for preventing internal initiation from cryptic promoters, and hence for fidelity of transcriptional initiation in vivo. First, Pol II density increased to abnormally high levels over extended 3′ portions of some, but not all, genes. Second, RNA levels increased at the 3′ portions and decreased at the 5′ portions of all three affected genes tested. Third, in the case of YLR454, we observed a novel and discrete-sized poly(A)-containing transcript as well as an optimal TATA element within an AT-rich region in the immediate vicinity of the presumed 5′ end of the novel transcript. Fourth, TBP occupancy increased in the vicinity of the optimal TATA element. These four lines of evidence convincingly demonstrate that initiation from cryptic Pol II promoters within mRNA coding regions occurs when FACT is inactivated; there is no other plausible interpretation that is consistent with all the data. Internal initiation within other genes has been observed in spt16 mutant strains (Craig Kaplan and Fred Winston, personal communication).
In addition to increased initiation within certain mRNA coding regions, loss of Spt16 activity resulted in reduced association of two essential components of the preinitiation complex, TBP and TFIIB, at all normal promoters tested. Furthermore, the level of de novo TBP recruitment to the GRE2 promoter upon osmotic stress was comparably reduced in FACT-deficient cells. The decreased TBP and TFIIB occupancy at normal promoters reflects a specific function of FACT and does not arise indirectly from a defect on transcription or Pol II occupancy in the vicinity of the promoter. Decreased association of preinitiation complex components was observed with two different kinds of conditional Spt16 alleles under inactivation conditions identical to those used to analyze the direct functions of many other essential transcription factors. More importantly, decreased TBP occupancy was not observed in Kin28-deficient cells, a condition in which Pol II transcription is essentially abolished (12) and Pol II occupancy in the vicinity of the promoter is significantly reduced (44) (Fig. 4). Thus, FACT is important for wild-type levels of preinitiation complexes and presumably for transcriptional initiation at natural promoters.
Our results indicate that FACT has an important role in the fidelity of transcriptional initiation. We suspect that increased internal initiation and decreased preinitiation complexes and transcription at normal promoters in FACT-deficient cells represent two sides of the same molecular phenomenon. Specifically, we suggest that the number of functional preinitiation complexes is limiting in vivo, such that increased internal initiation will be directly linked to decreased initiation at normal promoters due to competition for preinitiation complexes. mRNA coding regions are approximately five times larger than promoters (1,500 versus 300 bp), and the vast majority of yeast genes are poorly expressed (12, 13) and presumably have weak promoters. If internal initiation in FACT-deficient cells occurs at 20% of the level observed at most yeast promoters, the number of Pol II initiation events would effectively double. This estimate is plausible (and perhaps even low) given that internal initiation was observed at a significant proportion of the genes tested and often occurs at a level higher than that observed at normal promoters (as defined by Pol II density and RNA levels). In this view, FACT does not directly contribute to transcriptional initiation per se but rather affects transcriptional fidelity and hence the distribution of preinitiation complexes throughout the genome.
The model that FACT contributes to the fidelity, but not the mechanism, of transcriptional initiation is consistent with the results of biochemical experiments indicating a role after preinitiation complex formation (34) and the suggestion that FACT associates with active genes just downstream from the promoter (Fig. 3) (John Lis and Danny Reinberg, personal communication). In addition, this model explains the apparent contradiction that TBP occupancy decreases upon inactivation of FACT, but not Kin28, even though FACT association with active genes is drastically reduced in Kin28-deficient cells. In the absence of Kin28, transcription is blocked at an early step; hence, if FACT is important at a later step in the transcription process, its association with the gene is irrelevant under Kin28-deficient conditions. In contrast, inactivation of FACT does not disrupt preinitiation complexes but rather redistributes them towards mRNA coding regions and away from normal promoters.
FACT contributes to transcriptional fidelity by linking initiation and elongation.
As FACT specifically associates with active Pol II genes, we favor the idea that inhibition of internal initiation is linked to the function of FACT during the elongation process. Together with the biochemical (34, 36) and genetic (11) interactions of FACT with chromatin, we suggest a model in which FACT-containing Pol II elongation complexes generate or maintain a chromatin structure that inhibits preinitiation complex formation or stability within mRNA coding regions. In the absence of FACT, elongating Pol II complexes disrupt chromatin structure, thereby making mRNA coding regions more permissive for initiation. As exemplified by the novel YLR454 transcript, internal initiation sites would depend on the presence and relative location of otherwise cryptic TATA elements, initiator elements, and activator binding sites (and perhaps other sequence-dependent features) within the coding region. The likelihood of such cryptic promoter elements should increase in accord with the length of the coding region, and indeed, the internal transcripts observed here were generally in large mRNA coding regions.
It is important to note that increased internal initiation in spt16 mutant strains occurs at the expense of initiation at promoters, and it can occur at poorly expressed genes. We therefore suggest that, in the absence of FACT, the disruption of chromatin structure by elongating Pol II persists long enough to permit preinitiation complex formation and initiation within mRNA coding regions. Based on measurements of initiation frequency in vivo (13), we estimate that weak promoters initiate transcription once every 5 to 30 min. In addition, the GAL1 promoter used here is not completely inactive in glucose medium, because strains containing GAL1 promoter fusions to essential genes are often viable in glucose medium. Nevertheless, although we favor the view that FACT-dependent suppression of internal initiation is related to FACT's function during Pol II transcription, we cannot eliminate the possibility that it reflects a genome-wide, nontranscriptional function of FACT.
It is inevitable that the large and complex Pol II elongation machinery will disrupt nucleosome-DNA interactions as it travels across the gene. In principle, DNA in nucleosomes disrupted by elongating Pol II might be relatively accessible to nuclear proteins; hence, transcriptional elongation might undermine transcriptional fidelity. We suggest that FACT is important in reversing the nucleosome disruption that occurs upon elongation by Pol II, thereby preventing inappropriate access of transcription factors to mRNA coding regions. In this sense, elongation-related functions of FACT are linked to subsequent initiation events. If the number of functional preinitiation complexes is limiting yeast cells, an increase in internal initiation would concomitantly lead to a decrease in initiation at normal promoters by a simple competition model. Thus, by linking the processes of initiation and elongation, FACT increases transcription from the correct initiation site while minimizing initiation from internal sites, thereby improving the fidelity of Pol II transcription.
Acknowledgments
We thank Tim Formosa for Spt16 and Pob3 antibodies, Michael Keogh and Steve Buratowski for Tfb3 antibodies, Craig Kaplan, Fred Winston, John Lis, and Danny Reinberg for discussion of unpublished results, and Fred Winston and Rick Young for yeast strains. We thank Juliet Reid for the suggestion that polymerase redistribution in spt16 strains may be specific to distal portions of normal genes and Steve Buratowski, Michael Keogh, and Danny Reinberg for fruitful discussion.
This work was supported by an NIH postdoctoral fellowship to P.B.M. and a research grant from the National Institutes of Health to K.S. (GM 30186).
REFERENCES
- 1.Akhtar, A., G. Faye, and D. L. Bentley. 1996. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 15:4654-4664. [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, D. L., and M. Groudine. 1986. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature 321:702-706. [DOI] [PubMed] [Google Scholar]
- 3.Brewster, N. K., G. C. Johnston, and R. A. Singer. 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21:3491-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewster, N. K., G. C. Johnston, and R. A. Singer. 1998. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 273:21972-21979. [DOI] [PubMed] [Google Scholar]
- 5.Cismowski, M., G. Laff, M. Soloman, and S. Reed. 1995. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 7.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross, F. R. 1997. “Marker swap” plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647-653. [DOI] [PubMed] [Google Scholar]
- 9.Evans, D. R., N. K. Brewster, Q. Xu, A. Rowley, B. A. Altheim, G. C. Johnston, and R. A. Singer. 1998. The yeast protein complex containing Cdc68 and Pob3 mediates core-promoter repression through the Cdc68 N-terminal domain. Genetics 150:1393-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson, Y. Yu, A. R. Rhoades, P. D. Kaufman, and D. J. Stillman. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway. Polymerase passage may degrade chromatin structure. Genetics 162:1557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 13.Iyer, V., and K. Struhl. 1996. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5208-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 15.Kang, S. W., T. Kuzuhara, and M. Horikoshi. 2000. Functional interaction of general transcription initiation factor TFIIE with general chromatin factor SPT16/CDC68. Genes Cells 5:251-263. [DOI] [PubMed] [Google Scholar]
- 16.Keogh, M. C., E. J. Cho, V. Podolny, and S. Buratowski. 2002. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komarnitsky, P., E.-J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomic approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krumm, A., L. B. Hickey, and M. Groudine. 1995. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 9:559-572. [DOI] [PubMed] [Google Scholar]
- 20.Kulish, D., and K. Struhl. 2001. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol. Cell. Biol. 21:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo, M.-H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 22.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 23.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-612. [DOI] [PubMed] [Google Scholar]
- 24.Li, X.-Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 25.Li, X.-Y., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399:605-609. [DOI] [PubMed] [Google Scholar]
- 26.Lieb, J. D., X. L. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 27.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lycan, D., G. Mikesell, M. Bunger, and L. Breeden. 1994. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7455-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mai, X., S. Chou, and K. Struhl. 2000. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol. Cell. Biol. 20:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone, E. A., C. D. Clark, A. Chiang, and F. Winston. 1991. Mutations in the SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5710-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNeil, J. B., H. Agah, and D. Bentley. 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 12:2510-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 382:188-191. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien, T., and J. T. Lis. 1993. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 13:3456-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 35.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471-475. [DOI] [PubMed] [Google Scholar]
- 36.Orphanides, G., W.-H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human Spt16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 37.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 38.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9:1307-1317. [DOI] [PubMed] [Google Scholar]
- 39.Rougvie, A. E., and J. T. Lis. 1990. Postinitiation transcriptional control in Drosophila melanogaster. Mol. Cell. Biol. 10:6041-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rougvie, A. E., and J. T. Lis. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54:795-804. [DOI] [PubMed] [Google Scholar]
- 41.Rowley, A., R. A. Singer, and G. C. Johnston. 1991. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol. 11:5718-5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlesinger, M. B., and T. Formosa. 2000. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155:1593-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 44.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, X., A. Finkelstein, A. J. Wolf, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sopta, M., R. W. Carthew, and J. Greenblatt. 1985. Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 260:10353-10360. [PubMed] [Google Scholar]
- 47.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with mRNA export. Nature 417:304-307. [DOI] [PubMed] [Google Scholar]
- 49.Struhl, K. 1996. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell 84:179-182. [DOI] [PubMed] [Google Scholar]
- 50.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell 5:1067-1072. [DOI] [PubMed] [Google Scholar]
- 51.Wada, T., T. Talagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wittmeyer, J., and T. Formosa. 1997. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17:4178-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittmeyer, J., L. Joss, and T. Formosa. 1999. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38:8961-8971. [DOI] [PubMed] [Google Scholar]
- 54.Wobbe, C. R., and K. Struhl. 1990. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol. Cell. Biol. 10:3859-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]
- 56.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]