Abstract
Chromatin assembly in a crude DEAE (CD) fraction from budding yeast is ATP dependent and generates arrays of physiologically spaced nucleosomes which significantly protect constituent DNA from restriction endonuclease digestion. The CD fractions from mutants harboring deletions of the genes encoding histone-binding factors (NAP1, ASF1, and a subunit of CAF-I) and SNF2-like DEAD/H ATPases (SNF2, ISW1, ISW2, CHD1, SWR1, YFR038w, and SPT20) were screened for activity in this replication-independent system. ASF1 deletion substantially inhibits assembly, a finding consistent with published evidence that Asf1p is a chromatin assembly factor. Surprisingly, a strong assembly defect is also associated with deletion of CHD1, suggesting that like other SNF2-related groups of nucleic acid-stimulated ATPases, the chromodomain (CHD) group may contain a member involved in chromatin reconstitution. In contrast to the effects of disrupting ASF1 and CHD1, deletion of SNF2 is associated with increased resistance of chromatin to digestion by micrococcal nuclease. We discuss the possible implications of these findings for current understanding of the diversity of mechanisms by which chromatin reconstitution and remodeling can be achieved in vivo.
Eukaryotes package their genomic DNA into a complex nucleoprotein structure referred to as chromatin. The fundamental repetitive element of chromatin is the nucleosome, which is composed of 146 bp of DNA wrapped around an octamer of histone proteins: two dimers of histone H2A and H2B and a tetramer of histones H3 and H4 (56). This chromatinized template DNA is the substrate for in vivo reactions such as transcription, replication, recombination, and repair. Indeed, proper chromatin assembly or modification is necessary for the accurate execution and regulation of these processes.
The cell uses a number of mechanisms to build nucleosomes. Nucleosomes are assembled in a DNA replication-coupled (RC) manner following the replication fork during S phase and are assembled onto DNA during gap-repair in response to DNA damage (28, 35). In both cases, nascent DNA is packaged into chromatin. Alternatively, it has been demonstrated that nucleosomes can be assembled or reassembled independently of DNA replication. Replication-independent (RI) assembly is not limited to the S phase but occurs continually throughout the cell cycle (2, 4), perhaps functioning as a backup to RC assembly (44). RI assembly can introduce specific histone variants, such as the H3 variant Cid, at centrosomes (1) or histone H3.3 in transcriptionally active regions of the genome (2). By extension it has been proposed that RI assembly may replace histones that have been irreversibly modified by methylation (23); otherwise, the only way to change the histone methylation signal would be by gradual dilution of methylated with unmethylated histones in the course of cell proliferation. RI assembly also occurs in nondividing cells. For example, in nerve cells of higher eukaryotes infected with herpes simplex virus, RI assembly quickly packages viral DNA into chromatin, causing the virus to become latent (13). Since the histone requirements for RC and RI chromatin assembly are distinct, it has been suggested that the two pathways use different nucleosome assembly machineries (2).
Chromatin assembly is effected in the cell by so-called chromatin assembly factors (CAFs). This diverse group of proteins includes histone modifiers, core histone binding factors and ATP-dependent chromatin remodeling factors (36). Histone modifiers such as acetylases, kinases, and methylases covalently alter the nucleosome; such alterations are thought to affect nucleosome packing and the interaction of chromatin with other proteins (24). Core histone-binding factors appear to act as histone chaperones and deliver the histones to the DNA for deposition (22). ATP-dependent chromatin remodeling factors are multisubunit complexes that contain an ATPase subunit belonging to the SNF2-like subfamily of nucleic acid-stimulated DEAD/H ATPases (29, 37). In an ATP-dependent manner, some members of the SNF2-like subfamily are able to space nucleosomes in the course of chromatin assembly or remodel chromatin in response to DNA-binding factors, for example, Gal4-VP16 (20, 53, 57). Based on their domain structures the SNF2-like subfamily of enzymes has been divided into three major groups (29). Members of the SWI2 group contain a bromodomain, those in the ISWI group contain a SANT domain, and chromodomain (CHD)-type enzymes are characterized by chromodomains. In other SNF2-like subfamily members the homology is limited to the ATPase domain. Saccharomyces cerevisiae has multiple core histone-binding factors and SNF2-like subfamily members; however, only two of the ATPases are essential for viability. It is not clear whether all of these factors or only a subset are involved in chromatin assembly or remodeling.
Highly purified and/or recombinant forms of known yeast assembly factors have been widely used to study chromatin metabolism (25, 34, 43). Crude yeast systems have been less extensively used for this purpose, even though biochemical analysis in crude systems has been a mainstay of chromatin research in metazoans, and numerous SNF2-like subfamily members and core histone-binding factors are conserved in S. cerevisiae. Our group has developed a whole-cell extract to study RI chromatin assembly in yeast. This whole-cell extract has provided unexpected insights into the regulation of histone metabolism (4), but its capacity to properly space nucleosomes is very low. Other crude systems typically used for studying RI chromatin assembly in vitro employ extracts of Drosophila embryos (6, 10) and Xenopus oocytes or eggs (38). These extracts likely have greater assembly capacity than crude yeast extracts because chromatin assembly proteins are stockpiled in oocytes and eggs to support early embryogenesis (which in flies and amphibians involves multiple rounds of genome replication and division without intervening gap phases; [17, 39]). However, we anticipated that, by using appropriate extraction methods and a single chromatography step, it would be possible to obtain a crude yeast system in which assembly of correctly spaced nucleosomes could be readily demonstrated by routine micrococcal nuclease digestion analysis. We further expected that the standard genetic approaches available in yeast would provide a simple alternative to biochemical methods for altering the protein composition of chromatin assembly extracts.
We describe here the preparation and use of a crude DEAE (CD) fraction from budding yeast cells which, when supplemented with core histones, supports ATP-dependent assembly of physiologically spaced nucleosome arrays on nonreplicating DNA. Compared to whole-cell extract of yeast (46), this fraction assembles extensive nucleosome arrays in which the DNA is substantially protected from cutting by restriction endonucleases. We performed a targeted screen for genes whose deletion affects chromatin assembly in the yeast system. Disruption of chromatin assembly activity was associated with the absence of Asf1p, a known core-histone-binding factor, and Chd1p, a SNF2-like ATPase previously thought to function only in chromatin remodeling (52).
MATERIALS AND METHODS
Strains and cell growth.
Strains are listed in Table 1. All deletion mutants (from Research Genetics) were created by the yeast gene disruption project by using the kanMX4 disruption cassette (58). Yeast cells are grown in YPD1%AS [1% yeast extract, 2% Bacto Peptone, 2% glucose, 1% (NH4)2SO4 (pH 6.5)] at 30°C.
TABLE 1.
Strains used in this study
| Strain | Doubling time (min) | Relevant genotypea |
|---|---|---|
| DSY904 | 92 | MATabar1 ade1 his2 leu2 trp1 ura3 pep4::URA3; parental strain BF264-15 (45) |
| BY4741 | 90 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| asf1Δ | 124 | asf1::kanMX4 |
| cac1Δ | 98 | cac1::kanMX4 |
| chd1Δ | 92 | chd1::kanMX4 |
| isw1Δ | 94 | isw1::kanMX4 |
| isw2Δ | 92 | isw2::kanMX4 |
| nap1Δ | 92 | nap1::kanMX4 |
| spt20Δ | 145 | spt20::kanMX4 |
| snf2Δ | 147 | snf2::kanMX4 |
| swr1Δ | 92 | swr1::kanMX4 |
| YFR038w | 90 | yfr038w::kanMX4 |
All kanMX4 deletion strains listed are isogenic to the wild-type strain BY4741.
PICs.
Dimethyl sulfoxide (DMSO) protease inhibitor cocktail (PIC) (1 M phenylmethylsulfonyl fluoride [PMSF], 5 mg of pepstatin A/ml, 25 mg of TPCK [tolylsulfonyl phenylalanyl chloromethyl ketone]/ml, and 2.5 mg of chymostatin/ml dissolved in DMSO and stored at −20°C) and aqueous PIC (10 mg of aprotinin/ml, 5 mg of leupeptin/ml, 1 M p-aminobenzamidine, and 1 M ɛ-aminocaproic acid dissolved in water and stored at −20°C) were used where indicated at a 1:1,000 dilution.
Preparation of the CD fraction.
Cells were harvested at an optical density at 600 nm (OD600) of 2 to 2.5, resuspended in 1/4 volume of fresh YPD1%AS prewarmed to 30°C and grown at 30°C for an additional 90 min. Tris-HCl (pH 8.0) and β-mercaptoethanol were added to final concentrations of 0.1 M and 65 mM, respectively, and the culture was stirred at room temperature for 15 min. The cells were harvested, resuspended in 3 ml of S buffer [1.1 M sorbital, 1% yeast extract, 1% (NH4)2SO4, 2% peptone, 2% glucose, 40 mM Tris-HCl (pH 7.5), 1 mM PMSF, 2 mM benzamidine, 2 mM sodium metabisulfite, 10 mM β-mercaptoethanol]/g of cells, and incubated at 30°C with 16,000 U of β-endoglucanase/g of cells with shaking until the OD600 of a 1/100 dilution in water decreased to 20 to 30% of the starting value. The β-endoglucanase was produced from Escherichia coli (unpublished data). All subsequent spins were done at 4°C in ice-cold buffers. The cells were pelleted and washed twice with 5 ml of wash buffer II (1.1 M sorbital, 10 mM Tris-HCl [pH 6.8], 2 mM EDTA, 1 mM PMSF, 2 mM benzamidine, 2 mM sodium metabisulfite)/g of cells. For a nuclear extract, the spheroplasts were resuspended in lysis buffer [18% Ficoll 400, 80 mM KH2PO4 (pH 6.8), 0.25 mM EDTA, 0.25 mM EGTA, 0.2 mM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride], 3 mM benzamidine, 3 mM dithiothreitol (DTT), and 2 mM sodium metabisulfite plus DMSO PIC and aqueous PIC] at 3 ml/g of spheroplasts and lysed by Dounce homogenization. Unbroken cells and debris were removed by centrifugation for 5 min (3,000 × g at 4°C), the supernatant retrieved and centrifuged for 30 min (21,000 × g at 4°C) to pellet the nuclei. Nuclei were resuspended in buffer A (25 mM HEPES-KOH [pH 7.5], 0.35 M NaCl, 1.5 mM magnesium acetate, 0.5 mM EGTA, 10% glycerol, 0.2 mM AEBSF, 3 mM benzamidine, 3 mM DTT, and 2 mM sodium metabisulfite plus DMSO PIC and aqueous PIC) at 3 ml/g of nuclei with gentle douncing and then incubated on ice for 10 min. For a spheroplast extract, the spheroplasts were resuspended directly in 2 ml of buffer A/g of spheroplasts, homogenized, and incubated on ice for 25 min. For both nuclear and spheroplast extracts, an additional 5.5 mM magnesium acetate was added to the extract, and the mixture was spun at 40,000 rpm in an SW41 rotor for 2 h. The supernatant was collected carefully, avoiding the pellet and fat layer. The conductivity of the supernatant was adjusted to 0.1 M NaCl either by dialysis against HEMG (20 mM HEPES [pH 7.5], 0.5 mM EGTA, 1.5 mM magnesium acetate, 10% glycerol, 2 mM DTT, 2 mM benzamidine, 2 mM sodium metabisulfite, 1 mM PMSF, 1 mM β-glycerol phosphate) until its conductivity was equal to 0.1 M NaCl-HEMG and then frozen in liquid nitrogen or by diluting previously frozen supernatant with HEMG to a final NaCl concentration of 0.1 M immediately prior to column chromatography. The diluted or dialyzed supernatant was applied at a ratio of 45 mg of protein/ml of resin to a DEAE-Sepharose fast flow (AP Biotech) column that had been preequilibrated with 0.1 M NaCl-HEMG. After it was loaded, the column was washed with six column volumes of 0.1 M NaCl-HEMG, and the assembly-competent fraction was step eluted with five column volumes of 0.4 M NaCl-HEMG. This fraction was dialyzed against yR buffer (10 mM HEPES [pH 7.5], 10 mM potassium acetate, 1.5 mM magnesium acetate, 0.5 mM EGTA, 10% glycerol, 2 mM DTT, 2 mM benzamidine, 2 mM sodium metabisulfite, 1 mM PMSF, 1 mM β-glycerol phosphate) in tubing (molecular weight cutoff, 6,000 to 8,000) twice for 2 h each time. After dialysis this CD fraction was frozen in liquid nitrogen in aliquots.
Chromatin assembly reactions.
A standard reaction contains CD fraction and Drosophila core histones (diluted together in yR buffer to 1 mg/ml and 7.5 μg/ml, respectively; the core histones lack H1), 6 mM MgCl2, 5 μg of pGIE0 plasmid DNA (41)/ml, and an ATP regeneration system of 3 mM ATP, 30 mM creatine phosphate (Sigma catalog no. P6502), and 6 μg of creatine kinase (Sigma catalog no. C3755)/ml. The CD fraction and histones were added to yR buffer supplemented with 2.4 mM MgCl2, followed by incubation at room temperature for 15 min. The ATP regeneration mix and plasmid DNA were added, and the reaction was incubated at 30°C for 1 to 3 h. Where indicated, yeast histones were used at a concentration of 9 μg/ml, and apyrase (Sigma catalog no. A6410) was added to a final concentration of 2 U/ml. Unless indicated otherwise, chromatin assembly reactions were performed with CD fractions prepared from the yeast strain DSY904.
RE access.
Assembly reactions (50 μl) were mixed with 25 μl of RNG (10 mM HEPES [pH 7.5], 10 mM KCl, 12 mM MgCl2). Aliquots (25 μl) of this mix were treated with either 10 U (HindIII and XbaI) or 5 U (BamHI, KpnI, and SphI) of restriction enzyme (RE) or no RE and then incubated at 37°C for 30 min. Digestion was stopped with 20 mM EDTA, the samples were deproteinized, and the DNA was precipitated. All DNA samples were subsequently digested with BglII and RNase A.
Yeast core histone isolation.
Yeast core histones (shown in Fig. 4A) were isolated according to (43) with the following modifications. Previously frozen cells were digested with only β-1,3-endoglucanase (34,000 U/g of cells) in spheroplasting buffer (1.1 M sorbital, 0.75% yeast extract, 1.5% peptone, 10 mM Tris-HCl [pH 7.5], 1 mM PMSF, 2 mM benzamidine, 2 mM sodium metabisulfite, 10 mM β-mercaptoethanol). When digestion was complete, the cells were harvested at 4°C and washed twice in 5 ml of ice-cold wash buffer II/g. The cells were resuspended in 2 ml of ice-cold lysis buffer plus polyamines (0.5 mM spermidine and 0.15 mM spermine)/g and lysed with a polytron (Kinematica; distributed by Brinkman Instruments) on setting 7 for six 1-min intervals on ice. Unbroken cells and cell debris were removed by centrifugation at 3,000 × g for 10 min at 4°C. The supernatant was recovered, and the nuclei were spun down at 21,000 × g for 30 min at 4°C. The pellet was resuspended in ice-cold nuclear storage buffer (100 mM Tris acetate [pH 7.9], 50 mM potassium acetate, 20% glycerol, 2 mM EDTA, 0.2 mM AEBSF, 3 mM benzamidine, 3 mM DTT, and 2 mM sodium metabisulfite plus DMSO PIC and aqueous PIC) at 0.5 ml/g of cells and frozen in liquid nitrogen in roughly 18-ml aliquots.
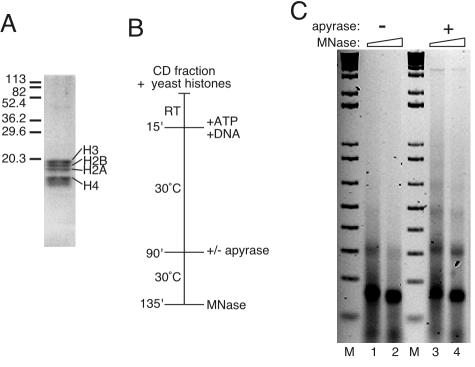
FIG. 4.
Yeast core histones form extensive nucleosomal arrays when ATP-dependent nucleosomal mobilization is inhibited. (A) Coomassie blue staining of yeast histones resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The migration of molecular mass markers is indicated on the left in kilodaltons. (B) Flow diagram of reactions, which were performed by using the standard reaction cocktail. After 75 min, apyrase was added to half of the samples (lanes 3 and 4 in panel C), and all of the reactions were incubated for an additional 45 min. RT, room temperature; MNase, micrococcal nuclease. (C) Assembly products were assayed by micrococcal nuclease digestion, followed by agarose gel electrophoresis. The DNA was visualized with ethidium bromide. M, 1 kbp plus DNA ladder.
Then, 200 μl of freshly made RNase A (10 mg/ml), 20 μl of 0.2 M AEBSF, and 18 μl each of 1,000× DMSO and 1,000× aqueous PIC was added to 18 ml of nuclei, while they were thawing. The nuclei were prewarmed to 37°C and then digested with 72 μl of 500 U of micrococal nuclease/ml for 5 min at 37°C. Digestion was stopped with addition of 1.2 ml of 0.5 M EDTA (to a final concentration of 30 mM), and the nuclei were placed on ice. A total of 36 μl of 0.5 M sodium metabisulfite and 1.87 ml of 5 M NaCl (final NaCl concentration of 0.5 M) was added, and the nuclei were pelleted (25,000 × g, 15 min, 4°C). The supernatant was removed and loaded onto a 320-ml Sephacryl S300 column (Amersham Pharmacia Biotech) that had been preequilibrated with 0.5 M gel filtration buffer (0.5 M NaCl, 100 mM Tris acetate, 50 mM potassium acetate, 2 mM EDTA, 10% glycerol, 3 mM DTT, 2 mM benzamidine, 2 mM sodium metabisulfite, 1 mM PMSF). Peak fractions were pooled and diluted to 0.3 M NaCl with 0 M gel filtration buffer and supplemented with DMSO PIC and aqueous PIC and AEBSF to a final concentration of 0.2 mM. The A260 of the chromatin fractions was determined, and the chromatin loaded onto Macro-Prep Ceramic Hydroxyapatite Type I (40 μm; Bio-Rad) in a XK-16 column at a ratio of 2.5 mg of chromatin/ml of resin and at a flow rate of 0.2 ml/min. The resin was preequilibrated with 0.3 M NaCl-HAP buffer (80 mM NaPi, 10% glycerol, 3 mM DTT, 2 mM benzamidine, 2 mM sodium metabisulfite, 1 mM PMSF). The column was washed with one column volume of 0.3 M NaCl-HAP buffer, the flow rate was increased to 1 ml/min, and the column was washed with a five-column gradient of 0.3 to 0.8 M NaCl, followed by five column volumes of 0.8 M NaCl-HAP buffer. The core histones were eluted with a 2.5 M NaCl step, and 1-ml fractions were collected. The peak fractions of histones were identified, and 1 μl each of 0.2 M AEBSF, DMSO PIC, aqueous PIC, and 10% NP-40 was added. The fractions were transferred to dialysis tubing (6,000 to 8,000 molecular weight cutoff), concentrated against polyethylene glycol (molecular weight, 8,000) for 2.5 h at 4°C, and then dialyzed against core histone storage buffer (10 mM HEPES [pH 7.5], 1 mM EDTA, 10 mM potassium acetate, 10% glycerol, 1 mM DTT, 2 mM benzamidine, 2 mM sodium metabisulfite, 0.1 mM PMSF, 0.1 mM AEBSF, 0.01% NP-40, aqueous PIC) twice for 2 h each time.
Other procedures.
The supercoiling and micrococcal nuclease digestion assays were performed as described previously (42). Micrococcal digestions were stopped with EDTA, the samples were treated with RNase A, and the digestion products were isolated.
RESULTS
A CD fraction that supports chromatin assembly.
We have developed a new in vitro chromatin assembly system for budding yeast. Figure 1A outlines the procedure for preparing the critical component of this system, a CD fraction of the 190,000 × g supernatant (S-190) from an extract of spheroplasts or nuclei. Chromatography on DEAE was chosen for enrichment of chromatin assembly and remodeling factors in the S-190 fraction because many known assembly factors bind to DEAE and elute between 0.1 and 0.4 M NaCl (e.g., dCAF1 [9, 10], nucleoplasmin-like protein [21], dNAP1 [19], ACF [20], hACF [31], FACT [40], and RSF [32]). In addition, we found that fractionation of S-190 on DEAE removes any contaminating chromosomal DNA and concentrates the assembly activity (data not shown). Because the CD fractions from spheroplast and nuclear S-190s behave identically in all assays, our reference to the CD fraction below indicates that the results apply to both spheroplast- and nucleus-derived CD fractions.
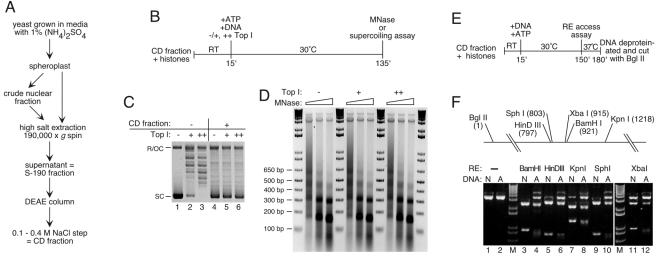
FIG. 1.
(A) Flow diagram for preparation of the CD fraction. (B) Chromatin assembly using the CD fraction: outline of experiment. Plasmid DNA was relaxed with topoisomerase I (Top I, + and ++) prior to assembly or not (−). RT, room temperature; MNase, micrococcal nuclease. (C) Assembly with the CD fraction results in supercoiling of previously relaxed templates. After assembly, plasmid DNA was reisolated, resolved by 1% agarose gel electrophoresis, and stained with ethidium bromide. SC, supercoiled plasmid; R/OC, relaxed/open circular plasmid. Lanes 1 to 3 show the input DNA for the assembly reactions in lanes 4 to 6. (D) Assembly of physiologically spaced nucleosomal arrays does not require exogenous topoisomerase. The reaction products were treated with 0.37, 1.1, or 3.3 U of micrococcal nuclease/ml, resolved by agarose gel electrophoresis, and visualized with ethidium bromide. M, 1 kbp plus DNA ladder (Stratagene). (E) RE protection assay: outline of experiment. (F) DNA assembled into chromatin by using the CD fraction is protected from RE digestion. The diagram shows the relative positions of cutting sites for enzymes used. Products from reactions with naked (N) or assembled (A) DNA were resolved by agarose gel electrophoresis and visualized with ethidium bromide. The 3.3-kbp linear plasmid is shown in lanes 1 and 2. M, 1 kbp plus DNA ladder (Stratagene).
Chromatin assembly was performed according to a standard reaction protocol (Fig. 1B [varied as indicated]) and monitored by plasmid supercoiling, digestion with micrococcal nuclease, and testing RE accessibility. The CD fraction supports supercoiling of previously relaxed plasmid DNA (Fig. 1C) by a mechanism that generates arrays of nucleosomes with a repeat length of ca. 160 bp (Fig. 1D), close to the physiological value for log-phase yeast (56). Assembled DNA is protected from RE cutting, as reported for other biological assembly systems (for an example, see reference 54). Chromatin was incubated with one of five REs (BamHI, HindIII, KpnI, SphI, or XbaI), and the DNA purified and cut with BglII at a known distance from the recognition site of the first enzyme (Fig. 1E and F). BglII generates a 3.3-kbp linear molecule from previously uncut plasmids and two smaller DNA fragments if the first enzyme had cut the nucleosomal template. Under the conditions used, naked DNA is efficiently cleaved by BamHI and SphI (Fig. 1F, lanes 3 and 9; note the substantial loss of 3.3-kbp fragment), whereas HindIII, KpnI, and XbaI cutting is incomplete because these enzymes function less efficiently in assembly buffer (Fig. 1F, lanes 5, 7, and 11). Even so, assembly into nucleosomes, as judged by the diminished yield of the smallest double-digestion products, protects all tested sites in chromatinized templates from RE access (Fig. 1F, lanes 4, 6, 8, 10, and 12). This protection is also evident in the increased yield of the 3.3-kbp linear molecule.
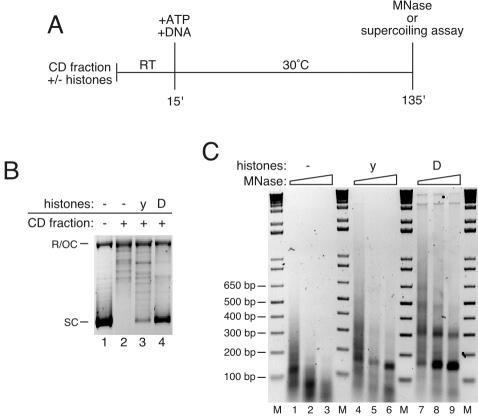
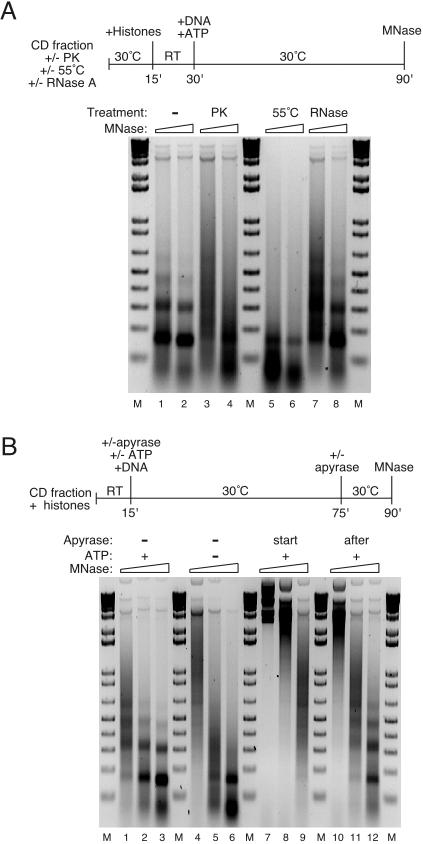
Under standard conditions, assembly is not stimulated by exogenous topoisomerase (Fig. 1C and D). The supercoiling and nucleosomal assembly activities observed are dependent on supplementation with exogenous histones (Fig. 2B and C). Array formation is abolished in reactions with proteinase K-treated (Fig. 3A, compare lanes 1 and 2 to lanes 3 and 4) or heat-treated (Fig. 3A, compare lanes 1 and 2 to lanes 5 and 6) CD fraction but not in reactions with CD fraction pretreated with RNase A (Fig. 4A, lanes 7 and 8). Interestingly, none of these treatments prevents mononucleosome formation, which occurs to the same low extent in reactions containing just DNA and histones (not shown). Furthermore, the extent of assembly is dependent on the amount of CD fraction protein used (see asf1Δ and chd1Δ results below). We conclude that although mononucleosomes can be generated spontaneously in the absence of additional protein factors, proteins in the CD fraction facilitate efficient nucleosome deposition in physiologically spaced arrays. A hallmark of biological systems that assemble extensive arrays of physiologically spaced nucleosomes is their dependence on ATP. We sought to determine whether the yeast assembly system shares this property. Figure 3B shows that nucleosomal ladders are not assembled in reactions from which ATP, creatine phosphate, and creatine kinase have been omitted (compare lanes 1 to 3 with lanes 4 to 6). Apyrase also strongly inhibits ladder formation when preincubated with the CD fraction and the ATP regeneration system (Fig. 3B, lanes 7 to 9). We conclude that assembly of nucleosome arrays by the CD fraction requires ATP. In contrast to its effect when preincubated before assembly, apyrase addition after 1 h of assembly in the presence of ATP does not inhibit the formation of nucleosome arrays (Fig. 3B, compare lanes 1 to 3 to lanes 10 to 12).
FIG. 2.
Chromatin assembly requires exogenous histones. (A) Outline of experiment. Plasmid DNA was incubated with only CD fraction or with CD fraction and yeast (y) or Drosophila (D) core histones. RT, room temperature; MNase, micrococcal nuclease. (B) Plasmid supercoiling. After assembly, plasmid DNA was reisolated, resolved by 1% agarose gel electrophoresis and stained with ethidium bromide. SC, supercoiled plasmid; R/OC, relaxed/open circular plasmid. (C) Micrococcal nuclease digestion assay. The product of the micrococcal nuclease digestions were resolved by agarose gel electrophoresis and visualized with ethidium bromide. M, 1 kbp plus DNA ladder.
FIG. 3.
Assembly of nucleosome arrays is protein dependent and requires ATP. (A) Assembly of nucleosome arrays is protein dependent. Reactions were performed as summarized in the flow diagram. The CD fraction was pretreated for 15 min at 30°C by itself (lanes 1 and 2), with proteinase K (PK, 0.3 μg/μl; lanes 3 and 4), or with RNase A (0.12 μg/μl; lanes 7 and 8) or was incubated at 55°C for 15 min (lanes 5 and 6). The proteinase inhibitor AEBSF (15 mM) was added to the proteinase K-digested sample after pretreatment. The pretreated samples were then mixed with core histones for assembly. Micrococcal digestion assays were performed and analyzed by agarose gel electrophoresis, and the DNA was visualized with ethidium bromide. RT, room temperature; MNase, micrococcal nuclease; M, 1 kbp plus DNA ladder. (B) Assembly of nucleosome arrays is ATP dependent. Assembly reactions were performed according to the flow diagram, in the absence (−) or presence (+) of ATP, creatine phosphate, and creatine kinase. All reactions contained 6 mM MgCl2. The ATP regeneration system and the CD fraction were pretreated with apyrase (start, lanes 7 to 9), or apyrase was added to the assembly reaction for 45 min after 1 h of assembly (after, lanes 10 to 12). Micrococcal digestions were performed, and the products were resolved by agarose gel electrophoresis and visualized with ethidium bromide. RT, room temperature; MNase, micrococcal nuclease; M, 1 kbp plus DNA ladder.
Surprisingly, bulk yeast histones are less suitable substrates for assembly under standard conditions than are Drosophila histones (the purified yeast histones used are shown in Fig. 4A). Using native yeast histones, supercoiling is less efficient than in reactions with the Drosophila proteins (Fig. 2B, lanes 2 to 4), and whereas nucleosomal monomers and dimers are readily generated by micrococcal nuclease digestion (Fig. 2C), trimers are barely detectable even when the amount of yeast histones is increased (not shown).
Addition of S-adenosylmethionine (to promote protein methylation) or trichostatin A (to inhibit histone deacetylases) or the use of recombinant yeast histones instead of native yeast histones did not improve assembly (data not shown). On the other hand, the addition of apyrase after 75 min of assembly and 45 min prior to reaction termination had a dramatic effect on array formation. Specifically, ATP depletion during the final 45 min of the 2 h reaction improved the definition and extent of the arrays, with tetramers being readily detected (Fig. 4C, lanes 3 and 4). Furthermore, when assembly reactions with yeast histones were digested with a higher concentration of micrococcal nuclease for a much shorter period of time, the nucleosomal ladders improved (K. M. Robinson and M. C. Schultz, data not shown). We conclude that native yeast histones can be efficiently assembled into nucleosomal arrays in this yeast system. Such arrays, compared to those formed by native Drosophila histones, may, however, be better substrates for ATP-dependent nucleosome mobilizing activities which are possibly active in the CD fraction.
A targeted screen of deletion mutants for effects on chromatin assembly activity in vitro.
Using strains with deletions of known CAFs or their homologues, we determined the effect on assembly of eliminating selected individual proteins from the CD fraction. Each CD fraction from wild-type and mutant strains was assayed repeatedly by plasmid supercoiling and partial micrococcal nuclease digestion. The results were highly reproducible with negligible variation in activity between different CD fractions produced from the same strain. Although chromatin assembly may be performed by redundant activities in yeast, we could detect substantial effects on activity of the CD fraction from some single mutants.
Deletion of known histone binding factors affects assembly in vitro.
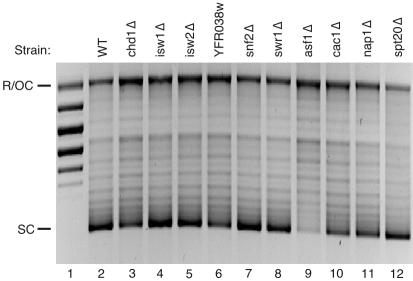
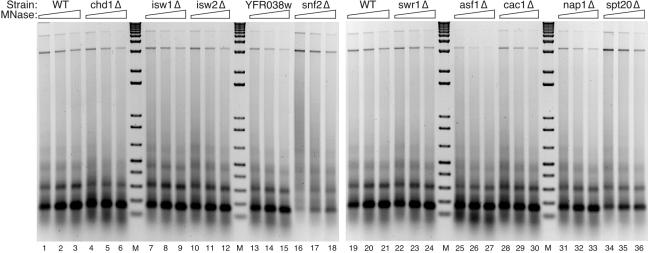
Among histone binding factors we tested NAP1, CAC1 (a subunit of the CAF-I complex), and ASF1. Even though none of these genes is essential, each has CAF function in vitro (19, 34, 43, 47), and the phenotypes of the respective null mutants suggest that Asf1p and Cac1p have a role in chromatin assembly in vivo (25, 55). Initial screening indicated that deletion of NAP1 or CAC1 causes slight inhibition of plasmid supercoiling (Fig. 5, compare WT in lane 2 to cac1Δ in lane 10 and nap1Δ in lane 11) but has little or no effect on the pattern of digestion by micrococcal nuclease (Fig. 6, compare WT in lanes 19 to 21 to cac1Δ in lanes 28 to 30 and nap1Δ in lanes 31 to 33).
FIG. 5.
Screening of CD fraction from 10 deletion mutants for chromatin assembly activity (plasmid supercoiling). Chromatin templates were assembled with CD fractions prepared from wild-type cells (WT; BY4741) or 10 isogenic strains harboring deletions of the indicated genes. After assembly reactions, the plasmid DNA was reisolated, resolved by agarose gel electrophoresis, and stained with ethidium bromide. In Fig. 6 and 7, reactions were performed with the final concentration of extract at 1.2 mg/ml. The activities of mutant and wild-type (lane 2) samples were compared. Relaxed plasmid DNA was run in lane 1. SC, supercoiled plasmid; R/OC, relaxed/open circular plasmid.
FIG. 6.
Screen of capacity to assemble nucleosome arrays. Two wild-type (WT; BY4741) CD fractions are shown (lanes 1 to 3 and 19 to 21). Array assembly was monitored by partial micrococcal nuclease digestion (MNase) followed by agarose gel electrophoresis and staining with ethidium bromide. M, 1 kbp plus DNA ladder.
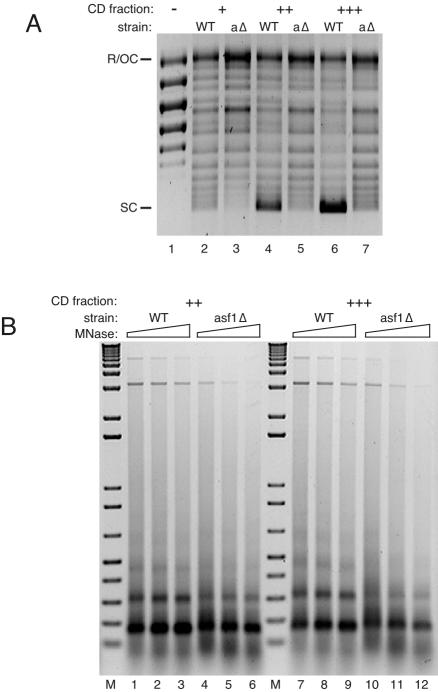
Deletion of ASF1 was associated with strong inhibition of supercoiling (Fig. 5, compare lanes 2 and 9). The CD fraction from the asf1Δ strain was defective in supercoiling relative to the wild type over a range of CD fraction concentrations (Fig. 7A; also, data not shown). Because the lowest amount of wild-type or asf1Δ CD fraction has enough topoisomerase activity to completely relax supercoiled input plasmid (data not shown), the supercoiling defect of asf1Δ CD fraction was not expected to be due to a deficiency in topoisomerase activity. Consistent with this idea, assembly was defective in asf1Δ reactions performed in the absence of exogenous topoisomerase (Fig. 5 and data not shown) or with previously relaxed template (Fig. 7A). The formation of nucleosomal arrays, as shown in Fig. 6, was also impaired in asf1Δ extract compared to wild type (lanes 19 to 21); the dimer and particularly the trimer were less distinct in the asf1Δ assemblies (lanes 25 to 27; see also the side-by-side experiment in Fig. 7B). Nucleosomal array formation activity was also impaired in asf1Δ CD fractions compared to wild type over increasing concentrations of CD fractions (Fig. 7B; only supercoiling is shown for the lowest concentration reactions because when the input of CD fraction is low, nucleosomal spacing activity is defective even in wild-type reactions [data not shown]). These results indicate that nucleosome assembly activity in this in vitro system is partly dependent on expression of Asf1p.
FIG. 7.
Chromatin assembly in the CD fraction from asf1Δ cells. (A) Supercoiling assay of reactions with increasing amounts of CD fraction from wild-type (WT; lanes 2, 4, and 6) and asf1Δ (aΔ; lanes 3, 5, and 7) strains. Reactions were done with CD fraction at final concentrations of 0.7 (+), 1.0 (++), and 1.4 (+++) mg/ml. After assembly, the plasmid DNA was reisolated, resolved by agarose gel electrophoresis, and stained with ethidium bromide. To ensure that the supercoiling defect was not due to a lack of topoisomerase activity, the plasmid DNA was relaxed prior to assembly, and the reactions were supplemented with topoisomerase I. The relaxed input DNA was run in lane 1. SC, supercoiled plasmid; R/OC, relaxed/open circular plasmid. (B) Micrococcal nuclease digestion assay of chromatin assembled with CD fraction from wild-type (WT; lanes 1 to 3 and lanes 7 to 9) and asf1Δ (lanes 4 to 6 and lanes 10 to 12) strains, with the amount of CD fraction varied as in panel A (++ and +++). After micrococcal digestion, the DNA was reisolated, resolved by agarose gel electrophoresis, and stained with ethidium bromide. MNase, micrococcal nuclease; M, 1 kbp plus DNA ladder.
Deletion of two SNF2-like ATPases affects assembly in vitro.
Six members of the SNF2-like subfamily of DEAD/H ATPases were also screened for involvement in chromatin assembly in vitro. Deletion of two SNF2-like homologues of unknown function, YFR038w and SWR1, had little or no effect on assembly (Fig. 5, compare lane 2 to lanes 6 and 8; Fig. 6, compare lanes 1 to 3 to lanes 13 to 15 and compare lanes 19 to 21 to lanes 22 to 24). Loss of either Isw1p or Isw2p, closely related ISWI group members, was also not associated with a loss of definition of micrococcal nuclease ladders (Fig. 6, compare lanes 1 to 3 to lanes 7 to 9 and lanes 10 to 12) or a decrease in supercoiling activity (Fig. 5, compare lanes 2 to 4 and lane 5). This result is surprising since Isw1p and Isw2p are found in separate multisubunit complexes and are the only SNF2-like homologues in yeast known to have nucleosomal spacing activity in vitro (16, 54). On the other hand, null mutation of two other SNF2-like ATPases did reproducibly affect the assembly reaction.
Snf2p, the founding member of the SNF2-like subfamily, is part of the SWI/SNF complex, which has been shown to disrupt nucleosome structure and aid in transcription factor binding to nucleosomal DNA (5, 12). Deletion of SNF2 does not affect plasmid supercoiling (Fig. 5, compare lanes 2 to 7) or the presence of trimers and tetramers in micrococcal nuclease digestion assays (Fig. 6, compare lanes 1 to 3 to lanes 16 to 18). Comparison of the micrococcal nuclease digestion products, however, reveals that snf2Δ chromatin (Fig. 6, lanes 16 to 18) is more resistant to digestion than wild type (lanes 1 to 3). Deletion of SNF2 therefore somehow affects the micrococcal nuclease accessibility of DNA in chromatin assembled by the CD fraction.
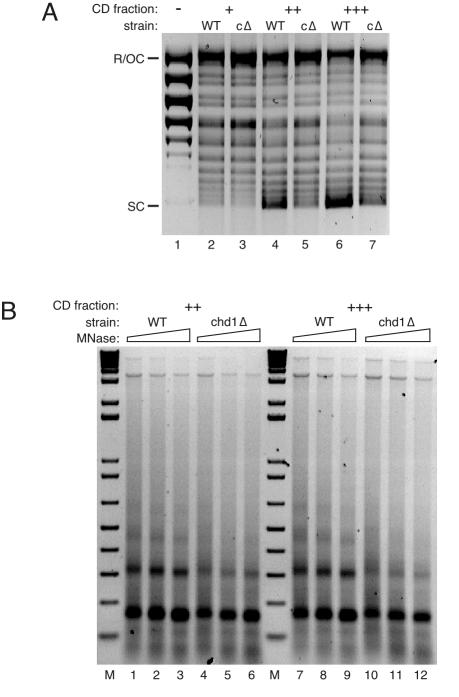
Chd1p has activity that affects DNA-histone interactions within the nucleosome and is the only CHD protein in yeast (52). Its loss decreased the extent of chromatin assembly in vitro more than the elimination of any other SNF2-like protein tested. Under standard conditions in the CD fraction from chd1Δ cells supercoiling is marginally decreased (Fig. 5, compare lanes 2 and 3), and the formation of nucleosomal arrays is defective (Fig. 7, compare lanes 1 to 3 to lanes 4 to 6). The difference in supercoiling activity between the wild-type and chd1Δ CD fractions was accentuated when reactions were performed with prerelaxed (Fig. 8A) as opposed to supercoiled template (Fig. 5; also, data not shown). We do not know why the difference in activity between the wild-type and chd1Δ CD fractions is sensitive to the topology of the input template, except that it is not due to a lack of topoisomerase activity in the mutant CD fraction (all mutant fractions relax the template as efficiently as wild type, even at the lowest amount of fraction used; data not shown). Although supercoiling and array reconstitution are improved when more chd1Δ CD fraction is used, assembly by the mutant CD fraction is always poor in comparison to the same amount of wild-type fraction (Fig. 8). This is particularly evident in dimer recovery after micrococcal nuclease digestion, which is consistently severalfold lower for chd1Δ chromatin (Fig. 8B).
FIG. 8.
Chromatin assembly in the CD fraction from chd1Δ cells. (A) Supercoiling assay of reactions with increasing amounts of CD fraction from wild-type (WT; lanes 2, 4, and 6) and chd1Δ (cΔ; lanes 3, 5, and 7) strains. Reactions were done with CD fraction at final concentrations of 0.7 (+), 1.0 (++), and 1.4 (+++) mg/ml. After assembly the plasmid DNA was reisolated, resolved by agarose gel electrophoresis, and stained with ethidium bromide. The relaxed input DNA was run in lane 1. SC, supercoiled plasmid; R/OC, relaxed/open circular plasmid. (B) Micrococcal nuclease digestion assay with chromatin assembled with CD fraction from wild-type (WT; lanes 1 to 3 and lanes 7 to 9) and chd1Δ (lanes 4 to 6 and lanes 10 to 12) strains, with the amount of CD fraction varied as in panel A (++ and +++). After micrococcal digestion, the DNA was reisolated and resolved by agarose gel electrophoresis. The DNA was visualized with ethidium bromide. MNase, micrococcal nuclease; M, 1 kbp plus DNA ladder.
DISCUSSION
We describe here the preparation and use of a crude fraction from yeast cells that supports RI chromatin assembly. Several key observations indicate that the assembly reaction occurs by a biologically relevant mechanism. (i) Assembly requires CD fraction proteins and exogenous histones. (ii) The reaction is ATP dependent. (iii) The reaction generates physiologically spaced arrays of nucleosomes in which the DNA is significantly protected from digestion by restriction endonucleases. Assay of the CD fraction from strains harboring individual deletions of genes known or suspected to function in chromatin metabolism further verified the utility of the system for studies of chromatin assembly and has implicated two members of the SNF2-like subfamily of ATPases in this process.
In addition to chromatin assembly, the CD fraction supports Gal4-VP16-dependent chromatin remodeling. This activity will be the subject of a separate study (unpublished data).
The validity of screening CD fractions from mutants for effects on chromatin assembly.
Screening of CD fractions from 10 null mutants created by the Saccharomyces Genome Deletion project yielded wild-type or near-wild-type assembly activity in preparations from seven mutants and impaired or altered activity in the CD fractions from three others. Therefore, the strategy used in the Genome Deletion project to generate null mutants does not have a dominant effect on assembly in vitro. None of the mutations resulted in a complete loss of chromatin assembly activity. Considering the number of possible SNF2-like ATPase chromatin remodeling factors and core histone binding factors in yeast and the fact that none of the null mutations examined is lethal, the removal of only one factor would be expected to have only a subtle effect on chromatin assembly in vitro. Thus, the degree of impairment we see in chd1Δ and asf1Δ mutant CD fractions is rather surprising.
It is possible that altered assembly activity in the CD fractions from the null mutants is due either wholly or partly to indirect in vivo effects on other proteins in chromatin metabolism. However, the in vitro reaction uses exogenous histones. Therefore, effects on assembly in vitro must be independent of in vivo effects on histone metabolism. Furthermore, microarray analysis of the three mutant strains with decreased or altered chromatin assembly activity—i.e., the chd1Δ (52) asf1Δ (J. S. Williams and M. C. Schultz, unpublished), and SWI/SNF mutant strains (18, 51)—does not reveal altered mRNA expression of other nonhistone genes known to function in chromatin assembly. Additionally spt20Δ cells yield near-wild-type assembly activity (Fig. 4A, lane 14, and B, lanes 37 to 39), although SPT20 deletion is associated with transcriptional misregulation of 16% of scored genes (data in supplementary material for reference 30). Therefore, misregulation of such genes is unlikely to account for the in vitro effects we observe. The assembly capacity of a CD fraction is also not a reflection of cell growth rate (Table 1). For example, assembly was impaired in the CD fraction from chd1Δ cells that grow at wild-type rate, and yet the CD fraction from the spt20Δ mutant, the slowest-growing strain, had near-wild-type activity. Therefore, differences in chromatin assembly activity are not caused by secondary effects of slower growth rates in the null mutants.
Activity of known CAFs in the CD fraction.
Loss of Asf1p impairs assembly in the CD fraction. The strong reduction of activity in the CD fraction lacking Asf1p, a known and well-characterized CAF, validates the system and suggests that it will be useful for further identification and characterization of assembly factors. Near-wild-type activity was supported in CD fractions lacking Cac1p, a highly conserved subunit of CAF-I (25). This is a surprising result because CAF-I, like Asf1p, is involved in RC and RI chromatin assembly (25, 47, 49, 55). The distinct assembly phenotypes conferred by ASF1 and CAC1 deletions may reflect the different roles that Asf1p and CAF-I could have in nucleosome reconstitution. Mello and Almouzni (35) have speculated that Asf1p has a more critical and global role in chromatin assembly than CAF-I because Asf1p delivers histones to either DNA or to CAF-I, which subsequently transfers the histones to DNA. This idea is consistent with the relative severity of the transcriptional silencing and DNA damage sensitivity phenotypes of asf1Δ and cac1Δ cells (14, 25, 55) and is supported by biochemical studies with purified factors (47). Our observation that the ASF1 deletion is associated with a more severe defect in activity in the CD fraction than the CAC1 deletion also supports this proposal.
Evidence that chromatin assembly involves members of the SNF2-like subfamily of ATPases.
SNF2-like ATPases have been broadly implicated in chromatin metabolism, including RI chromatin assembly (20, 33, 53, 57). We find that chromatin assembled in the CD fraction from snf2Δ cells has normal spacing but is more resistant to micrococcal nuclease digestion than wild-type chromatin. Among other possibilities this suggests that Snf2p is involved in processes that oppose chromatin “condensation.” In support of this contention, Krebs and colleagues (26) have presented evidence that activation of late mitotic genes requires disruption of condensed mitotic chromatin by the SWI/SNF complex, of which Snf2p is a critical subunit.
The yeast genome encodes only one member of the CHD class of SNF/SWI-related ATPases. Deletion of this family member (CHD1) is associated with strong inhibition of chromatin assembly in vitro. As revealed by micrococcal nuclease digestion analysis, this effect is equal to or greater than the effect of ASF1 deletion. On the other hand, CHD1 deletion has a modest affect on supercoiling compared to ASF1 deletion. A reasonable interpretation of this behavior is that, unlike Asf1p (47), Chd1p has nucleosome spacing activity but is not needed for histone deposition.
The assembly defect in chd1Δ is surprising because Chd1p has not previously been implicated in nucleosome reconstitution. It does, however, have similarities to other chromatin remodeling factors. For example, the biochemical properties of Chd1p (52) overlap with those of Snf2p and the ISWI complexes. Free and nucleosomal DNA stimulates the ATPase activity of Chd1p and the SWI/SNF and ISWI complexes (7, 11, 52, 54). Chd1p also interacts with nucleosomes in vitro, as do ISWI (8) and the yeast Isw2 complex (16). A role for CHD1 in nucleosome assembly in vivo is also suggested by its synthetic interactions with other genes involved in chromatin metabolism, namely, SWI1 and SWI2 (52), ISW1 and ISW2 (54) and, in an isw1Δ background, ITC1 (16). Thus, the published evidence is consistent with a role for Chd1p in RI chromatin assembly.
Chd1p has been implicated in transcription elongation. It localizes to regions of high transcriptional activity on polytene chromosomes in Drosophila (50) and to the transcribed regions of some yeast genes (27, 48). Deletion of yeast CHD1 is associated with resistance to 6-azauracil, a phenotype that indicates relief of transcriptional inhibition (59). Finally, two polymerase II transcription elongation factor complexes of yeast, Spt16p (Cdc68p)-Pob3p and Spt4p-Spt5, and a member of a third elongation complex, the Paf1 complex, interact with Chd1p (27, 48). Chd1p may also play a role in transcriptional termination (3).
These findings suggest that Chd1p contributes to chromatin remodeling during transcription elongation and/or termination; however, the mechanism of Chd1p action has not been deciphered. Our results raise the possibility that Chd1p has a role in the reassembly of nucleosomes in the wake of an elongating polymerase, reestablishing a repressive chromatin structure that obstructs the subsequent polymerase complex. An extension of this proposal is that Chd1p could be involved in RI histone replacement that is coupled to one or more steps in the transcription cycle (2). These are attractive possibilities in view of recent genetic evidence that the Spt16p-Pob3p complex has a role in chromatin assembly (15).
Summary and future prospects.
A simple yeast system that supports the assembly and remodeling of physiologically spaced arrays of nucleosomes is described here. The dependency of chromatin assembly on the known CAF Asf1p, validates this system. Using this system we provide the first evidence that Chd1p, a member of the CHD group of SNF2-like ATPases, may be a factor involved in nucleosome assembly. In the immediate future studies with the CD fraction are expected to shed further light on the role of Asf1p and Chd1p in RI chromatin assembly and the mechanisms of chromatin remodeling in yeast.
Acknowledgments
Some reagents used in this study were prepared by K.M.R. in the lab of Jim Kadonaga, whom we thank for advice, support, and provision of materials. We also thank Paul Laybourn for the β-endoglucanase gene and, along with John Pilon, for advice on yeast histone purification and the generous gift of recombinant yeast histones. In addition, we thank Dmitry Fyodorov, Mark Levenstein, Mike Pazin, and Jessica Tyler for useful discussions and David Stuart for the generous gift of yeast strain DSY904 and for critical reading of the manuscript.
This work was funded by an operating grant to M.C.S. from the Canadian Institutes for Health Research. M.C.S. is a Scientist of the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Alén, C., N. A. Kent, H. S. Jones, J. O'Sullivan, A. Aranda, and N. J. Proudfoot. 2002. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell 10:1441-1452. [DOI] [PubMed] [Google Scholar]
- 4.Altheim, B. A., and M. C. Schultz. 1999. Histone modification governs the cell cycle regulation of a replication-independent chromatin assembly pathway in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:1345-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazett-Jones, D. P., J. Côté, C. C. Landel, C. L. Peterson, and J. L. Workman. 1999. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol. Cell. Biol. 19:1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, P. B., T. Tsukiyama, and C. Wu. 1994. Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol. 44:207-223. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, L. A., C. Logie, E. Bonte, P. B. Becker, P. A. Wade, A. P. Wolffe, C. Wu, A. N. Imbalzano, and C. L. Peterson. 2000. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 275:18864-18870. [DOI] [PubMed] [Google Scholar]
- 8.Brehm, A., G. Längst, J. Kehle, C. R. Clapier, A. Imhof, A. Eberharter, J. Muller, and P. B. Becker. 2000. dMi-2 and ISWI chromatin remodeling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulger, M., T. Ito, R. T. Kamakaka, and J. T. Kadonaga. 1995. Assembly of regularly spaced nucleosome arrays by Drosophila chromatin assembly factor 1 and a 56-kDa histone-binding protein. Proc. Natl. Acad. Sci. USA 92:11726-11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulger, M., and J. T. Kadonaga. 1994. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet. 5:241-262. [Google Scholar]
- 11.Corona, D. F., G. Längst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 12.Côté, J., C. L. Peterson, and J. L. Workman. 1998. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. USA 95:4947-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshmane, S. L., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enomoto, S., P. D. McCune-Zierath, M. Gerami-Nejad, M. A. Sanders, and J. Berman. 1997. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 11:358-370. [DOI] [PubMed] [Google Scholar]
- 15.Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson, Y. Yu, A. R. Rhoades, P. D. Kaufman, and D. J. Stillman. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162:1557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glover, D. M. 1989. Mitosis in Drosophila. J. Cell Sci. 92:137-146. [DOI] [PubMed] [Google Scholar]
- 18.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 21.Ito, T., J. K. Tyler, M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J. Biol. Chem. 271:25041-25048. [DOI] [PubMed] [Google Scholar]
- 22.Ito, T., J. K. Tyler, and J. T. Kadonaga. 1997. Chromatin assembly factors: a dual function in nucleosome formation and mobilization? Genes Cells 2:593-600. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 24.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman, P. D., R. Kobayashi, and B. Stillman. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11:345-357. [DOI] [PubMed] [Google Scholar]
- 26.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 27.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krude, T., and C. Keller. 2001. Chromatin assembly during S phase: contributions from histone deposition, DNA replication and the cell division cycle. Cell. Mol. Life Sci. 58:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Längst, G., and P. B. Becker. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci. 114:2561-2568. [DOI] [PubMed] [Google Scholar]
- 30.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 31.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 32.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900-1904. [DOI] [PubMed] [Google Scholar]
- 33.MacCallum, D. E., A. Losada, R. Kobayashi, and T. Hirano. 2002. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell 13:25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McQuibban, G. A., C. N. Commisso-Cappelli, and P. N. Lewis. 1998. Assembly, remodeling, and histone binding capabilities of yeast nucleosome assembly protein 1. J. Biol. Chem. 273:6582-6590. [DOI] [PubMed] [Google Scholar]
- 35.Mello, J. A., and G. Almouzni. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 36.Morales, V., C. Giamarchi, C. Chailleux, F. Moro, V. Marsaud, S. Le Ricousse, and H. Richard-Foy. 2001. Chromatin structure and dynamics: functional implications. Biochimie 83:1029-1039. [DOI] [PubMed] [Google Scholar]
- 37.Muchardt, C., and M. Yaniv. 1999. ATP-dependent chromatin remodeling: SWI/SNF and Co. are on the job. J. Mol. Biol. 293:187-198. [DOI] [PubMed] [Google Scholar]
- 38.Murray, A. W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581-605. [PubMed] [Google Scholar]
- 39.O'Farrell, P. H., B. A. Edgar, D. Lakich, and C. F. Lehner. 1989. Directing cell division during development. Science 246:635-640. [DOI] [PubMed] [Google Scholar]
- 40.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 41.Pazin, M. J., J. W. Hermann, and J. T. Kadonaga. 1998. Promoter structure and transcriptional activation with chromatin templates assembled in vitro. A single Gal4-VP16 dimer binds to chromatin or to DNA with comparable affinity. J. Biol. Chem. 273:34653-34660. [DOI] [PubMed] [Google Scholar]
- 42.Pazin, M. J., and J. T. Kadonaga. 1998. Transcriptional and structural analysis of chromatin assembled in vitro, p. 173-194. In H. Gould (ed.), Chromatin: a practical approach. Oxford University Press, Oxford, England.
- 43.Pilon, J., A. Terrell, and P. J. Laybourn. 1997. Yeast chromatin reconstitution system using purified yeast core histones and yeast nucleosome assembly protein-1. Prot. Expr. Purif. 10:132-140. [DOI] [PubMed] [Google Scholar]
- 44.Ray-Gallet, D., J. P. Quivy, C. Scamps, E. M. Martini, M. Lipinski, and G. Almouzni. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091-1100. [DOI] [PubMed] [Google Scholar]
- 45.Richardson, H. E., C. Wittenberg, F. Cross, and S. I. Reed. 1989. An essential G1 function for cyclin-like proteins in yeast. Cell 59:1127-1133. [DOI] [PubMed] [Google Scholar]
- 46.Schultz, M. C. 1999. Chromatin assembly in yeast cell-free extracts. Methods 17:161-172. [DOI] [PubMed] [Google Scholar]
- 47.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 48.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, S., and B. Stillman. 1991. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stokes, D. G., K. D. Tartof, and R. P. Perry. 1996. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. USA 93:7137-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromo domain protein Chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 19:2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsukiyama, T., C. Daniel, J. Tamkun, and C. Wu. 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140-kDa subunit of the nucleosome remodeling factor. Cell 83:1021-1026. [DOI] [PubMed] [Google Scholar]
- 54.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 56.van Holde, K. E. 1988. Chromatin. Springer-Verlag, Inc., New York, N.Y.
- 57.Varga-Weisz, P. D., M. Wilm, E. Bonte, K. Dumas, M. Mann, and P. B. Becker. 1997. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388:598-602. [DOI] [PubMed] [Google Scholar]
- 58.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, and R. W. Davis. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 59.Woodage, T., M. A. Basrai, A. D. Baxevanis, P. Hieter, and F. S. Collins. 1997. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA 94:11472-11477. [DOI] [PMC free article] [PubMed] [Google Scholar]