Abstract
Twenty-four base pairs of the human antioxidant response element (hARE) are required for high basal transcription of the NAD(P)H:quinone oxidoreductase1 (NQO1) gene and its induction in response to xenobiotics and antioxidants. hARE is a unique cis-element that contains one perfect and one imperfect AP1 element arranged as inverse repeats separated by 3 bp, followed by a “GC” box. We report here that Jun, Fos, Fra, and Nrf nuclear transcription factors bind to the hARE. Overexpression of cDNA derived combinations of the nuclear proteins Jun and Fos or Jun and Fra1 repressed hARE-mediated chloramphenicol acetyltransferase (CAT) gene expression in transfected human hepatoblastoma (Hep-G2) cells. Further experiments suggested that this repression was due to overexpression of c-Fos and Fra1, but not due to Jun proteins. The Jun (c-Jun, Jun-B, and Jun-D) proteins in all the possible combinations were more or less ineffective in repression or upregulation of hARE-mediated gene expression. Interestingly, overexpression of Nrf1 and Nrf2 individually in Hep-G2 and monkey kidney (COS1) cells significantly increased CAT gene expression from reporter plasmid hARE-thymidine kinase-CAT in transfected cells that were inducible by β-naphthoflavone and tert-butyl hydroquinone. These results indicated that hARE-mediated expression of the NQO1 gene and its induction by xenobiotics and antioxidants are mediated by Nrf1 and Nrf2. The hARE-mediated basal expression, however, is repressed by overexpression of c-Fos and Fra1.
NAD(P)H:quinone oxidoreductase1 (NQO1) catalyzes reductive detoxification of quinones and its derivatives and protect the cells against redox cycling and oxidative stress (1). Twenty-four base pairs of the human antioxidant response element (hARE) were characterized previously in the promoter of the human NQO1 gene that are essential for high basal transcription and induction in response to xenobiotics and antioxidants including β-naphthoflavone (β-NF), tert-butyl-hydroquinone (t-BHQ), and hydrogen peroxide (2–4). Similar cis-elements have also been characterized in the promoters of the genes encoding rat NQO1, rat and mouse glutathione S-transferase (GST) Ya subunit, and rat GST P (4, 5). The current hypothesis is that coordinated induction of several defensive gene products may provide the required protection for cells against environmental insults and oxidative stress (4).
The NQO1 gene hARE contains one perfect and one imperfect AP1 binding site arranged as inverse repeats separated 3 bp apart followed by “GC” box (3, 4). The AP1 binding site [also known as 12-O-tetradecanoylphorbol-13-acetate (TPA) response element or TRE] was characterized previously in the promoter regions of the collagenase and metallothionein genes, and is required to activate their expression in response to TPA (6). The products of protooncogenes Jun (c-Jun, Jun-D, and Jun-B) form homodimers among themselves and heterodimers with c-Fos and Fra (Fra1 and Fra2) proteins and bind to the TRE (a single AP1 binding site) and regulate the expression of target genes (6). Nrf1 and Nrf2 are recently identified NF-E2-related factors that bind to the sequence GCTGAGTCATGATGAGTCA (7, 8). The Nrf1 and Nrf2 contain 742 and 589 amino acids, respectively. Both these proteins display marked homology to the NF-E2, which regulates expression of globin genes in developing erythroid cells through interaction with upstream AP1-like recognition sites (9). Like Jun and Fos, Nrf1 and Nrf2 are bZIP proteins (7, 8).
hARE is a unique element. This is because, the hARE (two AP1- or AP1-like elements and a GC box) and not the TRE (a single AP1 element) is responsive to xenobiotics and antioxidants (3). The Jun and Fos transcription factors are known to bind to the human NQO1 gene hARE but not with rat NQO1 gene ARE even though both elements contained highly conserved sequences (4, 5). Therefore, the actual proteins that bind to both hARE and rat ARE and mediate the expression and induction of the NQO1 gene in human and rat tissues were predicted to be other than Jun and Fos proteins (4, 5). We reasoned that these other proteins have to be AP1-related because both hARE and rat ARE contain AP1 and AP1-like elements. This prompted us to search the literature on AP1-related proteins. Interestingly, we found that tissue specific expression of Nrf1 and Nrf2 transcription factors was identical to the tissue specific expression of NQO1 gene regulated by hARE (7, 8, 10). In addition, we found that the binding site for Nrf1 and Nrf2 was very similar to hARE (4, 7, 8). These results encouraged us to determine the role of Nrf1 and Nrf2 proteins in the regulation of hARE-mediated expression of NQO1 and other detoxifying enzyme genes.
In the present report, we demonstrate that overexpression of combinations of nuclear proteins Jun and Fos or Jun and Fra1 down regulated hARE-mediated gene expression. We also show that downregulation of hARE-mediated chloramphenicol acetyltransferase (CAT) gene expression in transfected cells is due to overexpression of c-Fos and Fra1, but not due to Jun proteins. We further demonstrate that the nuclear protein Nrf1 binds to the hARE. The overexpression of Nrf1 and Nrf2 proteins individually and in combination upregulated both the expression of hARE-mediated CAT gene expression and induction by β-NF and t-BHQ.
MATERIALS AND METHODS
Plasmid Construction.
The construction of hARE-, mutant hARE- and TRE–thymidine kinase (tk)–CAT plasmids have been described (3). In addition, the plasmid pNQO1CAT0.837 are also described (2). The plasmid eukaryotic expression vector (LNCX), LNCX–c-Jun, LNCX–Jun-D, LNCX–Jun-B, LNCX–c-Fos and LNCX–Fra1 were obtained from Kevin Ryder (Fox Chase Cancer Center, Philadelphia) (11). These plasmids were all derived by inserting the respective murine genes into the polylinker region of LNCX, so that each gene is expressed via the cytomegalovirus promoter. The human Nrf1 and mouse Nrf2 cDNA were gift from Jefferson Y. Chan (University of California, San Francisco). The 2.6-kb EcoRI insert of Nrf1 cDNA was subcloned at HpaI site of LNCX to generate LNCX–Nrf1R and LNCX–Nrf1C. The Nrf1 cDNA was also subcloned at the EcoRI site of pMT2 to generate pMT2–Nrf1R and pMT2–Nrf1C. The Nrf1 cDNA subcloned in the pMT2 vector is transcribed by major adenovirus late promoter upon transfection in COS1 cells (12). The 1.5-kb SalI–NheI fragment of mouse Nrf2 cDNA was subcloned in expression vector LNCX to generate LNCX–Nrf2R and LNCX–Nrf2C. In the above described expression plasmids, R denotes the reverse orientation of the cDNA insert, and C denotes the correct orientation of the cDNA insert.
Cell Culture.
Mouse hepatoma (Hepa-1), mouse embryonal carcinoma (F9), human hepatoblastoma (Hep-G2), human cervical carcinoma (HeLa), and monkey kidney (COS1) cells were grown in monolayer cultures as described (2, 3).
Supershift Assays.
The nuclear extracts from various cells were prepared according to the procedure of Kadonaga and Tjian (13). The supershift assays with hARE, nuclear extracts from various cell lines and Jun, Fos, and Nrf1 antibodies were performed by procedures as described (2, 3). The supershifted Jun, Fos, and Nrf1 bands were observed in all the cell lines tested. However, their intensity varied depending on the degree of cross-species crossreactivity and abundance.
Cotransfection of Reporter and Expression Plasmids.
Ten micrograms of the reporter plasmids TRE–tk–CAT, hARE–tk–CAT, mutant hARE–tk–CAT, and pNQO1CAT0.837 were cotransfected alone or with various expression plasmids (Jun, Fos, Fra, and Nrf) individually and in several combinations to study the role of these transcription factors in the regulation of hARE-mediated NQO1 gene expression. Five micrograms of the Rous sarcoma virus (RSV)–β-galactosidase (β-gal) plasmid was included in each transfection to normalize the transfection efficiency. The transfected Hep-G2 and COS1 cells were harvested 48 hr after the transfection, homogenized by sonication and analyzed for β-gal and CAT activities (2, 3). In several experiments the transfected cells were treated with various concentrations of β-NF and t-BHQ for 12 hr prior to harvesting and enzymes analysis. The results are presented as mean ± SE of four independent transfection experiments.
In related experiments, the nuclear extracts from nontransfected and transfected cells were analyzed by Western blotting and probing with specific antibodies to determine the levels of expression of various transcription factors. The results indicated increase of 3- to 6-fold in the expression of the predicted respective proteins in transfected cells as compared with the vector-transfected controls as determined by densitometric analysis of Western blots (data not shown). The specific fold inductions were as follows: LNCX–c-Jun, 4.8-fold; LNCX–Jun-D, 4.1-fold; LNCX–Jun-B, 3.9-fold; LNCX–c-Fos, 5.1-fold; LNCX–Fra1R, 1.0-fold; LNCX–Fra1C, 3.7-fold; LNCX–Nrf1R, 1.0-fold; LNCX–Nrf1C, 3.2-fold; LNCX–Nrf2R, 1.0-fold; LNCX–Nrf2C, 4.3-fold; pMT2–Nrf1R, 1.0-fold; and pMT2-Nrf1C, 5.6-fold.
RESULTS
The nucleotide sequences of hARE, mutant hARE, and TRE used in the present report are shown in Fig. 1A. A complex of specific nuclear proteins bind to hARE (Fig. 1B). The supershift assays with hARE and Hepa-1 nuclear extracts indicated presence of Jun, Fos, and Nrf1 in the complex (Fig. 1B).
Figure 1.
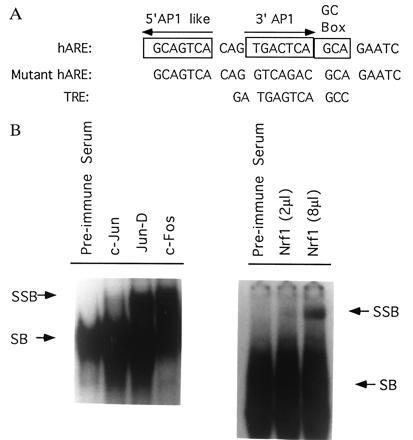
Band and supershift assays. (A) Nucleotide sequences of human NQO1 gene hARE, mutant hARE and human collagenase gene TRE are shown. The mutant hARE contained mutations in the 3′ AP1 element. This mutation is known to significantly reduce the hARE-mediated CAT gene expression and β-NF induction (3). (B) The nuclear extract from Hepa-1 cells was incubated with preimmune serum and antibodies against murine c-Jun, Jun-D, c-Fos, and human Nrf1 and supershift assays performed with the 32P-end labeled hARE. Only shifted (SB) and supershifted (SSB) bands are shown.
To determine the role of Jun, Fos, Fra1, Nrf1, and Nrf2 nuclear factors in the hARE-mediated regulation of NQO1 gene expression, we cotransfected hARE–tk–CAT plasmid with plasmids expressing these proteins individually and in combinations in Hep-G2 cells. The TRE–tk–CAT plasmid was also used in several cotransfection experiments as positive control of Jun and Fos regulation of gene expression. The cotransfection of TRE–tk–CAT with the LNCX vector led to a substantial amount of CAT gene expression in Hep-G2 cells that was more or less similar to the Hep-G2 cells transfected with TRE–tk–CAT alone (control) (Fig. 2A Left). The replacement of LNCX vector with expression plasmids (LNCX–c-Jun plus LNCX–c-Fos) in the transfection experiments resulted in 3.8-fold increase in CAT gene expression as compared with TRE–tk–CAT (control) transfected Hep-G2 cells (Fig. 2A Left). These results were expected as it is known that c-Jun and c-Fos proteins form heterodimers that bind to the TRE and increase the expression of target genes (6). The expression of the TRE-mediated CAT gene in Hep-G2 cells cotransfected with the LNCX vector was presumably mediated by endogenous c-Jun and c-Fos nuclear proteins. The upregulation of TRE-mediated CAT gene in Jun and Fos transfected Hep-G2 cells was due to overexpression of Jun and Fos proteins in Hep-G2 cells.
Figure 2.
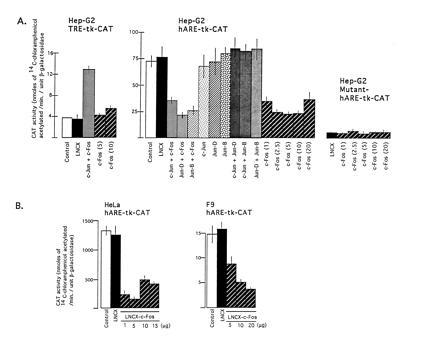
Effect of overexpression of Jun and Fos proteins on TRE-, hARE-, and mutant hARE-mediated CAT gene expression. (A) The Hep-G2 cells were cotransfected with 10 μg of reporter plasmids TRE–tk–CAT (Left), hARE–tk–CAT (Center), and mutant hARE–tk–CAT (Right) with 10 μg of expression plasmids LNCX, LNCX–Jun and LNCX–c-Fos individually and in combinations as shown in separate experiments. The LNCX–c-Fos was also used in different concentrations (μg). (B) HeLa and F9 cells were cotransfected with 10 μg of reporter plasmid hARE–tk–CAT and different concentration of c-Fos expression plasmid LNCX–c-Fos. Five micrograms of RSV–β-gal plasmid was included in each case as internal control of transfection efficiency. Forty-eight hours after transfection, the cells were analyzed for β-gal and CAT activities.
In similar experiments as described above, the hARE-mediated CAT gene expression from reporter plasmid hARE–tk–CAT in vector transfected Hep-G2 cells was 19.7-fold higher than the TRE–tk–CAT gene expression (Fig. 2A Left and Center, compare open boxes). Interestingly, the hARE-mediated CAT gene expression was significantly repressed in Hep-G2 cells overexpressing various combinations of Jun and Fos proteins (Fig. 2A Center, compare LNCX with c-Jun plus c-Fos, Jun-D plus c-Fos, and Jun-B plus c-Fos). This repression was not observed in Hep-G2 cells overexpressing individual and various combinations of Jun proteins alone (Fig. 2A Center). However, a significant amount of repression in hARE-mediated CAT gene expression was observed in Hep-G2 cells overexpressing different concentrations of c-Fos from plasmid LNCX–c-Fos (Fig. 2A Center). As compared with the hARE-mediated CAT gene expression, the mutant hARE–tk–CAT gene expression in the Hep-G2 cells was much lower and remained unaffected in response to overexpression of different concentrations of c-Fos protein (Fig. 2A Right). The c-Fos-mediated repression of hARE-regulated CAT gene expression was also observed in transfected HeLa and F9 cells (Fig. 2B). The replacement of c-Fos expression plasmids with Fra1 expression plasmids in cotransfection experiments in Hep-G2 cells revealed that overexpression of Jun and Fra1 combinations as well as Fra1 alone also repressed the hARE-mediated CAT gene expression (Fig. 3). The repression appeared to be more with Jun plus Fra1C, compared with Fra1C only (Fig. 3). It is noteworthy that treatment of transfected cells with β-NF and t-BHQ did not derepress the c-Fos- and Fra1-mediated repression of hARE regulated CAT gene expression in Hep-G2 cells (data not shown).
Figure 3.
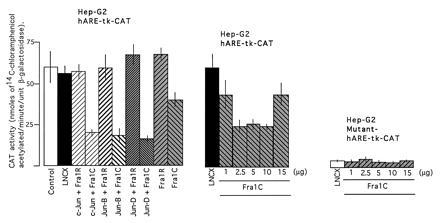
Effect of overexpression of Jun and Fra1 nuclear proteins on hARE-mediated CAT gene expression. The Hep-G2 cells were cotransfected with reporter plasmid hARE–tk–CAT and expression plasmids LNCX–Jun, LNCX–Fra1R, and LNCX–Fra1C individually and in combinations as shown (Left). In the related experiments, the reporter plasmids hARE–tk–CAT (Center) and mutant hARE–tk–CAT (Right) were cotransfected with different concentrations of LNCX–Fra1C plasmid. The LNCX–Fra1C and not LNCX–Fra1R produces Fra1 protein upon transfection in cells. RSV–β-gal plasmid was used in each case as internal control. The transfected cells were analyzed for β-gal and CAT activities.
Interestingly, cotransfection of Hep-G2 cells with reporter plasmid hARE–tk–CAT and expression plasmid LNCX–Nrf1C (Nrf1 in correct orientation) resulted in a several-fold increase in hARE-mediated CAT gene expression (Fig. 4A Left). The increase in hARE-mediated CAT gene expression was dependent on the amount of transfected LNCX–Nrf1C plasmid. The transfection of 10 μg of LNCX–Nrf1C produced a 4.8-fold increase in hARE-mediated CAT gene expression in Hep-G2 cells (Fig. 4A Left). The replacement of LNCX–Nrf1C with plasmid LNCX–Nrf1R (Nrf1 gene in reverse orientation) in cotransfection experiments did not increase the hARE-mediated CAT gene expression (Fig. 4A Left). In addition, cotransfection of mutant hARE–tk–CAT with LNCX–Nrf1C had no effect on mutant hARE regulated expression of CAT gene (Fig. 4A Center). However, as expected, the mutant hARE-mediated CAT gene expression in Hep-G2 cells was significantly lower than the hARE-mediated CAT gene expression (Fig. 4A, compare Center with Left). To determine if the Nrf1 also mediated upregulation of hARE controlled CAT gene expression in a second mammalian cell line, we used the COS1 cell system and pMT2 based Nrf1 expression plasmids (Fig. 4 A Right and B). The pMT2–COS1 system demonstratedtk identical results as LNCX–Hep-G2 cells experiments. The overexpression of Nrf1 from plasmid pMT2–Nrf1C significantly increased hARE-mediated CAT gene expression in concentration dependent manner (Fig. 4 A Right and B). The treatment of Hep-G2 and COS1 cells overexpressing the Nrf1 nuclear protein with β-NF and t-BHQ further increased the hARE-mediated CAT gene expression (Fig. 5 A and B Right). The induction of CAT gene expression by β-NF and t-BHQ was concentration dependent. Interestingly, the Hep-G2 and COS1 cells expressing endogenous (Nrf1R) and overexpressed (Nrf1C) levels of Nrf1 showed similar fold induction of hARE-mediated CAT gene expression upon treatment with β-NF and t-BHQ. The replacement of hARE–tk–CAT with reporter plasmid pNQO1CAT0.837 in experiments as described in Figs. 4 and 5 yielded similar results as with hARE–tk–CAT (data not shown).
Figure 4.
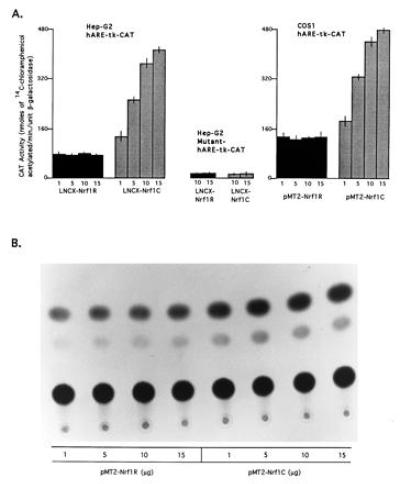
Effect of overexpression of nuclear protein Nrf1 on hARE-mediated CAT gene expression. (A Left and Center) The Hep-G2 cells were cotransfected with 10 μg of reporter plasmid hARE–tk–CAT (Left) and mutant hARE–tk–CAT (Center) and various concentrations (μg) of expression plasmids LNCX–Nrf1R (containing Nrf1 cDNA in reverse orientation) and Nrf1C (containing Nrf1 cDNA in correct orientation) in separate experiments as shown. (Right) The COS1 cells were cotransfected with 10 μg of reporter plasmid hARE–tk–CAT and different concentrations of expression plasmids pMT2–Nrf1R and pMT2–Nrf1C in separate experiments as shown. Five micrograms of RSV–β-gal plasmid were used in each case as control of transfection efficiency. Forty-eight hours after the transfection, the cells were analyzed for β-gal and CAT activities. (B) TLC autoradiogram of A Right.
Figure 5.
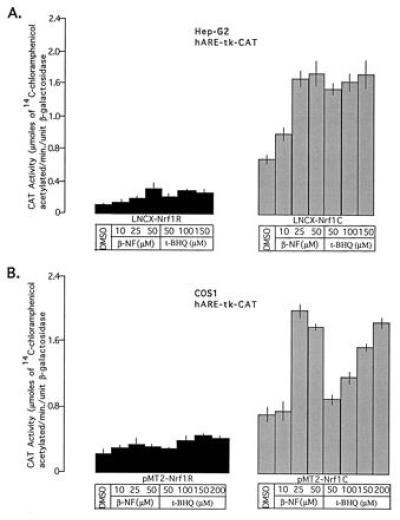
Effect of β-NF and t-BHQ on Nrf1-mediated hARE–tk–CAT expression. The Hep-G2 and COS1 cells were cotransfected with 10 μg of reporter plasmid hARE–tk–CAT and 10 μg of expression plasmids Nrf1R (Left) or Nrf1C (Right). Five micrograms of RSV–β-gal plasmid were used in each case as control for transfection efficiency. Thirty-six hours after transfection, the cells were treated with dimethyl sulfoxide (control) or different concentrations of β-NF and t-BHQ for 12 hr and analyzed for β-gal and CAT activities.
The replacement of Nrf1 with Nrf2 expression plasmids in cotransfection experiments also upregulated hARE-mediated CAT gene expression in Hep-G2 cells that was inducible in response to β-NF and t-BHQ (Fig. 6A Left). Interestingly, cotransfection of Nrf1 and Nrf2 expression plasmids with hARE–tk–CAT in Hep-G2 cells did not significantly increase the hARE-mediated CAT gene expression over the level observed with Nrf1 and Nrf2 alone (Fig. 6B). Various transcription factors that interact with the hARE and their effect on the hARE-mediated expression and induction of CAT gene are summarized in Fig. 6C.
Figure 6.
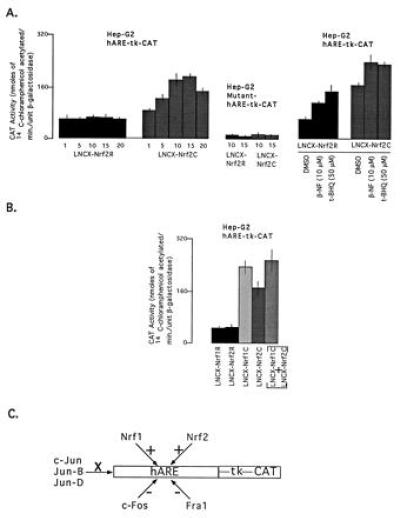
Effect of overexpression of Nrf2 on hARE-mediated CAT gene expression and induction by β-NF and t-BHQ. (A) The Hep-G2 cells were cotransfected with 10 μg of reporter plasmid hARE–tk–CAT (Left) or mutant hARE–tk–CAT (Center) and different concentrations (μg) of expression plasmid LNCX–Nrf2R or LNCX–Nrf2C. (Right) The reporter plasmid was cotransfected with 10 μg of LNCX–Nrf2R or LNCX–Nrf2C. (B) The Hep-G2 cells were cotransfected with 10 μg of hARE–tk–CAT and 10 μg of expression plasmids Nrf1R or Nrf1C or Nrf2R or Nrf2C or Nrf1C plus Nrf2C. In each case, 5 μg of RSV–β-gal was used as transfection standard. In the right panel, the cells were treated with either dimethyl sulfoxide (control), β-NF, or t-BHQ 12 hr prior to harvesting. The transfected cells were analyzed for β-gal and CAT activities. (C) Summary of transcription factors that bind the hARE and/or effect the hARE-mediated CAT gene expression in mammalian cells. +, Positive effect; −, negative effect; and X, no effect.
DISCUSSION
The detoxification of xenobiotics and carcinogens by detoxifying (phase II) enzymes including NQO1 and GST Ya play a major role in protecting the cells against chemically induced oxidative stress, cytotoxicity, mutagenicity, and carcinogenicity (1, 4, 5). There is sufficient evidence that the basal expression and xenobiotic and antioxidant induction of various detoxifying enzyme genes are coordinately regulated by a similar mechanism involving ARE (4, 5). Therefore, the nuclear proteins that bind to the ARE and regulate expression and induction of the detoxifying enzyme genes within the cells are of high significance. The supershift assays with hARE revealed that Jun, Fos, and Nrf1 nuclear proteins bind to the hARE. The binding of Fra1 to the hARE was predicted based on earlier report of binding of Fra1 to the mouse GST Ya subunit gene ARE (14).
The role of Jun, Fos, Fra1, Nrf1, and Nrf2 in the hARE-mediated regulation of NQO1 gene expression was determined in Hep-G2 and COS1 cells by overexpression of individual and combinations of various nuclear proteins. Overexpression of Jun plus Fos and Jun plus Fra1 combinations repressed the hARE-mediated CAT gene expression in Hep-G2 cells. Further experiments suggested that the repression in hARE-mediated CAT gene expression was due to c-Fos and Fra1 and not because of Jun nuclear proteins. The hARE contains a perfect copy of the AP1 element that is a high affinity binding site for Jun and Fos and Jun and Fra1 heterodimers. It is possible that overexpression of Fos and Fra1 results in increased concentration of these heterodimers at the AP1 site of the hARE that interfere with the binding of nuclear proteins that mediate the hARE regulated basal expression and xenobiotic and antioxidant induction of the NQO1 gene. Therefore, c-Fos and Fra1 at higher concentrations may negatively regulate the hARE-mediated NQO1 gene expression. The c-Fos repression of the NQO1 gene expression was not surprising because c-Fos is also known to act as repressor of c-Jun/ATF-2 regulated expression of human urokinase gene (15). The implications of negative regulation of hARE-mediated NQO1 gene expression by overexpression of c-Fos and Fra1 remains unknown.
Interestingly, the proteins that upregulate hARE-mediated transcription were identified as nuclear transcription factors Nrf1 and Nrf2. The overexpression of these proteins independently increased hARE-mediated CAT gene expression by several fold in Hep-G2 and COS1 cells. The upregulation of hARE-mediated CAT gene expression was not observed with Nrf1R and Nrf2R expression plasmids that contained the respective genes in reverse orientation and did not overexpress the cDNA derived Nrf1 and Nrf2 proteins. The role of Nrf1 and Nrf2 was specific to the hARE-mediated regulation because the mutant hARE failed to increase the CAT gene expression in cells overexpressing Nrf1 and Nrf2. The involvement of Nrf1 and Nrf2 in the hARE-mediated expression of NQO1 gene is also supported by two additional observations. Firstly, the Nrf1 and Nrf2 binding site as reported (7, 8) is very similar to the hARE (3). The Nrf1 binding site contained two AP1 elements arranged as inverse repeats at the interval of three bp followed by GC box, an arrangement similar as found in the NQO1 gene hARE. Secondly, the tissue specific expression of Nrf1 and Nrf2 among human tissues were found identical to that of tissue specific expression of human NQO1 gene (7, 8, 10). All the three genes (Nrf1, Nrf2, and NQO1) were found highly expressed in human kidney, skeletal muscle, and lung, moderately expressed in liver, placenta, and heart and least expressed in brain and pancreas. These results suggested that Nrf1 and Nrf2 may also regulate the variations in the tissue specific expression of NQO1 gene among various human tissues. Interestingly, coexpression of Nrf1 and Nrf2 did not significantly increase the hARE-mediated CAT gene expression over the levels of expression observed with individual Nrf1 and Nrf2 proteins. These results may indicate that Nrf1 and Nrf2 do not require heterodimerization for their function as positive regulators of hARE-mediated CAT gene expression and compete each other for binding to the available hARE sites. This conclusion is consistent with the earlier observations that Nrf1 and Nrf2 bind to their binding site independent of each other and upregulate the target genes (7, 8). However, it is possible that others as yet unknown nuclear proteins form heterodimers with Nrf1 and Nrf2 and further increase or decrease the hARE-mediated NQO1 gene expression. The upregulation of hARE-mediated CAT gene expression by Nrf1 and Nrf2 were significantly induced by β-NF and t-BHQ indicating the involvement of these proteins in the xenobiotic and antioxidant induction of NQO1 gene expression.
The finding that Nrf1 and Nrf2 regulate NQO1 gene expression, raised the question of involvement of Nrf1 and Nrf2 in the regulation of other detoxifying enzyme genes. It also raised the question of mechanism of signal transduction from xenobiotics and antioxidants to the Nrf1 and Nrf2 that bind to the hARE and mediate the expression and induction of NQO1 gene. It is expected that ARE-mediated coordinated induction of NQO1, GST and other detoxifying enzymes is mediated by Nrf1 and Nrf2 because ARE elements are highly conserved among various detoxifying enzyme genes (4, 5). The mechanism of signal transduction from xenobiotics and antioxidants to Nrf1 and Nrf2 proteins may be similar to the OxyR- and SoxRS-regulated expression of more than a dozen defense genes in bacteria (16). In this hypothesis, the xenobiotics and antioxidants are metabolically activated by cytochromes P450 and P450 reductase to generate electrophiles and reactive oxygen species (17). Either one or both of these reactive species directly or through intermediary proteins may activate the transcription of Nrf1 and Nrf2 or modify these proteins for increased binding and/or increased transcriptional activity resulting in the activation of NQO1 and other detoxifying enzyme genes expression. Several observations may support the hypothesis involving β-NF and t-BHQ induced modification of Nrf1 and Nrf2 proteins. These include the following. In the present report, the Hep-G2 and COS1 cells expressing endogenous and higher levels of Nrf1 and Nrf2 showed similar fold induction of hARE-mediated CAT gene expression. This is in spite of the fact that Nrf1 and Nrf2 overexpressing cells expressed several-fold higher basal expression of hARE-mediated CAT gene as compared with cells expressing endogenous levels of Nrf1 and Nrf2. The maintenance of similar fold induction of hARE-mediated CAT gene in response to β-NF and t-BHQ between cells expressing endogenous and higher levels of Nrf1 and Nrf2 could have been possible only if these proteins are modified for increased binding to the hARE and/or increased transcriptional activity of Nrf1 and Nrf2 resulting in induced hARE-mediated CAT gene expression. This is because β-NF and t-BHQ have no effect on cytomegalovirus promoter controlled expression of Nrf1 and Nrf2 produced from respective expression plasmids. In addition, the hARE-mediated CAT gene expression in Hepa-1 cells is highly sensitive to sulfhydryl agents (18). Furthermore, the Nrf1 and Nrf2 proteins like Jun and Fos proteins contain a cysteine residue in their DNA binding domain that may be the target of redox regulation by electrophiles and reactive oxygen species generated due to metabolic activation of xenobiotics and antioxidants (8).
In conclusion, we showed that Nrf1 and Nrf2 positively regulate the hARE-mediated basal expression and β-NF- and t-BHQ-mediated induction of the NQO1 gene expression. In addition, we demonstrated that overexpression of c-Fos and Fra1 repressed the hARE-mediated CAT gene expression that may presumably be due to interference in binding of Nrf1 and Nrf2 at the hARE binding site or due to titration of Nrf1 and Nrf2 by Fos and Fra1.
Acknowledgments
We thank Drs. James Sherely, Kevin Ryder, and Pius Joseph for helpful discussion and reading of the manuscript. This work was supported by National Institutes of Health Grant GM 47466.
Footnotes
Abbreviations: NQO1, NAD(P)H:quinone oxidoreductase1; ARE, antioxidant response element; hARE, human ARE; TRE, TPA (12-O-tetradecanoylphorbol-13-acetate) response element; β-NF, β-naphthoflavone; t-BHQ, tert-butyl hydroquinone; GST, glutathione S-transferase; RSV, Rous sarcoma virus; CAT, chloramphenicol acetyltransferase, TK, thymidine kinase; LNCX, eukaryotic expression vector; β-gal, β-galactosidase.
References
- 1.Riley R J, Workman P. Biochem Pharmacol. 1992;43:1657–1669. doi: 10.1016/0006-2952(92)90694-e. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Jaiswal A K. J Biol Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 3.Xie T, Belinsky M, Xu Y, Jaiswal A K. J Biol Chem. 1995;270:6894–6900. doi: 10.1074/jbc.270.12.6894. [DOI] [PubMed] [Google Scholar]
- 4.Jaiswal A K. Biochem Pharmacol. 1994;48:439–444. doi: 10.1016/0006-2952(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 5.Rushmore T H, Pickett C B. J Biol Chem. 1993;268:11475–11478. [PubMed] [Google Scholar]
- 6.Angel P, Karim M. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 7.Chan J Y, Han X, Kan Y W. Proc Natl Acad Sci USA. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moi P, Chan K, Asunis I, Cao A, Kan Y W. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moi P, Kan Y W. Proc Natl Acad Sci USA. 1990;87:9000–9004. doi: 10.1073/pnas.87.22.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal A K. J Biol Chem. 1994;269:14502–14508. [PubMed] [Google Scholar]
- 11.Francis M K, Phinney D G, Ryder K. J Biol Chem. 1995;270:11502–11513. doi: 10.1074/jbc.270.19.11502. [DOI] [PubMed] [Google Scholar]
- 12.Shaw P M, Reiss A, Adesnik M, Nebert D W, Schembri J, Jaiswal A K. Eur J Biochem. 1991;195:171–176. doi: 10.1111/j.1432-1033.1991.tb15691.x. [DOI] [PubMed] [Google Scholar]
- 13.Kadonaga J T, Tjian R. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshioka K, Deng T, Cavigelli M, Karin M. Proc Natl Acad Sci USA. 1995;92:4972–4976. doi: 10.1073/pnas.92.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Cesare D, Vallone D, Caracciolo A, Sassone-Corsi P, Nerlov C, Verde P. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- 16.Demple B. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 17.De Long M J, Santamaria A B, Talalay P. Carcinogenesis. 1987;8:1549–1553. doi: 10.1093/carcin/8.10.1549. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Jaiswal A K. Eur J Biochem. 1994;226:31–39. doi: 10.1111/j.1432-1033.1994.tb20023.x. [DOI] [PubMed] [Google Scholar]


