Abstract
Fresh isolates of lentiviruses are characterized by an outstanding resistance to antibody-mediated neutralization. By investigating the changes that occurred in a neutralization-sensitive tissue culture-adapted strain of feline immunodeficiency virus after it was reinoculated into cats, a previous study had identified two amino acid positions of the surface glycoprotein (residues 481 and 557) which govern broad neutralization resistance (BNR) in this virus. By extending the follow-up of six independently evolving in vivo variants of such virus for up to 92 months, we now show that the changes at the two BNR-governing positions not only were remarkably stereotyped but also became fixed in an ordered sequential fashion with the duration of in vivo infection. In one variant, the two positions were also seen to slowly alternate at determining BNR. Evidence that evolution at the BNR-governing positions was accompanied, and possibly driven, by changes in the antigenic makeup of the viral surface brought about by the mutations at such positions is also presented.
Most primary isolates of human immunodeficiency virus (HIV) type 1 (HIV-1) and other lentiviruses display an extreme resistance to inhibition by immune sera in vitro. This is in striking contrast to what is observed with many tissue culture-adapted (TCA) strains of the same viruses, which are instead readily neutralized (for reviews, see references 4 and 25). The difference is a major focus of interest because the global or broad neutralization resistance (BNR) of wild-type lentiviruses is considered instrumental in their ability to persist in infected hosts and a major obstacle to the development of effective vaccines. The structural bases of BNR remain poorly understood, however (5, 25, 32, 35). For example, the amino acid sequences of neutralization-resistant primary isolates and neutralization-sensitive TCA derivatives of HIV-1 may differ at very few positions of the envelope (Env) glycoproteins (24). Yet, it is not clear how such limited amino acid divergence determines the striking differences in neutralization phenotypes and whether the differential amino acids remain constant or evolve over time of persistence in vivo.
Feline immunodeficiency virus (FIV) is a useful model for studying aspects of the natural history, pathogenesis, and control of immunosuppressive lentiviruses in a natural host species as well as an important pathogen of domestic cats (8, 26, 30). In previous work addressing the molecular basis of BNR (3, 6), we showed that upon reinoculation into specific-pathogen-free (SPF) cats, an exquisitely neutralization-sensitive TCA strain of the prototype Petaluma isolate of FIV (27) changed its neutralization properties by following a constant pattern. More specifically, soon after seroconversion and concomitant with a period of high rates of mutation in the env gene, the viral populations growing in different animals evolved into neutralization escape mutants that resisted inhibition by autologous antibodies (which were in the serum of the specific cat in which the virus was growing) but not by most other immune sera. Subsequently, after postinoculation (p.i.) periods that in individual hosts ranged from 4 to 20 months, the same viral populations invariably became resistant to all or most immune sera; i.e., they reverted to the BNR phenotype typical of primary isolates. Similar observations have been made for TCA strains of HIV-1 and chimeric simian immunodeficiency virus-HIV (2, 7, 12). Interestingly, reversion to the BNR phenotype of TCA FIV generally occurred when Env mutation rates had already declined relative to early times p.i. and corresponded temporally with a change of the Lys at position 481 in the variable 4 (V4) domain of the viral surface glycoprotein (SU) to either an Asn or Glu. These findings, together with analysis of the neutralization properties of appropriate biological and molecular clones, led to the clear identification of position 481 as a key determinant of the BNR phenotype. On the other hand, analysis of variants of the same inoculum of TCA virus that had grown for 3 years in other cats identified an Ser→Asn change at position 557 in the V5 region of the SU as another determinant of BNR (although it was less powerful, since it required the collaboration of a second amino acid change) (3). By extending such studies, we show here that the BNR-conferring change at position 557 was a subsequent event relative to the one at position 481 and that both positions tended to evolve in a stereotyped fashion during the course of infection. The possibility that such evolution was driven by antigenic changes in the viral surface induced by the BNR-inducing mutations was also examined.
MATERIALS AND METHODS
Inoculum virus, animals, and cells.
The female SPF cats (Iffa Credo, L'Arbresle, France) included in the study were housed in our climatized animal facility under the conditions required by European Community law and were infected with FIV when they were 7 to 12 months old. The cats were infected by intravenous injection of 1 ml of supernatant harvested from FL4 cells (33) chronically infected with the Petaluma strain of FIV (generous gift of Janet K. Yamamoto) at cell passage 181 (cat 3524 in group 1 and all cats in group 2) or 193 (all other cats in group 1). MBM cells are nontransformed feline CD3+, CD4−, and CD8− T lymphocytes (20) and are routinely cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 5 μg of concanavalin A per ml, and 20 U of recombinant human interleukin-2 per ml.
Reisolation of virus from infected animals.
Reisolation of virus from cats was performed by standard coculture of peripheral blood mononuclear cells with MBM cells. The times the cultures were first positive and the levels of reverse transcriptase (RT) produced varied only slightly. Three to five rapid passages in MBM cells were sufficient to accumulate viral stocks of suitable titer. The culture fluids were clarified by centrifugation at 350 × g for 15 min and were stored in 1-ml aliquots in liquid nitrogen until use.
SU sequencing and analysis.
DNA was extracted from 6 × 106 infected MBM cells with phenol-chloroform and checked for integrity on an ethidium bromide-stained 0.8% agarose gel and for amplification ability by using feline tumor necrosis factor alpha-specific primers. Nested PCR amplification and cycle sequencing of the SUs were performed as described previously (3). The nucleotide sequences were edited and translated with BioEdit software (version 5.0.9) (11). Selected regions were resequenced to confirm the presence of the substitutions that distinguished the viral variants.
The DNA sequences obtained from this and a previous study (3) were aligned by using the program ClustalX (version 1.8) (28) and were manually refined with GENEDOC software (version 2.6.002 [22]). Genetic distances were generated by using the DAMBE program (version 4.0.98 [31]) and Kimura's two-parameter method by correcting for superimposed substitutions and a transition/transversion ratio of 2.0 (nucleotide sequences) and Poisson P value by correcting for multiple hits (amino acid sequences). Phylogenies for the SU amino acid sequences were inferred by median-joining network analysis (version 3.1.1.1 alpha [1]). An input file that contained the amino acid positions that were found to be mutated in the study was created. The tree was built by calculating the weighted Hamming distances by using the explicit parameter (ɛ), which was set equal to 2; the default frequency (F) for calculation of the minimal spanning network (i.e., the most parsimonious tree among species); and the mutated position option, which displays the amino acid substitution events along the evolutionary path. The robustness of the tree was assessed by repeating the calculation with manually bootstrapped sequences (100 reiterations).
Construction and characterization of molecular clones.
The Asn-Asn clone was produced specifically for this study exactly as described previously (3), while the others had been developed earlier. Before use, the amino acid sequences and neutralization phenotypes of all clones were checked or rechecked and were found to have the desired features. All clones were propagated in MBM cells. As discussed below, the clones were also compared for replication capacity in an in vitro multicycle replication assay in the presence or absence of antiviral antibodies, as described previously (10). In brief, the clones were seeded at a multiplicity of infection of 0.0001 onto 24-well plates containing 2 × 106 MBM cells in complete growth medium supplemented with 2% pooled normal cat serum (NCS) or selected immune sera. After 24 h of incubation, the cells were washed three times, and the cultures were continued for 25 days with periodic determination of the amount of RT released in the supernatant, followed by substitution of the respective media.
Adsorption and measurement of antivirion surface antibodies.
Adsorption and measurement of antivirion surface antibodies were carried out with intact virions of the molecular clones captured onto microwells by means of Galanthus nivalis lectin (GN) and evaluation of their ability to adsorb anti-FIV Env antibodies from the serum of a cat infected over the long term. As reported previously (9), GN has the ability to effectively capture FIV virions through their Env glycoproteins. The immune serum used was obtained from cat 275 at month 55 of infection (binding titers for FIV Env, 1,600 to 3,200, irrespective of the viral clone used as a source of antigen). Precoating of the wells with GN was carried out by incubation with 5 μg of GN (Sigma, St. Louis, Mo.) overnight at room temperature, followed by a saturation step with bovine serum albumin. The subsequent capture of virions was carried out by incubating the GN-containing wells twice for 6 h each time with freshly cloned viral preparations normalized for p25 content to saturate all the lectin, and this was followed by a postcoating step with skim milk at room temperature for 1 h. Wells thus prepared and wells mock coated with GN, which were coated with skim milk, were used both to adsorb the immune serum with the viral clones under scrutiny and to measure the Env-binding activity that resisted such adsorption. Adsorption of immune serum and control NCS was carried out by repeatedly incubating aliquots diluted 1:200 in fresh virion-coated or mock-coated wells. The Env-binding activities of virion-adsorbed and mock-adsorbed aliquots of serum to virion-coated wells were measured by an enzyme-linked immunosorbent assay, exactly as described previously (9).
RESULTS
The two positions that govern BNR evolved sequentially in stereotyped fashion.
Previous studies (3, 6) had shown that reversion occurred 4 to 20 months p.i. in four SPF cats injected with the study virus (group 1; Fig. 1) and investigated until the viral variants that they harbored reverted to BNR and was mediated by substitution of an Asn or Glu for Lys-481. However, amino acid 481 was substituted (Lys→Glu) in only one of the viral revertants harbored by three additional SPF cats that were first examined 36 months p.i. (group 2; Fig. 1), while all revertants presented the mutation Ser→Asn at position 557. This led us to postulate that the 557Ser→Asn substitution represents an event temporally subsequent to the change at position 481 that possibly emerged due to the need for the virus to resist an evolving immune response. However, because no samples from the earlier time points were available from the group 2 animals, the possibility that the amino acid at position 557 of the latter variants had changed before the amino acid change occurred at position 481 could not be excluded; i.e., they had reverted to BNR through a pathway different from that for the group 1 variants.
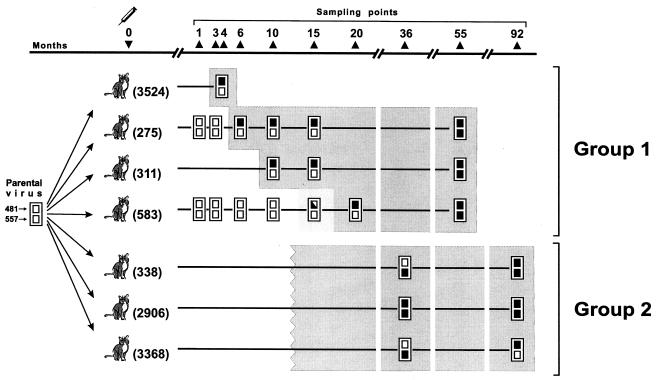
FIG. 1.
Evolution of two SU amino acid positions that govern BNR in the study of TCA FIV in seven infected cats, as observed in this and previous studies (3, 6). The boxes at each sampling time point represent amino acid positions 481 and 557, as indicated under the parental TCA virus used as the inoculum, and are solid when these positions are replaced by BNR-conferring residues. The shaded area includes the samples with virus exhibiting a BNR phenotype. Note that the precise timing of reversion to BNR is known only for group 1 cats, since group 2 cats were first tested 36 months p.i. Cat 311 yielded the first virus-positive cultures at month 10 p.i. The virus in the sample obtained from cat 583 at 15 months p.i. exhibited intermediate neutralization resistance and yielded two viral sequences, in only one of which was the amino acid at position 481 replaced by a BNR-conferring amino acid (3). The complete sequences of all variants are available from the authors by e-mail.
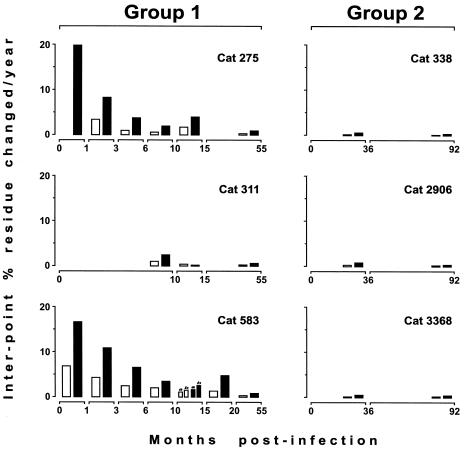
In an attempt to discriminate between the two possibilities described above, in the present study we extended the follow-up of the viral populations of group 1 cats. Thus, at 55 months p.i. new viral samples were cultured from the peripheral blood mononuclear cells of three animals in that group (the fourth cat had been euthanized due to a loss of appetite and emaciation), and their SUs were sequenced and analyzed within three to five tissue culture passages. As schematically illustrated in Fig. 1, the three independently growing variants were found to have maintained the BNR-conferring residues at position 481; but despite that, the rates of nucleotide and amino acid changes had dropped considerably relative to those at early times p.i. (Fig. 2), and all variants had also acquired the 557Ser→Asn mutation, which had not been detected in any of the previous samples obtained from the same animals. Of the total of 30 amino acid replacements detected among the 432 residues of the SUs of these variants relative to the sequences in the samples obtained 35 to 40 months earlier, only 557Ser→Asn had become fixed in all variants (Table 1). The likelihood that the same position might have become substituted by chance with the same amino acid in all three independently evolving SUs is extremely low (P < 0.00005, permutation test). Thus, it was evident that the 557Ser→Asn replacement represented a subsequent event relative to the BNR-inducing changes at position 481. It was also apparent that the BNR-inducing positions had evolved in a remarkably orderly and stereotyped fashion in viruses from all infected cats.
FIG. 2.
Rates of genetic divergence between sampling points in the SUs of the six viral variants described in Fig. 1 that were monitored over the long term. The results are expressed as the percentage of nucleotide (open bars) and amino acid (closed bars) changes per year over the time interval between sampling points. The two viral sequences that were obtained from cat 583 at 15 months and that have position 481 unsubstituted and substituted with a BNR-conferring amino acid (see the legend to Fig. 1) are marked a and b, respectively.
TABLE 1.
Amino acid changes found in the SUs of the viral samples obtained at 55 months p.i. relative to previous ones
| Cat no. | Change at the following amino acid positiona:
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 184 | 195 | 196 | 197 | 249 | 279 | 381 | 392 | 409 | 416 | 417 | 421 | 452 | 469 | 471 | 482 | 485 | 546 | 557 | 558 | 559 | 560 | 562 | 603 | |
| 275 | S→W | Q→D | G→C | R→K | T→S | E→G | Q→P | C→G | V→I | N→T | S→N | K→E | ||||||||||||
| 311 | I→V | Q→H | K→T | L→I | A→V | S→N | S→G | I→T | A→T | |||||||||||||||
| 583 | V→I | Q→H | L→I | K→E | P→S | A→V | S→N | A→T | M→L | |||||||||||||||
The changes are in relation to the sequences in samples collected 40 months earlier.
In one viral variant the two positions determining BNR slowly alternated.
Resampling and resequencing of FIV isolates from group 2 cats 92 months p.i. also yielded interesting findings (Fig. 1). By this time, the SUs of these viral variants had undergone very limited changes relative to those that had occurred over the previous 4.5 years, when the previous sampling had been performed (Fig. 2). Nonetheless, in the two variants that had previously conserved the Lys of the inoculum virus at position 481, this residue was replaced by an Asn residue, thus again showing a surprising uniformity of change. Possibly even more interestingly, one of such variants (from cat 3368) had also retromutated at position 557 (Asn→Ser), hence reacquiring the original residue present in the neutralization-sensitive inoculum virus. Thus, in this variant amino acid positions 481 and 557 had alternated in determining BNR.
Impact of BNR-conferring mutations on overall virus evolution.
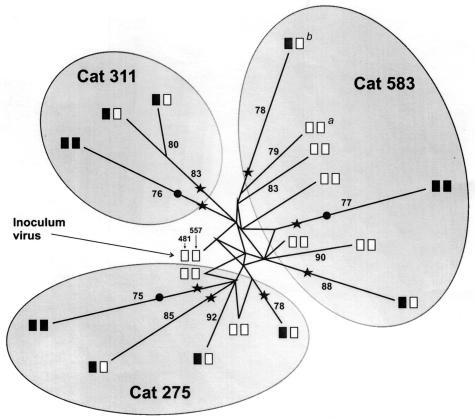
All the SU amino acid sequences obtained from three group 1 cats, from which we had the most samples, were used to analyze the evolution of viral variants. Because the genetic distance-based methods that were used in the first approach (i.e., the neighbor-joining and Fitch-Margoliash methods) led to polytomies, the recently described median-joining network algorithm of the discrete-character method was used to construct the most parsimonious tree. This method infers the most probable evolutionary routes from hypervariable regions by highlighting character conflicts, such as homoplasy or superimposed sequences, in the form of reticulations and likely evolutionary routes by assigning a different weight for the most informative characters (1). The robustness of the tree was assessed by repeating the calculation with manually bootstrapped sequences (100 reiterations). As depicted in Fig. 3, the tree topology was well resolved, and reticulates resulting from conflicting phylogenetic signals were present only in the early variants, as may be expected for viral populations that were mutating at a rapid rate under the pressure of the multiple selecting forces encountered by the TCA inoculum virus in the “new” in vivo environment. On the contrary, the variants carrying BNR-conferring residues at position 481 or 557, or both, were the ones that diverged the most from the inoculum TCA virus. In addition, as a further indication of the important impact exerted by the latter mutations on overall virus evolution, these replacements were constantly located close to the nodes from which the most divergent variants originated.
FIG. 3.
Median-joining network for the SU amino acid sequences of the viruses in all samples from three group 1 cats (Fig. 1). Solid and empty boxes are as described in the legend to Fig. 1. Stars and circles, substitutions conferring BNR at positions 481 and 557, respectively. Bootstrap values above 75% are indicated along the branches. The reticular pattern at the center depicts character conflicts. a and b, the two viral sequences obtained from cat 583 at 15 months p.i. in which position 481 was unsubstituted and substituted with a BNR-conferring amino acid, respectively. The considerable differences in branch lengths between these two sequences confirm the strong impact exerted by such a substitution on virus evolution.
Changes at BNR-conferring positions greatly facilitated virus replication in the presence of immune sera.
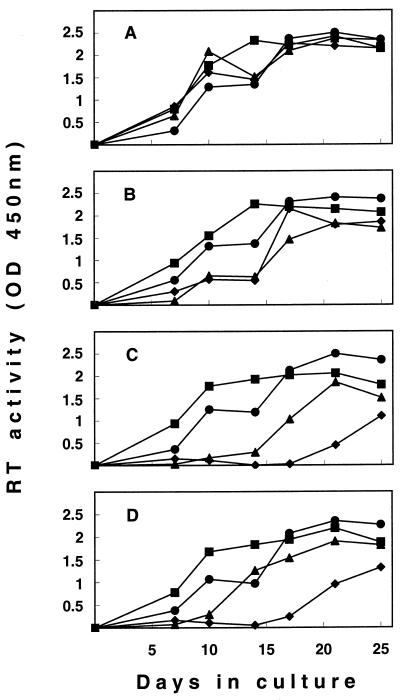
Studies have shown that modifications in the neutralization phenotype of HIV-1 and simian immunodeficiency virus can be accompanied by changes in virus growth efficiency in vitro, most likely resulting from alterations in the Env functions mediating virus entry into cells (14, 17, 23, 24, 25). It was therefore of interest to verify whether the BNR-conferring changes at positions 481 and 557 had similar effects on FIV. For this purpose, we compared the replication properties of molecular clones that had the same residues as the inoculum TCA virus at both positions (clone Lys-Ser) or that had Asn substitutions at either (clones Asn-Ser and Lys-Asn) or both (clone Asn-Asn) positions. This was done in a multicycle replication assay with MBM cells as the substrate, both in the absence of interfering immunological factors (cultures supplemented with NCS) and in the presence of sequential immune sera from a cat in which the TCA FIV isolate had evolved from neutralization sensitivity to BNR. More specifically, the immune sera were obtained from cat 275 before the sequence of the infecting virus had changed at positions 481 and 557 (month 3 p.i.), after Asn had become fixed at position 481 (month 15), and after position 557 had also become fixed with Asn (month 55). Figure 4 shows how the clones replicated during 25 days of culture. In the presence of immune sera the replication curves for the four clones were as expected on the basis of their genotypes and the time p.i. at which the selecting serum had been harvested (Fig. 4B to D). Thus, the growth of clone Lys-Ser was more or less profoundly impaired by all immune sera, the growth of clone Asn-Ser was only slightly delayed and was delayed only by the immune serum from the cat that had been infected over the longer term, the growth of clone Lys-Asn was moderately inhibited by all immune sera, and the growth of clone Asn-Asn was essentially unaffected regardless of the immune serum used. In contrast, in the cultures supplemented with NCS, the four clones exhibited comparable high levels of replication (Fig. 4A). Taken together, these findings indicate that the BNR-conferring mutations did not affect the basal replication capacities of the virus (19) in lymphoid cells but conferred on it a considerable selective advantage if antiviral antibodies were present in the environment.
FIG. 4.
Growth curves in MBM cell cultures of molecularly cloned FIV variants Lys-Ser (♦), Asn-Ser (•), Lys-Asn (▴), and Asn-Asn (▪). The cultures were supplemented with 2% NCS (A) or immune sera (B to D) during the entire duration of the experiment. Immune sera were obtained from cat 275 when the infecting virus had not yet developed changes at position 481 and 557 (month 3 p.i.) (B), after Asn had become fixed at position 481 (month 15 p.i.) (C), and after position 557 had also become fixed with Asn (month 55 p.i.) (D). Virus growth is expressed as levels of RT activity in the supernatants. OD, optical density.
Changes at BNR-conferring positions appreciably modified the binding epitopes exposed on the virion surface.
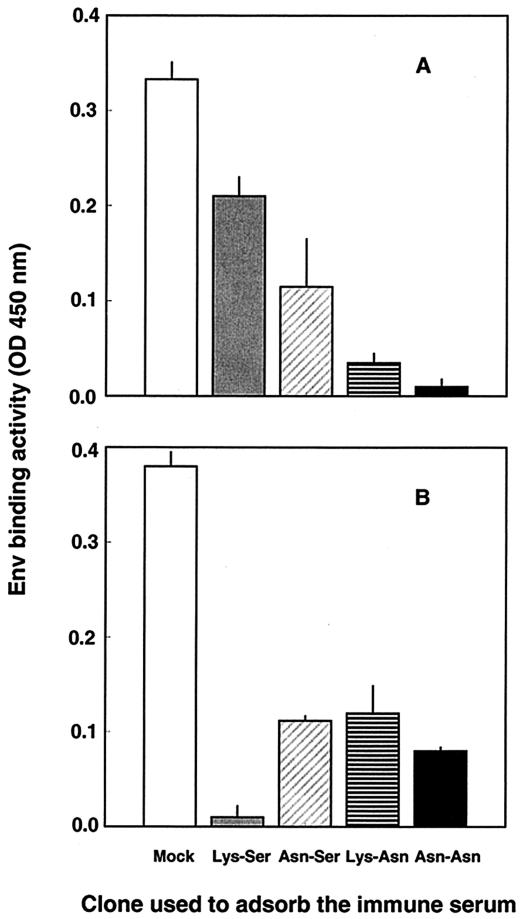
We investigated whether changes at the BNR-conferring positions could appreciably modify the antigenic makeup of the surface of the study virus. This was approached by comparing the abilities of the molecular clones described above to adsorb the Env-binding antibodies present in immune serum from a cat that had been infected over the long term, which presumably contained a large repertoire of such antibodies. Thus, aliquots of an appropriate dilution of the serum obtained from cat 275 at month 55 p.i., i.e., after the infecting virus had developed Asn at both position 481 and position 557 (Fig. 1), were repeatedly adsorbed with intact virions of each of the four clones until their binding activities for the specific adsorbing clones were completely depleted and then tested for residual binding to the clone carrying BNR-conferring residues at both positions, clone Asn-Asn. Because this protocol minimized the effects of possible changes in the density of Env molecules on the virions (5), any residual binding activity observed in serum adsorbed with a given clone clearly pointed to the existence of an epitope(s) in the Env of clone Asn-Asn that was absent from the Env of the clone used for adsorption. In all serum-clone combinations, four cycles of adsorption completely or nearly completely depleted the reactivity of the serum for the adsorbing clone (Fig. 5). However, the serum aliquots adsorbed with clone Asn-Ser and especially those absorbed with clone Lys-Ser still exhibited substantial binding activities for clone Asn-Asn (Fig. 5A), thus indicating that clones Asn-Ser and Lys-Ser had few binding epitopes or did not efficiently expose the binding epitope(s) that was instead well represented in clone Asn-Asn. Similarly, although the differences were less striking, when the adsorbed serum aliquots were examined for residual binding to clone Lys-Ser, i.e., the clone that carried the same residues as the inoculum TCA virus, it was found that clones Asn-Ser, Lys-Asn, and Asn-Asn had left some reactivity unadsorbed (Fig. 5B), thus again arguing for the existence of subtle but appreciable differences between the clones in terms of the binding epitopes exposed on the virion surfaces.
FIG. 5.
Abilities of intact virions of molecular clones Lys-Ser, Asn-Ser, Lys-Asn, and Asn-Asn to adsorb Env-binding antibodies from immune serum from a cat that had been infected over the long term. Aliquots of serum obtained from cat 275 at 55 months p.i. were repeatedly adsorbed with virions of the indicated clones until the reactivity for the adsorbing clone was exhausted and were then tested for residual binding activity against clones Asn-Asn (A) and Lys-Ser (B). Absorption and binding tests were carried out with wells precoated with the lectin GN, as described in Materials and Methods. Immune serum adsorbed with wells mock coated with GN served as a control. NCS, which was run in parallel, was constantly devoid of binding activity. The results presented here were obtained with a final dilution of the adsorbed and unadsorbed serum aliquots of 1:200. The optical density (OD) generated by the serum aliquots adsorbed with clone Asn-Ser or Lys-Asn when they were reacted with the homologous clone were uniformly less than 0.02. Bars represent standard deviations for triplicate wells. The experiment was repeated twice, with comparable results.
DISCUSSION
The BNR of wild-type HIV-1 and other lentiviruses has attracted considerable attention because of its conceptual and practical implications (for reviews, see references 5 and 25). By investigating the Env changes undergone by a neutralization-sensitive TCA strain of FIV while readapting to cats, previous studies (3) identified amino acids 481 and 557 of the SU as key positions for the control of BNR in this viral strain. By extending the analysis of the viral populations growing in the infected cats for up to over 7 years, we have now found that changes at such positions not only were a feature that emerged in all the animals but also became fixed in an ordered sequential fashion during virus persistence. In particular, the change at position 557 was seen to be a subsequent event relative to the time of occurrence of the one at position 481 and occurred long after the initial p.i. phase of rapid Env mutation had ended. Furthermore, with increasing time of in vivo infection, the viral population of one animal (cat 3368) was seen to slowly alternate the positions (positions 481 and 557) determining the state of BNR. Notably, as determined by constructing the most parsimonious tree with the SU sequences retrieved from the three animals from which large enough numbers of samples were available, the BNR-conferring mutations were found to exert a considerable impact on virus evolution. The mechanism(s) by which such mutations may have led to BNR were discussed elsewhere (3) and will not be further discussed here, since the experiments reported here have little bearing on this aspect. It is worth recalling, however, that two mutations (481Lys→Asn and 557Ser→Asn) introduced potential N-linked glycosylation sites, whereas the third mutation (481Lys→Glu) led to a modification of charge polarity. Thus, at least in FIV the acquisition of a single N-glycosylation site as well as other single amino acid changes of the SU, which most likely act by modifying the conformation of Env (15, 16), can have much more drastic repercussions on the accessibility or availability of neutralization epitopes than just the production of escape variants capable of resisting autologous antibodies but not others (29).
Relative to a molecular clone with the same residues as the TCA virus used in the original inoculum, clones that differed only by having BNR-conferring residues at position 481 or 557, or both, replicated with similar efficiencies in lymphoid cultures supplemented with NCS but exhibited variously pronounced selective advantages when they were propagated in the presence of sequential immune sera from a cat in which the TCA virus had reverted to BNR. Also, the same molecular clones showed differences in terms of the ability of intact lectin-captured virions to remove Env-binding antibodies from immune serum from a cat that had been infected over the long term, thus showing that the impacts of the changes responsible for BNR acquisition and maintenance were also detectable at the level of antibody binding by the viral surface. In vitro immune selection studies showed that antibodies in the sera of cats in which the reversion of virus to BNR had occurred drove BNR and amino acid changes in the study virus similar to those that viruses in infected cats underwent (10). Together with those findings, the present findings support a model in which the uniform and sequentially ordered fixation of BNR-conferring amino acids at positions 481 and 557 was driven by antibody specificities that developed in the same temporal order in all animals, although at different times p.i. In turn, the antibodies that drove the change at position 481 were probably elicited by a viral epitope(s) that was already present in the infecting TCA virus, while the ones that drove the change at position 557 were most likely evoked by an epitope(s) that had become functional as a result of the fixation of BNR-conferring substitutions at position 481. Indeed, the kinetics of the replication curves exhibited by the mutated clones in the presence of sequential infected cat sera, as discussed above (Fig. 4), support this view.
Amino acid substitutions have also been seen to become fixed in an orderly fashion in the pol gene of HIV-1 as a result of the treatment of infected patients with some viral enzyme-inhibiting drugs (18, 21). However, in our study we expected more diversity of change since (i) the selecting force was not a single drug but the much more complex and variegated humoral immune response of the hosts; (ii) the study cats were not inbred or siblings; (iii) in the early months p.i., the cats that were studied for this aspect had developed partially different repertoires of virus-neutralizing antibodies; and (iv) in individual animals the TCA virus had reverted to BNR at widely different times p.i., in all probability as a reflection of unevenly efficacious immune responses (3). Thus, the most likely explanation for the observed uniformity of evolution of the BNR-conferring positions is that very few Env amino acid positions, possibly only positions 481 and 557, had the potential to confer BNR to the study virus. In light of this finding, the alternation of these two positions in conferring the BNR observed in the viral variant from cat 3368 could be interpreted as the only strategy that this variant had at its disposal to maintain a state of BNR in the presence of a particularly effective, continuously evolving antibody response.
An independent study has recently shown that the same TCA strain of FIV studied here was strongly attenuated in its in vivo replicative and immunosuppressive properties but that once it was reinoculated into cats, it slowly (34 months) reacquired a more virulent profile (13). Thus, studies like the present one may not only lead to a better understanding of how FIV evades immune responses but also help shed light on the virus-host interactions that permit full expression of the pathogenic potential of FIV. At this time, it is not known how the present findings may be representative of the situation in other lentiviruses. However, a strain of simian immunodeficiency virus-HIV encoding the Env of a TCA HIV-1 strain was reinjected in vivo and was recently seen to become neutralization resistant and increasingly virulent by changing its Env at the same amino acid positions in individual hosts of different primate species (12), thus suggesting that the amino acid positions of Env that played a key role in permitting in vivo persistence were also few in this virus. The delineation of such key positions within a given lentivirus species could assist in the development of protective vaccines by indicating which viral epitopes the vaccines should incorporate in order to elicit immune responses that anticipate the antigenic changes that the virus tends to undergo in immunized hosts, thus reducing the likelihood that the virus will take or at least reducing the severity of infection.
Acknowledgments
This work was supported by Ministero della Salute-Istituto Superiore di Sanità, “Programma per l'AIDS” and Ministero della Istruzione, Università e Ricerca Scientifica “Programma Cofinanziamento 2001.” S.G. was the holder of fellowships from Ministero della Salute, Rome, Italy.
REFERENCES
- 1.Bandelt, H.-J., P. Forster, and A. Röhl. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16:37-48. [DOI] [PubMed] [Google Scholar]
- 2.Beaumont, T., A. van Nuenen, S. Broersen, W. A. Blattner, V. V. Lukashov, and H. Schuitemaker. 2001. Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J. Virol. 75:2246-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendinelli, M., M. Pistello, D. Del Mauro, G. Cammarota, F. Maggi, A. Leonildi, S. Giannecchini, C. Bergamini, and D. Matteucci. 2001. During readaptation in vivo, a tissue culture-adapted strain of feline immunodeficiency virus reverts to broad neutralization resistance at different times in individual hosts but through changes at the same position of the surface glycoprotein. J. Virol. 75:4584-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, D. R., and P. W. Parren. 2000. Vaccines and the induction of functional antibodies: time to look beyond the molecules of natural infection? Nat. Med. 6:123-125. [DOI] [PubMed] [Google Scholar]
- 5.Burton, D. R., E. O. Saphire, and P. W. Parren. 2001. A model for neutralization of viruses based on antibody coating of the virion surface. Curr. Top. Microbiol. Immunol. 260:109-143. [DOI] [PubMed] [Google Scholar]
- 6.Cammarota, G., D. Matteucci, M. Pistello, E. Nicoletti, S. Giannecchini, and M. Bendinelli. 1996. Reduced sensitivity to strain-specific neutralization of laboratory-adapted feline immunodeficiency virus after one passage in vivo: association with amino acid substitutions in the V4 region of the surface glycoprotein. AIDS Res. Hum. Retrovir. 12:173-175. [DOI] [PubMed] [Google Scholar]
- 7.Cayabyab, M., G. B. Karlsson, B. A. Etemad-Moghadam, W. Hofmann, T. Steenbeke, M. Halloran, J. W. Fanton, M. K. Axthelm, N. L. Letvin, and J. G. Sodroski. 1999. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2). J. Virol. 73:976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elder, J. H., and T. R. Phillips. 1995. Feline immunodeficiency virus as a model for development of molecular approaches to intervention strategies against lentivirus infections. Adv. Virus Res. 45:225-247. [DOI] [PubMed] [Google Scholar]
- 9.Giannecchini, S., D. Matteucci, and M. Bendinelli. 1998. Effect of enzymatic deglycosylation on feline immunodeficiency virus sensitivity to antibody-mediated neutralization. AIDS Res. Hum. Retrovir. 14:199-204. [DOI] [PubMed] [Google Scholar]
- 10.Giannecchini, S., D. Matteucci, A. Ferrari, M. Pistello, and M. Bendinelli. 2001. Feline immunodeficiency virus-infected cat sera associated with the development of broad neutralization resistance in vivo drive similar reversions in vitro. J. Virol. 75:8868-8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Hofman-Lehmann, R., J. Vlasak, A.-L. Chenine, P.-L. Li, T. W. Baba, D. C. Montefiori, H. M. McClure, D. C. Anderson, and R. M. Ruprecht. 2002. Molecular evolution of human immunodeficiency virus env in humans and monkeys: similar patterns occur during natural disease progression or rapid virus passage. J. Virol. 76:5278-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosie, M. J., B. J. Willett, D. Klein, T. H. Dunsford, C. Cannon, M. Shimojiama, J. C. Neil, and O. Jarrett. 2002. Evolution of replication efficiency following infection with a molecularly cloned feline immunodeficiency virus of low virulence. J. Virol. 76:6062-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. H. I. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 16.Labrijn, A. F., and P. W. H. I. Parren. 1999. Neutralizing epitopes of HIV-1, p. 18-34. In B. Korber, C. Brander, B. F. Haynes, J. P. Moore, R. Koup, B. Walker, and D. I. Watkins (ed.), HIV molecular immunology database. Los Alamos National Laboratory, Los Alamos, N.M.
- 17.Lerner, D. L., and J. H. Elder. 2000. Expanded host cell tropism and cytopathic properties of feline immunodeficiency virus strain PPR subsequent to passage through interleukin-2-independent T cells. J. Virol. 74:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loveday, C. 2001. Nucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26:S10-S24. [DOI] [PubMed] [Google Scholar]
- 19.Maldarelli, F. 2003. HIV-1 fitness and replication capacity: what are they and can they help in patient management? Curr. Infect. Dis. Rep. 5:77-84. [DOI] [PubMed] [Google Scholar]
- 20.Matteucci, D., P. Mazzetti, F. Baldinotti, L. Zaccaro, C. Giannelli, and M. Bendinelli. 1995. The feline lymphoid cell line MBM and its use for feline immunodeficiency virus isolation and quantitation. Vet. Immunol. Immunopathol. 46:71-82. [DOI] [PubMed] [Google Scholar]
- 21.Miller, V. 2001. Resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26:S34-S50. [DOI] [PubMed] [Google Scholar]
- 22.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 23.Park, E. J., M. K. Gorny, S. Zolla-Pazner, and J. V. Quinnan, Jr. 2000. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J. Virol. 74:4183-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, E. J., and G. V. Quinnan, Jr. 1999. Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J. Virol. 73:5707-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13:S137-S162. [PubMed] [Google Scholar]
- 26.Pedersen, N. C. 1993. Feline immunodeficiency virus infection, p. 181-228. In J. A. Levy (ed.), The retroviridae, vol. 2. Plenum Press, New York, N.Y.
- 27.Pedersen, N. C., E. W. Ho, M. L. Brown, and J. K. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science 235:790-793. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 30.Willett, B. J., J. N. Flynn, and M. J. Hosie. 1997. FIV infection of the domestic cat: an animal model for AIDS. Immunol. Today 18:182-189. [DOI] [PubMed] [Google Scholar]
- 31.Xia, X. 2000. Data analysis in molecular biology and evolution. Kluwer Academic Publishers, Boston, Mass.
- 32.Xiao, Y., X. Dong, and Y. H. Chen. 2002. Neutralizing antibodies mechanism of neutralization and protective activity against HIV-1. Immunol. Res. 25:193-200. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto, J. K., C. D. Ackley, H. Zochlinski, H. Louie, E. Pembroke, M. Torten, H. Hansen, R. Munn, and T. Okuda. 1991. Development of IL-2-independent feline lymphoid cell lines chronically infected with feline immunodeficiency virus: importance for diagnostic reagents and vaccines. Intervirology 32:361-375. [DOI] [PubMed] [Google Scholar]
- 34.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]