Abstract
Two Drosophila tumor necrosis factor receptor-associated factors (TRAF), DTRAF1 and DTRAF2, are proposed to have similar functions with their mammalian counterparts as a signal mediator of cell surface receptors. However, their in vivo functions and related signaling pathways are not fully understood yet. Here, we show that DTRAF1 is an in vivo regulator of c-Jun N-terminal kinase (JNK) pathway in Drosophila melanogaster. Ectopic expression of DTRAF1 in the developing eye induced apoptosis, thereby causing a rough-eye phenotype. Further genetic interaction analyses revealed that the apoptosis in the eye imaginal disc and the abnormal eye morphogenesis induced by DTRAF1 are dependent on JNK and its upstream kinases, Hep and DTAK1. In support of these results, DTRAF1-null mutant showed a remarkable reduction in JNK activity with an impaired development of imaginal discs and a defective formation of photosensory neuron arrays. In contrast, DTRAF2 was demonstrated as an upstream activator of nuclear factor-κB (NF-κB). Ectopic expression of DTRAF2 induced nuclear translocation of two Drosophila NF-κBs, DIF and Relish, consequently activating the transcription of the antimicrobial peptide genes diptericin, diptericin-like protein, and drosomycin. Consistently, the null mutant of DTRAF2 showed immune deficiencies in which NF-κB nuclear translocation and antimicrobial gene transcription against microbial infection were severely impaired. Collectively, our findings demonstrate that DTRAF1 and DTRAF2 play pivotal roles in Drosophila development and innate immunity by differentially regulating the JNK- and the NF-κB-dependent signaling pathway, respectively.
Members of the tumor necrosis factor receptor (TNFR) superfamily can induce a wide spectrum of cellular responses, including cell proliferation, apoptosis, and differentiation (32, 51). Most of these functions are mediated by a family of intracellular TNFR-binding proteins, the TNFR-associated factors (TRAFs) (3, 51). In humans and mice, TRAF family consists of six members (TRAF1 to TRAF6), and these proteins have a conserved stretch of amino acids near their C termini termed the TRAF domain. The TRAF domain is required for binding of these signal-transducing adaptor proteins to TNFRs (3). Two additional functional domains, the zinc finger domain and the RING finger domain, are located at the N terminus of TRAF protein and are proposed to be essential for the activation of specific downstream signaling components (10, 11).
The involvement of TRAF family proteins in a variety of signal transduction pathways and cellular responses has been extensively studied by numerous cell culture-based studies (6, 35, 38, 45, 51) and several mouse genetic studies (31, 36, 54). In previous reports, mammalian TRAF2 and TRAF6 were found to regulate the transcription of downstream target genes through the activation of two different intracellular signaling pathways, c-Jun N-terminal kinase (JNK) and nuclear factor-κB (NF-κB) signaling pathways (6, 29, 35, 38, 45). Even though many attempts were made to distinguish the major TRAF-mediated signaling pathways and to deduce the in vivo function of each TRAF, it has been hampered by highly redundant roles of mammalian TRAFs in correlation with their signaling mechanisms (3, 32, 51).
In addition to the intensive studies of TRAFs in the mammalian system, there were some pioneering studies to reveal the function of TRAFs in Drosophila melanogaster (23, 56). Two Drosophila homologues of mammalian TRAFs, DTRAF1 and DTRAF2, have been identified, and the biochemical and cell culture-based studies with these proteins have shown that TRAF-dependent signaling pathways are indeed highly conserved in Drosophila (23, 30, 56). DTRAF2, like mammalian TRAF6, interacts with Drosophila ECSIT and Pelle, and consequently activates NF-κB in Schneider cells (23). DTRAF1 interacts with Drosophila Ste20 kinase (Misshapen, msn) and induces a synergistic activation of JNK in mammalian cultured cells (30). However, there was a contradictory report showing the functional interactions between DTRAF1 and the NF-κB signaling pathway in cell culture-based experiments (56). Despite these efforts, in vivo studies with a whole animal to confirm these in vitro experiments and to further dissect the specific signaling mechanisms of DTRAFs regulating developmental and immunological functions remain to be accomplished.
To better understand the in vivo functions of DTRAFs, it is necessary to conduct genetic studies with various TRAF mutants. Fortunately, only two TRAFs exist in the Drosophila genome that would provide us with a lower number of the signaling molecules and more simple phenotypes and mutants to investigate (14, 23, 30, 56). Using various convenient genetic systems, we were able to analyze the downstream signaling pathways of TRAFs under well-defined and physiologically relevant environments in Drosophila. Moreover, since the two major downstream signaling pathways for TRAFs, the mitogen-activated protein (MAP) kinase signaling cascades and the NF-κB pathway, are highly conserved between vertebrates and Drosophila, the genetic interactions between the DTRAFs and these downstream components can easily be confirmed in the fruit fly and applied to mammalian systems (2, 13, 15, 20, 23, 30, 39, 43, 48).
We identified here the downstream signaling pathways and physiological functions of DTRAF1 and DTRAF2 by using their gain-of-function and loss-of-function mutants. Our results indicate that DTRAF1 is essential for endogenous JNK activation and Drosophila development, whereas DTRAF2 is required for NF-κB signaling and activation of the antimicrobial immune system. Interestingly, DTRAF1 and DTRAF2 do not interfere in each other's signaling and consequent physiological activities. Therefore, we conclude that DTRAF1 and DTRAF2 have independent roles in Drosophila by selectively regulating different downstream signaling pathways.
MATERIALS AND METHODS
Fly strains.
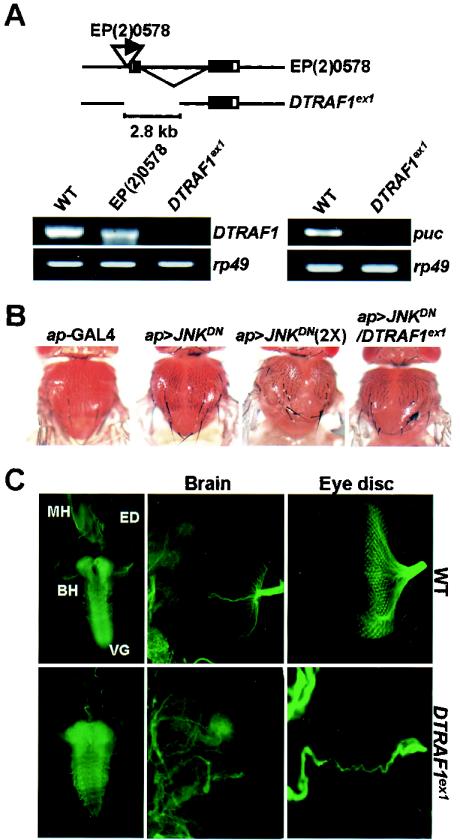
The GAL4 driver fly lines [glass multimer reporter (gmr)-, heat shock (hs)-, and apterous (ap)-GAL4] were obtained from the Bloomington Stock Center. Upstream activation sequence (UAS) fly lines for the JNK pathway [UAS-basket (bsk) and -hemipterous (hep)] (52) were gifts from M. Mlodzik (European Molecular Biology Laboratory, Heidelberg, Germany). UAS-JNKDN (JNKDN; encodes a dominant-negative form of Drosophila JNK) fly was obtained from Bloomington Stock Center. The hemipterous1 (hep1) fly line (13) was a gift from S. Noselli (Centre Nationale de la Recherche Scientifique [CNRS], Paris, France). Drosophila TAK1 mutant (DTAK11) and UAS-DTAK1 flies were provided by N. Ueno (National Institute for Basic Biology, Tokyo, Japan) (49). UAS fly lines for Drosophila p38-MAP kinase (D-p38b) and its dominant-negative allele (D-p38bDN) (2) were provided by T. Adachi-Yamada (Kobe University, Kobe, Japan). The puckered (puc)-LacZ reporter fly line (1) was also obtained from T. Adachi-Yamada. The drosomycin-green fluorescent protein (GFP) reporter fly line (12) was provided by D. Ferrandon (CNRS). The diptericin-LacZ reporter fly line (4) was obtained from M. Meister (Institut de Biologie Moleculaire, Paris, France). The relE20 mutant fly (17) was a gift from D. Hultmark (Umea University, Umea, Sweden). EP fly lines were obtained from the Szeged Drosophila melanogaster P Insertion Mutant Stock Center, Szeged, Hungary. DTRAF1ex1 and DTRAF2ex1, the imprecise excision alleles for EP(2)0578 and EP(X)1516, respectively, were generated by conventional P element excision method with the Δ2-3 transposase line.
Ectopic gene expression with the GAL4/UAS system.
To examine the phenotypes induced by overexpression of DTRAF1 and DTRAF2, we used the GAL4/UAS system (41). The EP lines, EP(2)0578 and EP(X)1516, developed for modular misexpression screening in Drosophila to detect tissue-specific phenotypes (44), were used in our study to ectopically express DTRAF1 and DTRAF2, respectively. The capabilities of the EP lines for ectopic expression of DTRAF1 and DTRAF2 by tissue-specific GAL4 drivers were tested by Northern blot analysis. Open reading frames for DTRAF1 and DTRAF2 of the EP lines were confirmed by genomic PCR and reverse transcription-PCR (RT-PCR) clonings and repeated sequencing of the PCR products.
Immunohistochemistry.
In order to detect the increase of JNK phosphorylation in eye imaginal discs, third-instar larval eye disks were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1 h at room temperature and then incubated first with anti-phospho-specific JNK antibody (1:200; Promega) and subsequently with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibody (1:200; Molecular Probes). Alexa Fluor 568 tyramide (Molecular Probes) was used as a substrate for the secondary antibody. The samples were examined under a fluorescence microscope.
For the photosensory neuron detection, the brain-eye disc complexes from third-instar larvae were dissected in PBS, and the conventional immunostaining procedure was performed with monoclonal antibody 22C10 (1:200; Developmental Studies Hybridoma Bank) (18).
In the case for fat body staining, the fat bodies from third-instar larvae or adult flies were dissected in cold PBS and quickly transferred to 4% paraformaldehyde in PBS in order to prevent bacterial contamination. Anti-Dorsal-related immunity factor (DIF) antibody (obtained from Y. Engstrom) (1:100; Stockholm University, Stockholm, Sweden) (19), anti-Relish antibody (1:100; S. Stoven, Umea University, Umea, Sweden) (47), and fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G secondary antibody (1:200; Molecular Probes) were used to detect the nuclear translocation of DIF and Relish.
X-Gal staining of eye discs and fat bodies.
For X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining, eye discs or fat bodies were fixed in 4% formaldehyde in PBS for 30 min, washed, and then incubated in the standard X-Gal staining solution [0.2% X-Gal, 3.1 mM K4Fe(CN)6, 3.1 mM K3Fe(CN)6, 1 mM MgCl2, 150 mM NaCl, 10 mM Na2HPO4, 10 mM NaH2PO4, 0.3% Triton X-100] overnight at 37°C prior to observation.
TUNEL assay for eye imaginal discs.
For TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay, third-instar larval eye discs were dissected in PBS and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature as previously described (8). The samples were then washed with PBS and permeabilized by incubation in a solution containing 0.1% sodium citrate and 0.1% Triton X-100 on ice for 2 min. After an extensive washing, the samples were further incubated in TUNEL reaction solution for 1 h in a 37°C chamber. After three rinses with PBS, the eye discs were observed by using a fluorescence microscope.
Microbial infection.
Microbial infection to induce immune responses was performed by pricking third-instar larvae with a thin needle that had been dipped into a concentrated culture of Escherichia coli. Infected larvae were further incubated at 25°C for 3 h in a petri dish containing the standard medium of Drosophila before subsequent experiments were conducted.
Northern blot analysis.
Total RNA, extracted by the Easy-Blue system (Intron, Seoul, Korea), was separated by electrophoresis on denaturing formaldehyde agarose gels in morpholinepropanesulfonic acid buffer, transferred onto a nylon membrane, and successively hybridized with nick-translated 32P-labeled cDNA probes. Hybridized probes were visualized by autoradiography.
RESULTS
Characterization of EP lines for Drosophila TRAF1 and TRAF2.
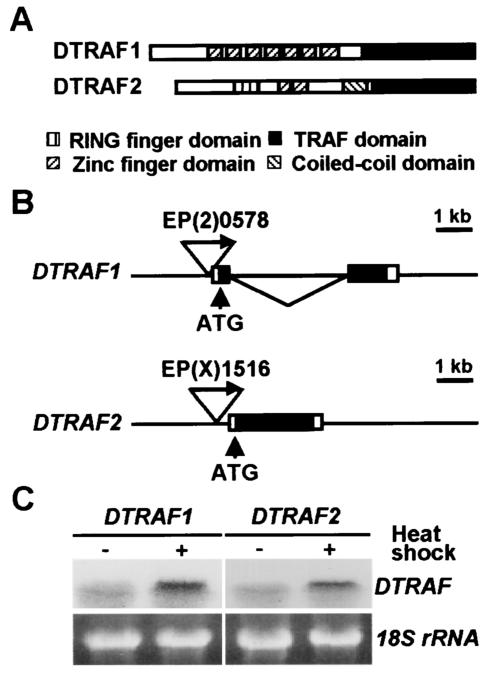
Previously, putative DTRAF1 (CG3048 and GenBank accession number AE119794) and DTRAF2 (CG10961 and GenBank accession number AE119793) were annotated by computational analyses of the Drosophila genome in the Berkeley Drosophila Genome Project Database. Both DTRAF1 and DTRAF2 have a TRAF domain at the C terminus, but DTRAF1 has seven repeated zinc finger domains at the N terminus, in contrast to DTRAF2, which has a single RING finger domain, as well as two zinc finger domains (Fig. 1A).
FIG. 1.
Characterization of DTRAF overexpression flies. (A) Schematic representation of the protein domains of DTRAF1 and DTRAF2. (B) EP fly lines for DTRAF genes. Two EP lines, EP(2)0578 and EP(X)1516, have a P-element in the 5′ flanking regions of DTRAF1 and DTRAF2, respectively. The triangle with an arrow represents the P-element, and ATG denotes the translational initiation site. Exons are indicated by boxes, and coding regions are highlighted by black boxes. (C) Inducible expression of DTRAFs in vivo. Using a GAL4/UAS system, ectopic expression of DTRAF1 or DTRAF2 was induced by heat shock at 37°C for 3 h, and their transcript levels were determined by Northern blot analysis. (Left panel) DTRAF1 mRNA from hs-GAL4/EP(2)0578; (right panel) DTRAF2 mRNA from EP(X)1516/X; hs-GAL4/+. 18S rRNA (18S rRNA) was used as a loading control.
To understand the physiological roles of DTRAF1 and DTRAF2, we decided to overexpress these genes by using tissue-specific GAL4 drivers and look for developmental abnormalities in Drosophila. While searching through the Berkeley Drosophila Genome Project P-element database, we found two P-element insertion lines, EP(2)0578 and EP(X)1516, with insertions at the 5′ upstream region of the DTRAF1 and DTRAF2 gene, respectively (Fig. 1B). We presumed that the insertion sites and directions of the EP elements in both EP fly lines are optimal for inducing DTRAF1 and DTRAF2 expression using tissue-specific GAL4 drivers.
In order to determine whether these EP lines indeed are capable of inducing the transcription of DTRAF1 and DTRAF2 genes, these lines were crossed with a hs-GAL4 line, and the mRNA levels for both DTRAFs after a heat shock were examined by Northern blot analysis. As expected, the DTRAF1 and DTRAF2 transcript levels were strongly increased by GAL4 inductions (Fig. 1C). These results confirmed that the EP(2)0578 and EP(X)1516 lines are appropriate for overexpressing DTRAF1 and DTRAF2, respectively, at a specific location and time using various GAL4 drivers.
Ectopic expression of DTRAF1 perturbs normal eye development.
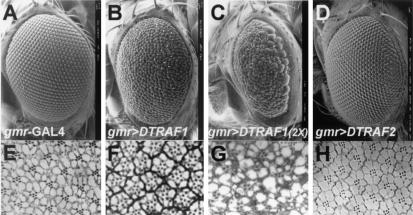
To investigate the consequences of ectopic expression of DTRAF1 in the developing Drosophila eye, we overexpressed DTRAF1 by using an eye-specific gmr-GAL4 driver. The eyes of adults carrying one copy each of both gmr-GAL4 and DTRAF1 showed a rough-eye surface with disorganized arrays of ommatidia (Fig. 2B), whereas the eyes of flies carrying either one copy of gmr-GAL4 or one copy of DTRAF1 alone appeared normal (Fig. 2A and data not shown). Examination of the retinal sections of adults carrying both gmr-GAL4 and DTRAF1 revealed the number of ommatidia to be reduced and the number and shape of the photoreceptor cells in each ommatidium also to be abnormal (Fig. 2F) compared to the control fly which carries only the gmr-GAL4 driver (Fig. 2E).
FIG. 2.
Effects of DTRAF1 on Drosophila eye development. Scanning electron micrographs of the compound eyes (A to D) and their tangential sections (E to H) are shown. (A and E) gmr-GAL4/±; (B and F) gmr-GAL4, EP(2)0578/+; (C and G) gmr-GAL4, EP(2)0578/EP(2)0578; (D and H) EP(X)1516/Y; gmr-GAL4/+. All pictures are shown with anterior to the left and dorsal to the top. Magnifications: A to D, ×200; E to H, ×1,000.
When two copies of DTRAF1 were overexpressed in the eye, it displayed a more severe phenotype and a reduced number of ommatidia, resulting in a size reduction of the compound eye, and some ommatidia were fused with each other (Fig. 2C and G). On the other hand, ectopically expressed DTRAF2 had no effect on the eye development; ommatidial array, bristles, and compound eye size were all found to be normal (Fig. 2D and H).
DTRAF1 interacts with the JNK pathway.
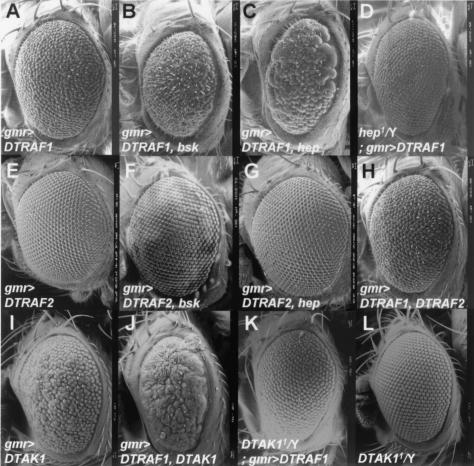
To determine which signaling pathway is activated and induces malformation of the optic system by ectopic expression of DTRAF1 in vivo, we tested the genetic interactions between mutants of various signaling pathways and a DTRAF1-overexpressing line (gmr>DTRAF1/+). Included in this screen were UAS lines that activate extracellular signal-regulated kinase (ERK), p38 MAP kinase, and JNK signaling pathway, respectively. Among the various overexpression lines tested, only Hemipterous (Hep; Drosophila homologue of MKK7 encoded by hemipterous [hep]) and Basket (Bsk; Drosophila homologue of JNK encoded by basket [bsk]) were found to interact genetically with DTRAF1 (Fig. 3). The UAS-hep or UAS-bsk itself under the control of gmr-GAL4 driver had no effect on eye morphogenesis (data not shown). Coexpression of Bsk with DTRAF1 increased the disturbance of the ommatidial array in the compound eye (Fig. 3B) in comparison to the eye phenotype resulting from one copy overexpression of DTRAF1 (Fig. 3A). In addition, when Hep was coexpressed with DTRAF1, the number of ommatidia and the size of the compound eye were reduced more dramatically (Fig. 3C), which is very similar to the DTRAF1 two-copy expression phenotype (Fig. 2C). To further examine whether DTRAF1 signaling is mediated by Hep, DTRAF1 was expressed under a hemizygous hep mutant background. As a result, the abnormal ommatidial array of the compound eye was recovered to the level of the wild-type eye (Fig. 3D).
FIG. 3.
Genetic interactions between DTRAF1 and the JNK signaling pathway components. Scanning electron micrographs of adult eyes are shown. (A) gmr-GAL4, EP(2)0578/+. (B) gmr-GAL4, EP(2)0578/UAS-bsk. (C) gmr-GAL4, EP(2)0578/UAS-hep. (D) hep1/Y; gmr-GAL4, EP(2)0578/+. (E) EP(X)1516/Y; gmr-GAL4/+. (F) EP(X)1516/Y; gmr-GAL4/UAS-bsk. (G) EP(X)1516/Y; gmr-GAL4/UAS-hep. (H) EP(X)1516/Y; gmr-GAL4, EP(2)0578/+. (I) gmr-GAL4/UAS-DTAK1. (J) gmr-GAL4, EP(2)0578/UAS-DTAK1. (K) DTAK11/Y; gmr-GAL4, EP(2)0578/+. (L) DTAK11/Y. Anterior to the left and dorsal to the top. Magnification: ×200.
However, DTRAF2, interestingly, did not display any interaction with the ERK or the p38 MAP kinase pathway components (data not shown), nor even with JNK pathway components (Fig. 3E to G). We also tested whether DTRAF2 exerts its effect on the eye development by interacting with DTRAF1. Coexpression of DTRAF2 with DTRAF1 in the Drosophila compound eye did not alter the rough-eye phenotype caused by a sole overexpression of DTRAF1 (Fig. 3H). These results strongly support the view that DTRAF1 can activate the JNK signaling cascade in vivo and that DTRAF2 is not correlated with DTRAF1 signaling at all, at least during eye development.
Based upon the result that DTRAF1 is involved in JNK signaling, we next attempted to find out the signaling components between DTRAF1 and Hep by genetic interaction studies. Various kinases, such as Misshapen (msn), Slipper (slpr), and Drosophila transforming growth factor β-activated kinase 1 (DTAK1), are known to be the upstream kinases for Hep in the eye development. Among these kinases, DTAK1 synergistically increased the roughness of the compound eye surface and also reduced the eye size when coexpressed with DTRAF1 (Fig. 3J). Moreover, DTAK1-null mutation (DTAK11), which has no effect on the eye morphology (Fig. 3L), was able to block the rough-eye phenotype caused by DTRAF1 overexpression (Fig. 3K). These data suggest that DTRAF1 activates the JNK signaling pathway via DTAK1 and Hep.
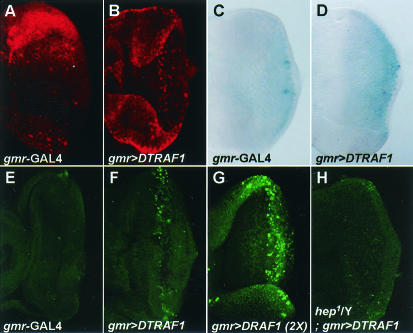
In order to further confirm that DTRAF1 activates the JNK kinase signaling pathway at a molecular level, we examined JNK activity by two different experimental approaches—an immunohistochemical assay with anti-phospho-specific JNK antibody and a puckered-LacZ reporter assay—in the eye discs. As shown in Fig. 4, JNK phosphorylation was highly induced by overexpression of DTRAF1 (Fig. 4B) compared to the control (Fig. 4A). In addition, expression of puckered (puc), a well-known downstream target of JNK, was also highly induced by DTRAF1 (Fig. 4D) compared to the control (Fig. 4C). Collectively, the results shown in Fig. 3 and 4 clearly demonstrated that DTRAF1 activates the JNK signaling pathway in vivo.
FIG. 4.
DTRAF1 induces activation of the JNK signaling pathway and apoptosis. (A and B) The eye imaginal discs were immunostained with an anti-phospho-specific JNK antibody as described in Materials and Methods. (A) gmr-GAL4/+. (B) gmr-GAL4, EP(2)0578/+. (C and D) puckered-LacZ reporter assays were also conducted in the eye discs. (C) gmr-GAL4/+; puckered-LacZ/+. (D) gmr-GAL4, EP(2)0578/+; puckered-LacZ/+. (E to H) DTRAF1-induced apoptosis in the eye discs was examined by TUNEL assays. (E) gmr-GAL4/+. (F) gmr-GAL4, EP(2)0578/+. (G) gmr-GAL4, EP(2)0578/EP(2)0578. (H) hep1/Y; gmr-GAL4, EP(2)0578/+.
DTRAF1 overexpression induces apoptosis.
It has been reported that apoptosis can be induced by the activation of the JNK pathway (33, 49). Because ectopic coexpression of DTRAF1 with Hep had a synergistic effect on the reduction of ommatidia number and eye size (Fig. 3C), the rough-eye phenotype induced by DTRAF1 overexpression seemed to be a result of the Hep/JNK signaling-dependent apoptotic cell death. Therefore, we investigated whether overexpression of DTRAF1 can induce apoptosis in the eye disc cells by using TUNEL assay. In the discs of wild-type third-instar larvae, there were few apoptotic cells (Fig. 4E). In contrast, the eye imaginal discs from transgenic flies overexpressing DTRAF1 revealed a highly increased number of apoptotic cells in the region posterior to the morphogenetic furrow in a gene dosage-dependent manner (Fig. 4F and G). However, ectopic expression of DTRAF2 failed to induce apoptotic cell death (data not shown), which is consistent with its inability to activate the JNK pathway.
Since hemizygous hep mutation (hep1/Y) was sufficient to suppress the rough-eye phenotype caused by DTRAF1 overexpression (Fig. 3D), we examined whether hep mutation can inhibit the DTRAF1-induced apoptotic cell death in the eye imaginal discs. When TUNEL assay was performed against the eye discs of hep1/Y; gmr>DTRAF1/± larvae, the number of apoptotic cells dramatically decreased to almost wild-type levels (Fig. 4H). These results proved that DTRAF1 overexpression can activate the Hep/JNK signaling pathway and consequently induce apoptosis.
Null mutation for DTRAF1 causes developmental defects and decreased JNK activities.
DTRAF1ex1, a loss-of-function allele for DTRAF1 gene, was generated through imprecise excision of the P-element in EP(2)0578 fly (Fig. 5A, upper panel). RT-PCR analysis (Fig. 5A, lower left panel) clearly demonstrated that the homozygous DTRAF1ex1 mutant failed to produce DTRAF1 mRNA, indicating that DTRAF1ex1 is a null allele for DTRAF1. We also examined the endogenous puckered transcription level in the mutant larvae by RT-PCR analysis, and it was found that amount of puckered gene transcript was severely decreased in DTRAF1ex1 mutant in compare to wild-type larvae (Fig. 5A, lower right panel), which strongly implies the reduced JNK activity in DTRAF1ex1 mutant.
FIG. 5.
Characterization of DTRAF1-null mutant. (A) Molecular characteristics of DTRAF1ex1 mutant. In the upper panel are shown genomic structures of original EP(2)0578 line and its derivative, DTRAF1ex1 mutant. Exons are indicated by boxes, and coding regions are highlighted by black boxes. The deleted region in DTRAF1ex1 is displayed as a gap. In the lower left panel, the RT-PCR result demonstrated that DTRAF1ex1 lacks DTRAF1 gene expression. In the lower right panel, the RT-PCR results showed the reduction of puckered (puc) transcription level in DTRAF1ex1. Ribosomal protein 49 (rp49) was used as an internal control. Lanes: WT, wild-type w1118; EP(2)0578, EP(2)0578/EP(2)0578; DTRAF1ex1, DTRAF1ex1/DTRAF1ex1. (B) Enhanced thorax closure defects of JNKDN transgenic flies by DTRAF1ex1 mutation. Subpanels: ap-GAL4 (ap-GAL4/+), ap>JNKDN (UAS-JNKDN/X; ap-GAL4/+), ap>JNKDN(2X) (UAS-JNKDN/UAS-JNKDN; ap-GAL4/+), ap>JNKDN/DTRAF1ex1 (UAS-JNKDN/X; ap-GAL4/DTRAF1ex1). (C) DTRAF1 is required for normal development of Drosophila optical system. Photosensory neurons spanning from imaginal eye discs into brain hemisphere were immunostained with monoclonal antibody 22C10 as described in Materials and Methods. WT = wild type, w1118; DTRAF1ex1 = DTRAF1ex1/DTRAF1ex1. The left panels show the brain-eye disc-mouth complex (magnification, ×70). The middle panels show the brain hemisphere (magnification, ×200). The upper right panels show the wild-type eye disc (magnification, ×200). The lower right panel shows the image for the eye imaginal disc of DTRAF1ex1 mutant was further magnified to obtain a better view (magnification, ×600). BH, brain hemisphere; ED, eye disc; MH, mouth hook; VG, ventral ganglia.
In order to confirm this, we examined the genetic interaction between the DTRAF1-null fly and a transgenic fly for JNK (ap>JNKDN) by observing the thorax closure phenotype. Thorax closure, the joining of the parts of the two wing imaginal discs during metamorphosis, is tightly controlled by the Drosophila JNK signaling pathway (57). When the activity of JNK pathway is downregulated by expressing a dominant-negative form of JNK on the thorax in ap>JNKDN flies, the joining process is impaired and a cleft is formed at the dorsal midline in a gene dosage-dependent manner (Fig. 5B, second and third panels). Strikingly, a reduction of DTRAF1 gene dosage in heterozygous DTRAF1ex1 dramatically enhanced the thorax closure defect in ap>JNKDN flies by expanding the cleft and also disrupting its notum structure (Fig. 5B, fourth panel), suggesting that DTRAF1 mutation leads to a more reduction of the endogenous JNK activity in ap>JNKDN flies. These data strongly support the critical roles of DTRAF1 in Drosophila development by positively modulating JNK signaling activities.
Indeed, DTRAF1ex1 mutant failed to develop into the pupal stage. In order to understand the cause of the lethality, the internal organ structures of DTRAF1ex1 larvae were examined. The brain-ventral ganglia complex of the mutant larva showed no apparent defects (Fig. 5C, lower left panel) compared to the wild-type control (Fig. 5C, upper left panel). However, interestingly, the mutant larva contained small-sized imaginal discs, especially the eye discs (Fig. 5C, lower right panel) in comparison to the wild-type discs (Fig. 5C, upper right panel). To examine the photosensory neuron projections in the brain hemisphere, eye-brain complexes were stained with monoclonal antibody 22C10, a well-characterized probe for sensory neurons in Drosophila (18). Axons from wild-type photoreceptors fanned out evenly upon leaving the optic stalk and formed a smooth neuronal array in the lamina (Fig. 5C, upper middle panel). On the other hand, the photoreceptor axons from DTRAF1ex1 mutant formed few axonal bundles and failed to defasciculate in the brain hemisphere (Fig. 5C, lower middle panel). These findings suggest that DTRAF1 is indispensable for the development of imaginal eye discs and the formation of a correct photosensory neuronal array in the brain hemisphere.
DTRAF2 mediates antimicrobial defense mechanisms.
Microbial infection studies have demonstrated the ability of Drosophila to detect pathogens and activate specific signaling pathways, Toll or Imd pathways, which lead to adapted immune responses (22, 28). In recent years, several families of antimicrobial peptides and their coding genes have been successfully identified: cecropins, attacins, diptericin, defensin, drosomycin, drosocin, and diptericin-like protein (dptlp) (5, 25, 26, 42, 53). Understanding the molecular mechanisms behind how microbial infection induces expression of these antimicrobial peptides has been the main question to answer in this field. Meanwhile, DTRAF2 have been identified as a downstream adaptor for Toll receptor (46) and, as mentioned above, Toll activation leads to immune responses. Therefore, we suspected that DTRAFs would be involved in this defense mechanism.
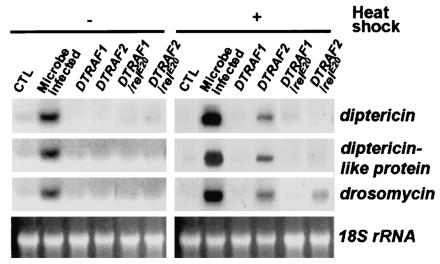
We have chosen three representative antimicrobial genes—diptericin, dptlp, and drosomycin—as probes to determine the activity of the antimicrobial defense system. To examine whether DTRAF1 and DTRAF2 have the ability to induce the transcription of diptericin, dptlp, and drosomycin, DTRAF1 or DTRAF2 was ectopically expressed in third-instar larvae by using hs-GAL4 driver, and the expression levels of diptericin, dptlp, and drosomycin were monitored by Northern blot analyses.
As shown in Fig. 6, the transcription of diptericin, dptlp, and drosomycin was increased by ectopic expression of DTRAF2 in the absence of microbial infection. However, the expression levels of diptericin, dptlp, and drosomycin were not altered by DTRAF1 overexpression (Fig. 6). In addition, DTRAF2-induced expression of diptericin and dptlp was completely inhibited in a relish (rel, Drosophila NF-κB)-null mutant background, whereas drosomycin expression was partially inhibited by the same mutation (Fig. 6). The partial inhibition of the drosomycin expression by rel mutation suggests that the possible involvement of another Drosophila NF-κB, such as DIF, in antimicrobial response gene transcription is consistent with the previous report (19). These results strongly suggest that DTRAF2, but not DTRAF1, functions downstream of microbial sensory receptors, Toll or Imd, and upstream of the NF-κBs to regulate Drosophila immune responses.
FIG. 6.
DTRAF2 overexpression induces antimicrobial gene expression. Using the hs-GAL4 driver, DTRAF1 or DTRAF2 were ectopically expressed in wild-type or relE20 homozygous mutant third-instar larvae (−, untreated control; +, heat-shocked at 37°C). Total RNA from each sample was prepared, and Northern blot analysis was completed to determine the expression of diptericin, diptericin-like protein, and drosomycin. Lanes: CTL (hs-GAL4/+, uninfected control,), Microbe infected (hs-GAL4/+, pricked with a concentrated culture of E. coli), DTRAF1 [hs-GAL4/EP(2)0578], DTRAF2 [EP(X)1516/X; hs-GAL4/+], DTRAF1/relE20 [hs-GAL4/EP(2)0578; relE20/relE20], DTRAF2/relE20 [EP(X)1516/X; hs-GAL4/+; relE20/relE20]. 18S rRNA (18S rRNA) was used as a loading control.
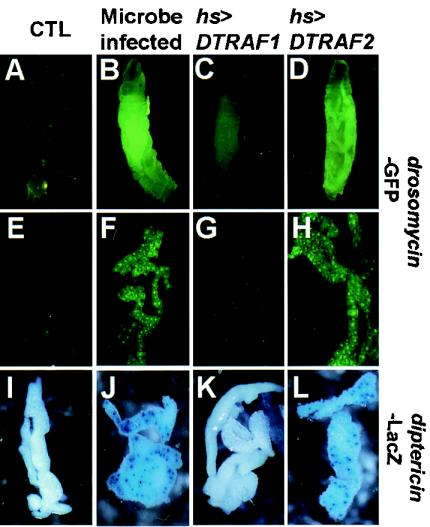
To further confirm the results, transgenic fly lines that have a GFP or a LacZ reporter gene fused to the drosomycin or the diptericin promoter, respectively, were used (4, 12), allowing observation of the reporter gene activity, which reflects the drosomycin or diptericin gene expression level. Consistent with a previous study (12), the drosomycin-GFP reporter activity was dramatically increased in the microbe-infected larva (Fig. 7B) compared to the uninfected control (Fig. 7A). As expected, DTRAF2 overexpression alone in the absence of microbial infection strongly induced drosomycin-GFP reporter gene activity (Fig. 7D). Further dissection analyses showed that drosomycin-GFP (compare Fig. 7F and H to the untreated control, Fig. 7E) and diptericin-LacZ reporter activities (compare Fig. 7J and L to the untreated control, Fig. 7I) were highly induced in the fat body, which is a representative target tissue for immune responses in Drosophila. However, DTRAF1 overexpression failed to induce the reporter activities in both whole larvae (Fig. 7C) and their fat bodies (Fig. 7G and K), further confirming the noninvolvement of DTRAF1 in the immune responses of Drosophila.
FIG. 7.
DTRAF2 overexpression induces antimicrobial gene expression in situ. Third-instar larvae of the designated genotypes were either infected with Escherichia coli or heat shocked at 37°C for 3 h as described in Materials and Methods. (A to D) The larvae were examined under a fluorescence microscope to locate the GFP-expressing tissues. (E to H) In addition, the fat bodies dissected from the larvae were examined by using a fluorescence microscope. (I to L) In the case of diptericin-LacZ reporter larvae, the fat bodies were X-Gal stained and observed under light microscope. (A, B, E, and F) drosomycin-GFP/X; hs-GAL4/+. (C and G) drosomycin-GFP/X; hs-GAL4/EP(2)0578. (D and H) EP(X)1516/drosomycin-GFP; hs-GAL4/+. (I and J) diptericin-LacZ/X; hs-GAL4/+. (K) diptericin-LacZ/X; hs-GAL4/EP(2)0578. (L) EP(X)1516/diptericin-LacZ; hs-GAL4/+. Columns: CTL, uninfected control samples; Microbe infected, E. coli-infected control samples; hs>DTRAF1, heat shock-induced DTRAF1-overexpressing samples; hs>DTRAF2, heat shock-induced DTRAF2-overexpressing samples.
The DTRAF2-induced immune responses are mediated by NF-κB.
In order to confirm that the DTRAF2-induced immune responses are mediated by DIF and Relish, which are Drosophila NF-κBs specifically activated by Toll and Imd pathways, respectively, we determined the subcellular localization of DIF and Relish by using their specific antibodies.
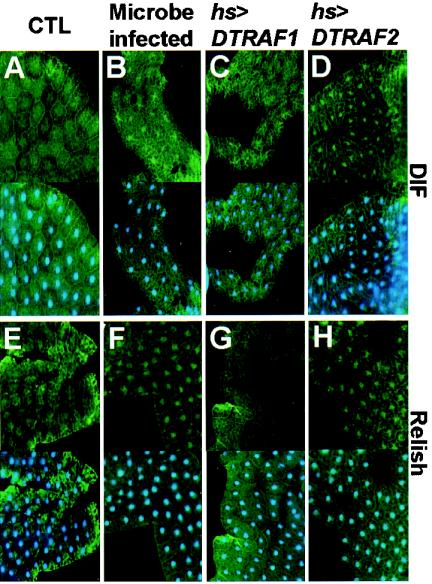
DIF and Relish were dispersed in the cytoplasm of fat body cells in the absence of microbial infection (Fig. 8A and E). On the contrary, either the microbial infection or overexpression of DTRAF2 fully induced the nuclear translocation of both DIF (Fig. 8B and D) and Relish (Fig. 8F and H), demonstrating that both DIF and Relish participate in the DTRAF2-mediated immune responses. However, the subcellular localization of DIF and Relish was not altered by DTRAF1 induction (Fig. 8C and G), further confirming that DTRAF1 is not involved in the NF-κB signaling pathway. These data clearly demonstrated that DTRAF2, but not DTRAF1, has the capability to induce transcriptional activation of immune response genes by specifically activating NF-κBs.
FIG. 8.
DTRAF2 overexpression induces nuclear translocation of DIF and Relish in fat bodies. (A to D) Immunohistochemcal analysis with anti-DIF antibody. (E to H) Immunohistochemcal analysis with anti-Relish antibody. (A, B, E, and F) hs-GAL4/+. (C and G) hs-GAL4/EP(2)0578. (D and H) EP(X)1516/X; hs-GAL4/+. The top row of panels show antibody staining only; the lower panels show merged images of the upper images with Hoechst-nucleus staining images. Columns: CTL, fat bodies of uninfected control larvae; Microbe infected, fat bodies of E. coli-infected control larvae; hs>DTRAF1, fat bodies of heat shock-induced DTRAF1-overexpressing larvae; hs>DTRAF2, fat bodies of heat shock-induced DTRAF2-overexpressing larvae.
DTRAF2 is critical for antimicrobial immune responses.
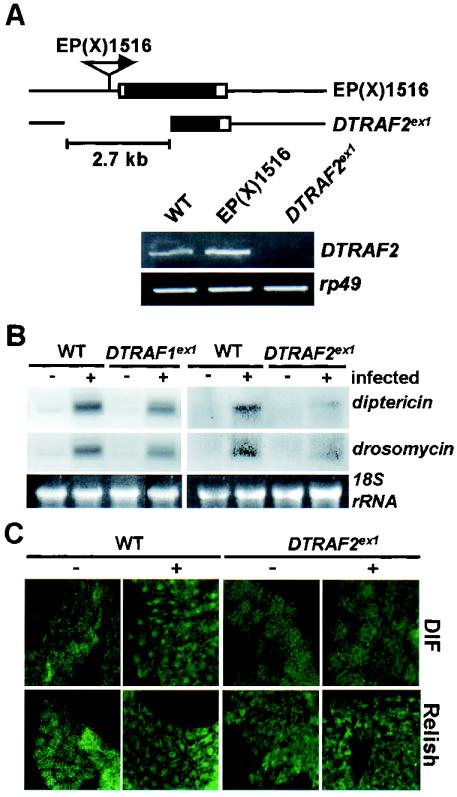
The DTRAF2-null mutant, DTRAF2ex1, was generated by P-element excision method (Fig. 9A, upper panel). RT-PCR analysis showed that the homozygous DTRAF2ex1 mutant failed to produce DTRAF2 mRNA (Fig. 9A, lower panel). Intriguingly, the mutant flies managed to develop into adult and showed no morphological defects. To determine whether the DTRAF2ex1 mutant shows a deficiency in immune responses, we examined the transcriptional induction level of diptericin and drosomycin after microbial infection. The null mutation of DTRAF2 drastically disrupted the transcriptional induction of diptericin and drosomycin compared to the wild-type control (Fig. 9B, right panel). However, DTRAF1-null mutation (DTRAF1ex1) had no effect on the induction of diptericin and drosomycin gene expression after microbial infection (Fig. 9B, left panel). We also examined the nuclear translocation of DIF and Relish in the DTRAF2-null mutant. Consistent with the Northern blot analysis shown in Fig. 9B, the nuclear translocation of DIF and Relish induced by microbial infections was impaired in the DTRAF2ex1 mutant (Fig. 9C). These results support the position that DTRAF2, but not DTRAF1, is critical for the NF-κB-mediated Drosophila innate immune responses.
FIG. 9.
Characterization of DTRAF2-null mutant. (A) Molecular characteristics of DTRAF2ex1 mutant. The upper panel shows genomic structures of original EP(X)1516 and its derivative, the DTRAF2ex1 mutant. The deleted region in DTRAF2ex1 is displayed as a gap. In the lower panel, RT-PCR results showed that DTRAF2ex1 lacks DTRAF2 transcription. (B) Impaired immune responses in DTRAF2ex1 mutant. Wild type (WT, w1118), DTRAF1ex1 mutant (DTRAF1ex1/DTRAF1ex1), and DTRAF2ex1 mutant (DTRAF2ex1/DTRAF2ex1) were infected with E. coli (+), and Northern blot analyses were performed to determine the induction of diptericin and drosomycin transcription. Control groups were not infected (−). 18S rRNA (18S rRNA) was used as a loading control. (C) Impaired nuclear-translocation of DIF and Relish in DTRAF2ex1 mutants. Immunohistochemical analyses were completed with anti-DIF (upper panels) and anti-Relish antibodies (lower panels) to localize DIF and Relish in the fat bodies from uninfected (−) and E. coli-infected (+) wild-type (WT, w1118) and DTRAF2ex1 (DTRAF2ex1/DTRAF2ex1) larvae.
DISCUSSION
DTRAF1 specifically activates the JNK signaling pathway.
We have shown here that DTRAF1 can specifically activate the JNK signaling cascade. This conclusion is based on four lines of evidence. First, ectopic expression of Bsk, Hep, or DTAK1 with DTRAF1 exerted a highly synergistic effect on the rough-eye phenotype of DTRAF1 overexpressing flies (Fig. 3B, C, and J). Second, disruption of hep or DTAK1 function sufficiently suppressed the DTRAF1-induced eye defects (Fig. 3D and K). Third, histochemical analysis with either an anti-phospho-specific JNK antibody or the puckered-LacZ reporter system provided direct molecular evidences that DTRAF1 can induce phosphorylation and consequent activation of JNK (Fig. 4B and D). Fourth, DTRAF1 deficiencies in DTRAF1ex1 mutant generated the same phenotypes detected in the loss-of-function mutants of the JNK signaling pathway, such as increased thorax closure defects (Fig. 5B) and reduced puckered transcription (Fig. 5A).
In addition to its role in the JNK signaling pathway, previous results from cell culture experiments have implicated the involvement of TRAF in other MAP kinase signaling pathways (16, 21). However, we found no evidence for the involvement of DTRAF1 in the p38- or the ERK-MAP kinase pathway in Drosophila. Specifically, overexpression of DTRAF1 with D-p38b or rlSem had no effect on the DTRAF1-induced eye phenotype (data not shown). Likewise, ectopic expression of DTRAF1 with a dominant-negative form of D-p38b, D-p38bDN, where Thr183 of the MAP kinase kinase target site is replaced with Ala, or the expression of DTRAF1 in a heterozygotic rl1 genetic background also had no effect on the DTRAF1-induced rough-eye phenotypes (data not shown). Taken together, these data strongly suggest that DTRAF1 specifically activates JNK signaling, but not the other MAP kinases (ERK- and p38-MAP kinase) signaling pathways, in Drosophila.
DTRAF1 overexpression induces apoptosis.
We have found that ectopic DTRAF1 expression in imaginal eye discs induces apoptosis. Is the DTRAF1-induced apoptosis mediated through the JNK pathway? There are two observations strongly supporting this possibility. First, phosphorylation of JNK and an increase in puckered gene expression were detected at the posterior region of eye discs (Fig. 4B and D), in which programmed cell death occurred most intensely by DTRAF1 expression (Fig. 4F). Second, the apoptosis induced by DTRAF1 overexpression is strongly suppressed by a loss-of-function allele of hep (Fig. 3D and 4H).
There are several supporting studies suggesting an involvement of the TRAF/JNK pathway in apoptosis in vertebrate cells (9, 24, 34, 37, 50, 55). In addition, the Drosophila JNK pathway has been implicated in regulating apoptosis upon deregulation of Decapentaplegic (dpp), Wingless (wg), and integrin/tensin (int/ten) signalings in the developing wing discs (1, 27). Moreover, it has been reported that overexpression of a constitutively activated form of Hep (hepAct) or Jun (junAsp) can lead to highly similar eye phenotypes as DTRAF1 overexpression (40, 52). Collectively, it is highly likely that TRAF regulates apoptosis through the JNK signaling pathway not only in vertebrates but also in flies.
DTRAF2 activates NF-κBs and stimulates the antimicrobial immune functions.
In the mammalian system, when interleukin-1 (IL-1) receptor, a Toll-like receptor, is stimulated by binding of its ligand, IL-1 receptor associated kinase (IRAK) is recruited to the IL-1 receptor complex and phosphorylated (6). Consequently, the receptor associated IRAK binds to TRAF6, which evokes a strong activation of the NF-κB signaling pathway. The importance of TRAF6 in the activation of this pathway has been confirmed by various experiments. For example, overexpression of TRAF6 can lead to NF-κB activation, and a dominant-negative mutant of TRAF6 inhibits IL-1-induced NF-κB activation (7).
Between the two Drosophila homologues of mammalian TRAFs, the TRAF domain of DTRAF2 is most closely related to that of mammalian TRAF6. Based on this structural similarity, there have been reports that DTRAF2 contributes to dorsal activation and immune responses by activating NF-κB in a cell culture system (23, 46). In agreement with these results, we demonstrated here that DTRAF2 can activate Drosophila NF-κBs (Fig. 8) and their downstream target genes diptericin, dptlp and drosomycin (Fig. 6 and 7). It has been also suggested that DTRAF1 is involved in the NF-κB-mediated immune response (56). However, in the present study, we clearly demonstrated that DTRAF1 does not induce NF-κB activation and the consequent NF-κB-dependent immune responses in vivo. These data suggest that DTRAF2 is a highly specific signal mediator activating the NF-κB signaling pathway.
Although overexpression of DTRAF2 was sufficient to activate NF-κB signaling pathway and induce innate immune responses, the DTRAF2-null mutation could not completely block the processes (Fig. 9B). This suggests the presence of other signaling pathway(s) that bypasses DTRAF2 to transmit the exogenous microbial signals to NF-κBs. Further studies with DTRAF2-null mutant are required to elucidate the unknown signaling mechanism.
DTRAF1 and DTRAF2 are involved in physiologically separate signaling pathways.
Interestingly, in mammals, TRAFs are thought to be involved in both JNK and NF-κB signalings to regulate various cellular responses (3, 7, 11, 16). On the other hand, our findings suggest that, in Drosophila, DTRAF1 and DTRAF2 are involved in the regulation of separate signaling pathways and correspondingly serve different physiological functions. We hypothesized that this difference between mammalian and Drosophila TRAFs is originated from structural differences of the N-terminal domains of TRAFs.
Cell culture-based studies have suggested that the structural differences among various TRAFs mainly exist in their RING finger and zinc finger domains, and these domains of the proteins must play essential roles in determining their downstream signaling pathways (10, 11). According to one of the studies, an intact RING finger domain is required for the TRAF-mediated NF-κB activation but is dispensable for JNK signaling (10). There was another report that zinc finger domains are responsible for membrane localization of TRAF and activation of the JNK pathway (11). A forced localization of TRAF3 (which is normally unable to activate the JNK pathway) to the cell membrane by substitution of zinc finger domains was sufficient to convert this molecule into an activator of JNK.
Intriguingly, the two Drosophila TRAFs are distinguished from each other by the presence of a complete zinc finger domain or RING finger domain (Fig. 1A). Therefore, it is quite possible that the absence of the RING finger domain in DTRAF1 prevents the protein from interacting with the NF-κB pathway. On the other hand, DTRAF2 has fewer zinc finger domains than DTRAF1 or mammalian TRAFs, and this may inhibit DTRAF2 from its membrane localization and/or consequent activation of the JNK pathway. Further studies at a molecular level with various domain-modified DTRAF proteins should resolve this proposition.
We have investigated here in vivo functions of two Drosophila TRAFs during development of D. melanogaster. Our results indicate that DTRAF1 can act as a component of the JNK signal transduction pathway. On the other hand, DTRAF2 is involved in the antibacterial immune responses mediated by NF-κB. Interestingly, we found no evidence for the presence of functional interactions between DTRAF1 and DTRAF2 in vivo. These results imply that the different downstream signaling events that activate JNK and NF-κB may bifurcate at the level of TRAF in Drosophila.
Acknowledgments
We acknowledge the gifts of fly stocks from T. Adachi-Yamada, Y. Engstrom, D. Ferrandon, D. Hultmark, M. Meister, M. Mlodzik, S. Stoven, S. Noselli, and N. Ueno. Monoclonal antibody 22C10 was obtained from the Developmental Studies Hybridoma Bank maintained by University of Iowa at Iowa City. We thank members of the Chung laboratory for advice and helpful discussions.
REFERENCES
- 1.Adachi-Yamada, T., K. Fujimura-Kamada, Y. Nishida, and K. Matsumoto. 1999. Distortion of proheximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 400:166-169. [DOI] [PubMed] [Google Scholar]
- 2.Adachi-Yamada, T., M. Nakamura, K. Irie, Y. Tomoyasu, Y. Sano, E. Mori, S. Goto, N. Ueno, Y. Nishida, and K. Matsumoto. 1999. p38 mitogen-activated protein kinase can be involved in transforming growth factor beta superfamily signal transduction in Drosophila wing morphogenesis. Mol. Cell. Biol. 19:2322-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arch, R. H., R. W. Gedrich, and C. B. Thompson. 1998. Tumor necrosis factor receptor-associated factors (TRAFs): a family of adapter proteins that regulates life and death. Genes Dev. 12:2821-2830. [DOI] [PubMed] [Google Scholar]
- 4.Braun, A., J. A. Hoffmann, and M. Meister. 1998. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc. Natl. Acad. Sci. USA 95:14337-14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulet, P., J. L. Dimarcq, C. Hetru, M. Lagueux, M. Charlet, G. Hegy, A. Van Dorsselaer, and J. A. Hoffmann. 1993. A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J. Biol. Chem. 268:14893-14897. [PubMed] [Google Scholar]
- 6.Cao, Z., W. J. Henzel, and X. Gao. 1996. IRAK: a kinase associated with the interleukin-1 receptor. Science 271:1128-1131. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Z., J. Xiong, M. I. Takeuchi, T. Kurama, and D. V. Goeddel. 1996. TRAF6 is a signal transducer for interleukin-1. Nature 383:443-446. [DOI] [PubMed] [Google Scholar]
- 8.Cho, K. S., J. H. Lee, S. Kim, D. Kim, H. Koh, J. Lee, C. Kim, J. Kim, and J. Chung. 2001. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc. Natl. Acad. Sci. USA 98:6144-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coso, O. A., M. Chiariello, J. C. Yu, H. Teramoto, P. Crespo, N. Xu, T. Miki, and J. S. Gutkind. 1995. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell 81:1137-1146. [DOI] [PubMed] [Google Scholar]
- 10.Dadgostar, H., and G. Cheng. 1998. An intact zinc ring finger is required for tumor necrosis factor receptor-associated factor-mediated nuclear factor-kappaB activation but is dispensable for c-Jun N-terminal kinase signaling. J. Biol. Chem. 273:24775-24780. [DOI] [PubMed] [Google Scholar]
- 11.Dadgostar, H., and G. Cheng. 2000. Membrane localization of TRAF3 enables JNK activation. J. Biol. Chem. 275:2539-2544. [DOI] [PubMed] [Google Scholar]
- 12.Ferrandon, D., A. C. Jung, M. C. Criqui, B. Lemaitre, S. Uttenweiler-Joseph, L. Michaut, J. M. Reichhart, and J. A. Hoffmann. 1998. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glise, B., H. Bourbon, and S. Noselli. 1995. Hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell 83:451-461. [DOI] [PubMed] [Google Scholar]
- 14.Grech, A., R. Quinn, D. Srinivasan, X. Badoux, and R. Brink. 2000. Complete structural characterization of the mammalian and Drosophila TRAF genes: implications for TRAF evolution and the role of RING finger splice variants. Mol. Immunol. 37:721-734. [DOI] [PubMed] [Google Scholar]
- 15.Han, Z. S., H. Enslen, X. Hu, X. Meng, I. H. Wu, T. Barrett, R. J. Davis, and Y. T. Ip. 1998. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol. Cell. Biol. 18:3527-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzoglou, A., J. Roussel, M. F. Bourgeade, E. Rogier, C. Madry, J. Inoue, O. Devergne, and A. Tsapis. 2000. TNF receptor family member BCMA (B cell maturation) associates with TNF receptor-associated factor (TRAF) 1, TRAF2, and TRAF3 and activates NF-kappa B, elk-1, c-Jun N-terminal kinase, and p38 mitogen-activated protein kinase. J. Immunol. 165:1322-1330. [DOI] [PubMed] [Google Scholar]
- 17.Hedengren, M., B. Asling, M. S. Dushay, I. Ando, S. Ekengren, M. Whilborg, and D. Hultmark. 1999. Relish, a central factor in the control of humoral but no cellular immunity in Drosophila. Mol. Cell 4:827-837. [DOI] [PubMed] [Google Scholar]
- 18.Hummel, T., K. Krukkert, J. Roos, G. Davis, and C. Klambt. 2000. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26:357-370. [DOI] [PubMed] [Google Scholar]
- 19.Ip, Y. T., M. Reach, Y. Engstrom, L. Kadalayil, H. Cai. S. Gonzalez-Crespo, K. Tatei, and M. Levine. 1993. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell 19:753-763. [DOI] [PubMed] [Google Scholar]
- 20.Ip, Y. T., and R. J. Davis. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK)-from inflammation to development. Curr. Opin. Cell Biol. 10:205-219. [DOI] [PubMed] [Google Scholar]
- 21.Kashiwada, M., Y. Shirakata, J. I. Inoue, H. Nakano, K. Okazaki, K. Okumura, T. Yamamoto, H. Nagaoka, and T. Takemori. 1998. Tumor necrosis factor receptor-associated factor 6 (TRAF6) stimulates extracellular signal-regulated kinase (ERK) activity in CD40 signaling along a ras-independent pathway. J. Exp. Med. 187:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khush, R. S., and B. Lemaitre. 2000. Genes that fight infection: what the Drosophila genome says about animal immunity. Trends Genet. 16:442-449. [DOI] [PubMed] [Google Scholar]
- 23.Kopp, E., R. Medzhitov, J. Carothers, C. Xiao, I. Douglas, C. A. Janeway, and S. Ghosh. 1999. ECSIT is an evolutionarily conserved intermediated in the Toll/IL-1 signal transduction pathway. Genes Dev. 13:2059-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuan, C. Y., D. D. Yang, D. R. Samanta Roy, R. J. Davis, P. Rakic, and R. A. Flavell. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22:667-676. [DOI] [PubMed] [Google Scholar]
- 25.Kylsten, P., C. Samakovlis, and D. Hultmark. 1990. The cecropin locus in Drosophila: a compact gene cluster involved in the response to infection. EMBO J. 9:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. H., K. S. Cho, J. Lee, J. Yoo, J. Lee, and J. Chung. 2001. Diptericin-like protein: an immune response gene regulated by the antibacterial gene induction pathway in Drosophila. Gene 271:233-238. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. B., K. S. Cho, E. Kim, and J. Chung. 2003. blistery encodes Drosophila tensin protein and interacts with integrin and the JNK signaling pathway during wing development. Development 130:4351-4361. [DOI] [PubMed] [Google Scholar]
- 28.Lemaitre, B., J. Reichhart, and J. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94:14614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leo, E., K. Welsh, S. Matsuzawa, J. M. Zapata, S. Kitada, R. Mitchell, K. Ely, and J. Reed. 1999. Differential requirements for tumor necrosis factor receptor-associated factor family proteins in CD40-mediated induction of NF-kappaB and Jun N-terminal kinase activation. J. Biol. Chem. 274:22414-22474. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H., Y. C. Su, E. Becker, J. Treisman, and E. Y. Skolnik. 1999. A Drosophila TNF-receptor-associated factor (TRAF) binds the Ste20 kinase Misshapen and activates Jun kinase. Curr. Biol. 9:101-104. [DOI] [PubMed] [Google Scholar]
- 31.Lomaga, M., W. C. Yeh, I. Sarosi, G. Duncan, C. Furlonger, A. Ho, S. Morony, C. Capparelli, G. Van, S. Kaufman, A. van der Heiden, A. Itie, A. Wakeham, W. Khoo, T. Sasaki, Z. Cao, J. Penninger, C. Paige, D. Lacey, C. Dunstan, W. Boyle, D. V. Goeddel, and T. Mak. 1999. TRAF6 deficiency results in osteoporosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magnusson, C., and D. L. Vaux. 1999. Signalling by CD95 and TNF receptors: not only life and death. Immunol. Cell Biol. 77:44-46. [DOI] [PubMed] [Google Scholar]
- 33.Mihaly, J., L. Kockel, K. Gaengel, U. Weber, D. Bohmann, and M. Mlodzik. 2001. The role of the Drosophila TAK homologue dTAK during development. Mech. Dev. 102:67-79. [DOI] [PubMed] [Google Scholar]
- 34.Minden, A., A. Lin, F. X. Claret, A. Abo, and M. Karin. 1995. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell 81:1147-1157. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, H., H. Oshima, W. Chung, L. Williams-Abbott, C. F. Ware, H. Yagita, and K. Okumura. 1996. TRAF5, an activator of NF-κB and putative signal transducer for the lymphotoxin-beta receptor. J. Biol. Chem. 271:14661-14664. [DOI] [PubMed] [Google Scholar]
- 36.Nakano, H., S. Sakon, H. Koseki, T. Takemori, K. Tada, M. Matsumoto, E. Munechika, T. Sakai, T. Shirasawa, H. Akiba, T. Kobata, S. Santee, C. Ware, P. D. Rennert, M. Taniguchi, H. Yagita, and K. Okumura. 1999. Targeted disruption of TRAF5 gene causes defects in CD40- and CD27-mediated lymphocyte activation. Proc. Natl. Acad. Sci. USA 96:9803-9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishina, H., C. Vaz, P. Billia, M. Nghiem, T. Sasaki, J. L. De la Pompa, K. Furlonger, C. Paige, C. C. Hui, K. D. Fischer, H. Kishimoto, T. Iwatsubo, T. Katada, J. R. Woodgett, and J. M. Penninger. 1999. Defective liver formation and liver cell apoptosis in mice lacking the stress signaling kinase SEK1/MKK4. Development 126:505-516. [DOI] [PubMed] [Google Scholar]
- 38.Nishitoh, H., M. Saitoh, Y. Mochida, K. Takeda, H. Nakano, M. Rothe, K. Miyazono, and H. Ichijo. 1998. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol. Cell 2:389-395. [DOI] [PubMed] [Google Scholar]
- 39.Noselli, S. 1998. JNK signaling and morphogenesis in Drosophila. Trends Genet. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 40.Paricio, N., F. Feiguin, M. Boutros, S. Eaton, and M. Mlodzik. 1999. The Drosophila STE20-like kinase Misshapen is required downstream of the Frizzled receptor in planar polarity signaling. EMBO J. 18:4669-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phelps, C. B., and A. H. Brand. 1998. Ectopic gene expression in Drosophila using GAL4 system. Methods 14:367-379. [DOI] [PubMed] [Google Scholar]
- 42.Reichhart, J. M., M. Meister, J. L. Dimarcq, D. Zachary, D. Hoffmann, C. Ruiz, G. Richards, and J. A. Hoffmann. 1992. Insect immunity: developmental and inducible activity of the Drosophila diptericin promoter. EMBO J. 11:1469-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riesgo-Escovar, J. R., M. Jenni, A. Fritz, and E. Hafen. 1996. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 10:2759-2768. [DOI] [PubMed] [Google Scholar]
- 44.Rorth, P. 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93:12418-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothe, M., V. Sarma, V. M. Dixit, and D. V. Goeddel. 1995. TRAF2-mediated activation of NF-κB by TNF receptor 2 and CD40. Science 269:32767-32770. [DOI] [PubMed] [Google Scholar]
- 46.Shen, B., H. Liu, E. Y. Scolnik, and J. Manley. 2001. Physical and functional interactions between Drosophila TRAF2 and Pelle kinase contribute to Dorsal activation. Proc. Natl. Acad. Sci. USA 98:8596-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silverman, N., R. Zhou, S. Stoven, N. Pandey, D. Hultmakr, and T. Maniatis. 2000. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 14:2461-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sluss, H. K., Z. Han, T. Barrett, R. J. Davis, and Y. T. Ip. 1996. A JNK signal transduction pathway that mediates morphogenesis and an immune response. Genes Dev. 10:2745-2758. [DOI] [PubMed] [Google Scholar]
- 49.Takatsu, Y., M. Nakamura, M. Stapleton, M. C. Danos, K. Matsumoto, M. B. O'Connor, H. Shibuya, and N. Ueno. 2000. TAK1 participates in c-Jun N-terminal kinase signaling during Drosophila development. Mol. Cell. Biol. 20:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. J. ones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 51.Wallach, D., E. E., Varfolomeev, N. L. Malinin, Y. V. Goltsev, A. V. Kovlenko, and M. P. Boldin. 1999. Tumor necrosis factor receptor and Fas signaling mechanisms. Rev. Immunol. 17:331-367. [DOI] [PubMed] [Google Scholar]
- 52.Weber, U., N. Paricio, and M. Mlodzik. 2000. Jun mediates Frizzled induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development 127:3619-3629. [DOI] [PubMed] [Google Scholar]
- 53.Wicker, C., J. M. Reichhart, D. Hoffmann, D. Hultmark, C. Samakovlis, and J. A. Hoffmann. 1990. Insect immunity: characterization of a Drosophila cDNA encoding a novel member of the diptericin family of immune peptides. J. Biol. Chem. 265:2493-22498. [PubMed] [Google Scholar]
- 54.Xu, Y., G. Cheng, and D. Baltimore. 1996. Targeted disruption of TRAF3 leads to postnatal lethality and defective T-dependent immune responses. Immunity 5:407-415. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X., R. Khosravi-Far, H. Y. Chang, and D. Baltimore. 1997. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zapata, J. M., S. Matsuzawa, A. Godzik, E. Leo, S. A. Wasserman, and J. C. Reed. 2000. The Drosophila tumor necrosis factor receptor-associated factor-1 (DTRAF1) interacts with pelle and regulates NF-κB activity. J. Biol. Chem. 275:12102-12107. [DOI] [PubMed] [Google Scholar]
- 57.Zeilinger, J., and D. Bohmann. 1999. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126:3947-3956. [DOI] [PubMed] [Google Scholar]