Abstract
Natural killer T (NKT) cells, coexpressing natural killer (NK) and T-cell receptors (TCR), are associated with immunity to viruses, tumors, and parasites. A well-characterized subclass of these NKT cells expresses biased TCR and recognizes glycolipids such as α-galactoceramide, which is found naturally only in marine sponges and presented by the cell surface glycoprotein CD1d. However, a larger number of T cells present in human blood coexpress the NK marker CD56 and unbiased TCR and do not appear to require CD1 for antigen presentation. Observing high frequencies of CD4 and CD8 coreceptor expression in human CD56+ T cells, we examined the potential role of major histocompatibility complex (MHC) class II molecules in the activation of these cells. Activation of mononuclear cells with bacterial superantigens presented by MHC class II molecules resulted in increased frequency of CD56+ T cells. Primarily, CD4+ cells within the CD56+-T-cell population responded to the bacterial superantigens, and cytokine expression profiles were Th1-like. Further, increased levels of T cells expressing CD56 were observed in mononuclear cell cultures responding to a Staphylococcus aureus vaccine or tetanus toxoid. Collectively, our data suggest that a significant number of CD56+ T cells recognize pathogen-associated ligands in association with MHC class II molecules.
Human natural killer (NK) inhibitory or activating receptors present on T-cell subsets include the lectin-like glycoprotein complexes of CD94 with NKG2A or NKG2C, which recognize HLA-E, and immunoglobulin-like receptors, which bind to classical major histocompatibility complex (MHC) class I molecules (6, 7, 19). The CD56 molecule (neural cell adhesion molecule) is a membrane glycoprotein belonging to the immunoglobulin superfamily expressed on neural and NK cells that also defines a major subclass of NKT cells. A small percentage (0.9 to 1.2%) of peripheral blood mononuclear cells (PBMC) coexpress NK cell markers, such as CD56, and a restricted T-cell receptor (TCR) repertoire consisting of an invariant Vα24 rearrangement with JαQ paired with Vβ11 in humans (12, 18, 23) or Vα14 with mixed Vβs that represent <2% of T lymphocytes in mice (5, 10, 21). These invariant TCR recognize ceramide-containing glycolipids that are presented by the cell surface glycoprotein CD1d. However, most NKT cells exhibit nonbiased TCR usage (11; R. Ulrich, T. Kissner, B. Dyas, and K. U. Saikh, Abstr. 11th Int. Congr. Immunol., abstr. AG. Wed. 4.8/782, 2001) and do not appear to require CD1 for activation (11, 16, 26).
A significant portion of CD8+ T cells in mice acquire NK cell-associated molecules during viral infections (16, 26), suggesting that these receptors may regulate MHC class I-dependent T-cell effector functions. Furthermore, many virus-specific CD8+ T cells express NK cell receptors in influenza A virus-infected CD1d−/− mice, implicating the role of classical MHC class I molecules and a variable TCR repertoire (2, 16). Evidence for the involvement of MHC class II recognition by NKT cells is less clear. Host resistance by CD4+ NKT cells against intracellular bacteria was reported in mice (14). In addition, a recent report suggests that NKT cells develop as a result of high avidity TCR-MHC class II interactions and that common signaling pathways are involved in positive selection of CD1-independent and conventional T cells (8). A small but significant number of human peripheral blood T cells express the NK marker CD56 and unbiased TCR (25, 29). Although CD56+ T cells appear to be associated with infections (33), their function is unknown. To investigate their role in cellular immunity, we examined recognition of MHC class II ligands by human CD56+ T cells from peripheral blood and their response to polypeptide vaccines.
MATERIALS AND METHODS
Reagents.
Human AB serum was obtained from Pel-Freez (Brown Deer, Wis.). Anti-MHC class II-coated microbeads and goat anti-mouse immunoglobulin (Ig)-coated M-450 Dynalbeads were purchased from Dynal (Hamburg, Germany). The fluorescein isothiocyanate (FITC)-conjugated Leu HLA-DR (anti-DR) and Leu-4 (anti-CD3) monoclonal antibodies (MAbs) and Ig isotype control antibodies were purchased from Becton Dickinson (San Jose, Calif.). Human anti-CD56 MAbs conjugated to phycoerythrin (PE) were purchased from Pharmingen, San Diego, Calif. The FITC-conjugated anti-CD4 and anti-CD8 MAbs and Ig isotype control antibodies were purchased from Becton Dickinson. Superantigens were purchased from Toxin Technology (Sarasota, Fla.). The [methyl-3H]thymidine was purchased from Amersham Life Sciences (Arlington Heights, Ill.). The experimental Staphylococcus aureus vaccine STEBvax (inactivated staphylococcal enterotoxin B [SEB]) was described previously (31). The purified recombinant C fragment of tetanus toxin was purchased from Boehringer Mannheim (Indianapolis, Ind.). The peptide (Tet peptide, LYNGLKFIIKRY) from the C fragment of tetanus toxin is described elsewhere (24). The rabbit anti-DR antibody used in this study was described elsewhere (4).
Cell isolation.
PBMC were obtained from healthy volunteers. Fifty milliliters of blood was obtained from each donor and was provided with fully informed consent. Mononuclear cells (MNC) were separated by Ficoll-Hypaque density gradient centrifugation and purified by a series of physical separation steps (plastic adherence and discontinuous Percoll gradient centrifugation) to obtain a lower density of cells. The isolation of CD56+ T cells was previously described (25). In brief, activated cells were eliminated from top fractions (low density) by paramagnetic beads that were coated with antibodies specific for HLA-DR (Dynal, Inc.). HLA-DR− cells were labeled with paramagnetic beads coated with anti-CD56 antibody (Miltenyi Biotech, Inc., Auburn, Calif.) and passed through a magnetic column. CD56+ cells were eluted from the column, and beads were released from the cells by enzymatic digestion. Cells were washed and incubated with paramagnetic beads coated with anti-CD3 antibody (Miltenyi Biotech, Inc.) and passed through the magnetic column, and CD3+ cells were then eluted from the column. The purity of CD56+-T-cell preparations used in all experiments was 95% or above.
Flow cytometry.
To characterize the cell surface markers of CD56+ T cells, cells were incubated (20 min at 4°C) with human IgG FcR-blocking reagent (Miltenyi Biotech, Inc.), washed twice with medium, and then incubated (30 min at 4°C) with FITC- and PE-labeled control or isotype-matched MAb. Unbound antibody was removed by washing the cells with medium (4°C). After two more washes, cells were fixed with 1% paraformaldehyde, and the cell-associated immunofluorescence was measured by flow cytometry (FACSort; Becton Dickinson).
Proliferation assay.
The CD56+-T-cell response to different superantigens was detected in a proliferation assay by measuring the incorporation of [3H]thymidine. CD56+ T cell (2 × 105 cells/well) were cultured (37°C, 5% CO2) with various concentrations (0.001 to 1 μM) of different superantigens and gamma-irradiated (1,500 rads) autologous PBMC (2 × 105 cells/well). The response to superantigens, peptides, or vaccines was measured in triplicate by incubating CD56+ T cells alone, equal numbers of CD56+ T cells and irradiated MNC, and MNC alone. After 60 h (superantigens) or 6 days (antigens) of culture, cells were pulsed with [3H]thymidine (1 μCi/well) for an additional 10 h. Cellular proliferation was detected by measuring incorporated radioactivity in a liquid scintillation counter (Packard, Meriden, Conn.). To examine the role of HLA-DR in superantigen presentation to CD56+ T cells, 2 × 105 cells/well were cultured (37°C, 5% CO2) with gamma-irradiated (1,500 rads) autologous PBMC as antigen-presenting cells (APC) (2 × 105 cells/well) in the presence of staphylococcal enterotoxin A (SEA) (0.36 nM) and various amounts of rabbit anti-HLA-DR or control preimmune antibody. After 60 h of culture, cells were pulsed with [3H]thymidine (1 μCi/well) for an additional 10 h. Cells were harvested, and proliferation was assessed by measuring the incorporation of radioactivity in a liquid scintillation counter (Packard).
Allo-response assay.
CD56+ T cells (2 × 105 cells) were cultured with gamma-irradiated PBMC (2 × 104 to 20 × 104) from HLA-DR-mismatched donors for 6 days (37°C, 5% CO2) and harvested after pulsing for 10 h with [3H]thymidine (1 μCi/well). Proliferation was assessed by measuring the incorporation of radioactivity in a liquid scintillation counter (Packard).
Cytokine assays.
Cytokine production was measured by an enzyme-linked immunosorbent assay (Predicta/Genzyme Corporation, Cambridge, Mass.) according to the manufacturer's instructions. Briefly, 100 μl of culture supernatant or control (standard) was added to 96-well microtiter plates precoated with MAb to each cytokine and incubated for 30 min (37°C). The wells were washed, and 100 μl of biotin-labeled rabbit anti-gamma interferon (IFN-γ), -IFN-α, -tumor necrosis factor alpha (TNF-α), or -interleukin 4 (IL-4) was added to detect bound cytokine. After a brief incubation (30 min at 37°C), the wells were washed and incubated (20 min at 37°C) with peroxidase-labeled streptavidin (100 μl). Unbound streptavidin was washed from the plates, substrate was added, and the absorbance of each well was measured at 450 nm. A standard curve was used to estimate the cytokine concentration.
Measurement of transcriptional activation of cytokines by reverse transcription-PCR.
Total cellular RNA was extracted from 1 × 106 to 2 × 106 cells by using TRI REAGENT (Molecular Research Center, Inc., Cincinnati, Ohio) according to the manufacturer's instructions. The concentration of purified RNA was estimated by measuring the absorbance at 260 nm. Single-stranded cDNA was synthesized from total RNA by using the Gene Amp RNA PCR kit (Applied Biosystems, Foster City, Calif.). Briefly, a 20-μl reaction mixture contained 200 ng of RNA template, 2.5 μM oligo(dT) primer, 5 mM MgCl2, 1× final concentration of 10× PCR buffer II, 1 mM deoxynucleoside triphosphates, 1 U of RNase inhibitor/μl, and 2.5 U of murine leukemia virus reverse transcriptase/μl. The cDNA was synthesized in a Stratagene Robocycler (La Jolla, Calif.) with the following conditions: 1 cycle of 42°C for 15 min, 99°C for 5 min, and 4°C for 5 min. Relative levels of specific gene transcripts were measured by PCR. The assay mixture consisted of 3 μl of cDNA in a final volume of 50 μl that contained 800 mM Tris-HCl (pH 8.9), 200 mM (NH4)2SO4, 50 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (Promega, Madison, Wis.), 0.2 μM concentrations of each primer, and 1.5 U of Platinum DNA Taq polymerase (Life Technologies, Gaithersburg, Md.). The primers used for PCR were as follows: TNF-α forward (5′-CGGGACGTGGAGCTGGCCGAGGAG-3′) and reverse (5′-CACCAGCTGGTTATCTCTCAGCTC-3′), amplifying a 354-bp product; IFN-α forward (5′-CAGGAGGAGTTTGATGGCAACCAG-3′) and reverse (5′-GACAACCTCCCA GGCACAAGGGC-3′), amplifying a 315-bp product; and IFN-γ forward (5′-AAAAGCTTATGAAATATACAAGTTATATCT-3′) and reverse (5′-GGGGATCCTTACTGGGATGCTCTTCGACCAA-3′), amplifying a 517-bp product. The PCR was done in a Stratagene Robocycler with an initial denaturation step of 94°C for 5 min followed by 35 cycles consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final elongation step of 72°C for 7 min. PCR products were separated on a 2% agarose gel containing ethidium bromide.
RESULTS
Analysis of CD56+ T cells in human peripheral blood.
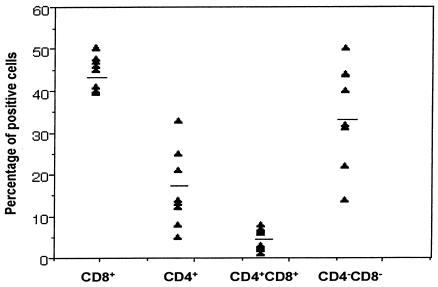
Flow cytometry distinguishes CD3+ CD56+ T cells from other lymphoid cells such as NK cells (CD56+) or T cells (CD56− CD3+). The frequency of these CD56+ T cells varied from 3 to 14% in healthy donors (n = 12), with an average of 6.5% ± 1.1% (standard error of the mean). The frequencies of coreceptor expression (Fig. 1) were as follows: CD8+, 44%; CD4−CD8−, 33%; CD4+, 20%; CD4+ CD8+, 4%. Almost equal numbers of CD45RA+ (naive) and CD45RO+ (memory) cells were found among these CD56+ T cells. In addition, 10 to 15% of the CD56+ T cells also expressed CD28 (data not shown).
FIG. 1.
Surface coreceptor expression of CD4 or CD8 on CD3+CD56+ T cells. For detecting cell surface expression of CD4 and CD8, purified CD56+ T cells were labeled with FITC-conjugated anti-CD4 or anti-CD8 and PE-conjugated anti-CD56 or isotype-matched control antibodies and analyzed by flow cytometry. Results are presented as frequencies of coreceptor expression (mean value), with CD8+ > CD4−CD8− > CD4+ > CD4+CD8+.
MHC class II-dependent activation of CD56+ T cells by bacterial superantigens.
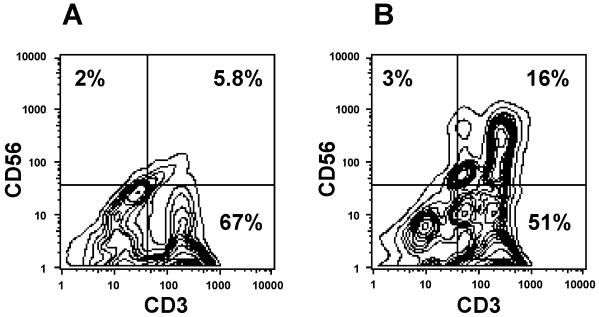
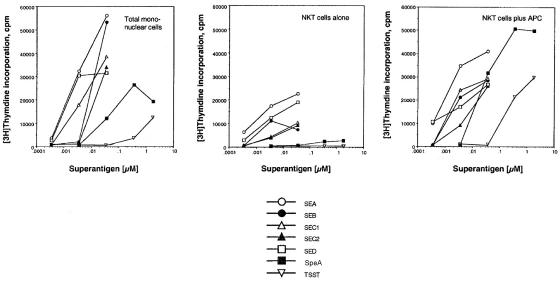
Our observations indicated that, in humans, numerous CD56+ T cells express the coreceptor CD4 (Fig. 1), suggesting an association with MHC class II-mediated activation. It is also known that T-cell activation by bacterial superantigens requires MHC class II molecules for presentation. To first examine functional activation of CD56+ T cells, we depleted MNC of CD56+ cells and then cultured them with SEB. The results shown in Fig. 2 indicate that SEB treatment induced an increase in the number of CD56+ T cells compared to cultures without SEB. Significant levels of CD56+-T-cell proliferation were observed in response to all bacterial superantigens tested (Fig. 3). In addition, because each superantigen activates a different cluster of T cells by coligating a distinct TCR Vβ subset, these data indicate that the responding CD56+ T cells expressed functional and nonbiased TCR. Most CD56+-T-cell responses were dependent on APC (Fig. 3). In the absence of APC, CD56+ T cells proliferated to all superantigens, except SpeA and toxic shock syndrome toxin 1, but at reduced levels, likely due to presentation by autologous HLA-DR expression. Our results suggested that activation was induced through HLA-DR binding (Fig. 3). Activation of CD56+ T cells by superantigens stimulated the secretion of Th1 but not Th2 cytokines (data not shown), consistent with responses obtained from conventional T cells (27).
FIG. 2.
Bacterial superantigen (SEB)-induced expansion of CD56+-T-cell subset. CD56-depleted MNC were cultured for 48 h (37°C, 5% CO2) alone (A) or with 0.1 μM SEB (B) and harvested, and expression of CD3 and CD56 antigens were measured by using FITC-conjugated anti-CD3 and PE-conjugated anti-CD56 antibodies, respectively. Cells were analyzed by flow cytometry. Results from one representative experiment of three are shown.
FIG. 3.
Proliferative response of CD56+ T cells to bacterial superantigens. CD56+ T cells (2 × 105 cells/well) were cultured (37°C, 5% CO2) with various concentrations (0.001 to 1 μM) of superantigens and with or without gamma-irradiated (1,500 rads) autologous PBMC (2 × 105 cells/well). After 60 h of culture, cells were then pulsed with [3H]thymidine (1 μCi/well) for an additional 10 h. Cellular proliferation was assessed by incorporation of radioactivity into DNA. Results are from a representative experiment (total of three experiments).
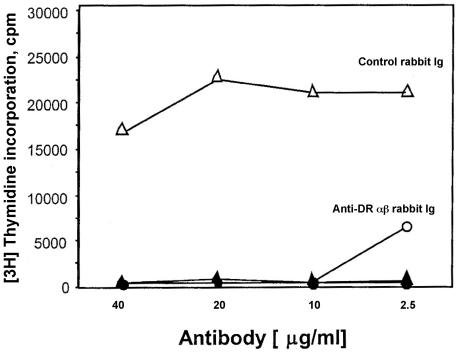
We next examined whether CD56+-T-cell recognition of superantigens was similar to that of conventional T cells by exhibiting dependence on presentation by MHC class II molecules. Our results showed that, compared to nonimmune sera (control rabbit antibody), the CD56+-T-cell response to SEA was completely inhibited by 10 μg of rabbit anti-HLA-DR antibody/ml, confirming that anti-HLA-DR antisera blocked CD56+-T-cell responses (Fig. 4). Collectively, these results showed that CD56+ T cells expressing heterogeneous TCR respond to bacterial superantigens presented by HLA-DR.
FIG. 4.
HLA-DR-dependent recognition of bacterial superantigens by CD56+ T cells. CD56+ T cells (2 × 105 cells/well) were cultured (37°C, 5% CO2) with gamma-irradiated (1,500 rads) autologous PBMC (2 × 105 cells/well) in the presence of SEA (0.36 nM) and various amounts of rabbit anti-HLA-DR or control antibody. After 60 h of culture, cells were pulsed with [3H]thymidine (1 μCi/well) for 10 h. Cells were harvested, and cellular proliferation was assessed by measuring incorporated radioactivity by liquid scintillation. Open symbols (circles and triangles) represent CD56+ T cells, autologous PBMC, and antibody; closed symbols (circles and triangles) represent CD56+ T cells and antibody without APC. The data are presented as mean counts per minute ± standard errors of the means of triplicate wells. Results presented in the figure were representative of three experiments.
Increase in CD56+-T-cell frequency in response to vaccines.
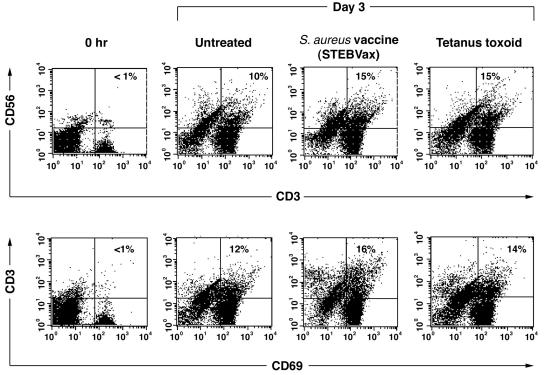
We next examined the CD56+-T-cell response to antigens that are presented by classical MHC molecules. MNC were obtained from a donor with previous immunity to tetanus toxoid and an experimental S. aureus vaccine (STEBVax). MNC were depleted of cells expressing cell surface CD56 and cultured in the presence of the recall antigens. The results shown in Fig. 5 indicated that NKT cell numbers increased upon exposure to the S. aureus vaccine or tetanus toxoid compared to those of untreated cells. These CD56+ T cells also expressed the activation marker CD69 (Fig. 5). We concluded from these results that a significant number of CD56+ T cells appeared from CD56− T cells as a consequence of activation by recall antigens. It is of interest that a small number of T cells spontaneously expressed NK markers in culture without antigen added. A rebound of this minor population of CD56+ T cells occurred despite repeated rounds of CD56 depletion (data not shown). This observation suggested that some CD56− T cells were committed to becoming CD56+ T cells. Indeed, intracellular staining of freshly isolated CD56+ T cells clearly demonstrated intracellular storage of CD56 (data not shown). There was no colocalization of CD3 with CD56 either within the cell or at the cell surface.
FIG. 5.
Increase in CD56+-T-cell frequency upon exposure to S. aureus vaccine (STEBvax) or tetanus toxoid. MNC from a tetanus toxoid- and S. aureus-immune donor were depleted of CD56+ cells and cultured (37°C, 5% CO2) with S. aureus vaccine (STEBVax) or tetanus toxoid (50 μg/ml). After 3 days of culture, cells were analyzed by flow cytometry. The dot plot representation shows the increase in the population of CD3+CD56+ and CD3+CD69+ T cells. The proportions of CD3+CD56+ cells were compared between the untreated group and the STEBVax and tetanus toxoid groups by chi-square test. Results showed that there was a significant difference in the proportion of positive cells between the STEBVax group and the untreated group [χ2(1) = 181.49; P < 0.0001]. Results also showed that there was a significant difference in the proportion of positive cells between the tetanus toxoid group and the untreated group [χ2(1) = 185.69; P < 0.0001]. The proportions of CD3+CD69+ cells were compared between the untreated group and the STEBVax and tetanus toxoid groups by chi-square test. Results also showed that there was a significant difference in the proportion of activated cells with recall antigens, i.e., between the STEBVax group and the untreated group [χ2(1) = 196.46; P < 0.0001] and between the tetanus toxoid group and the untreated group [χ2(1) = 69.43; P < 0.0001]. Results shown are from one representative experiment of three performed.
CD4+CD56+ T cells are the predominant cells responding to SEB.
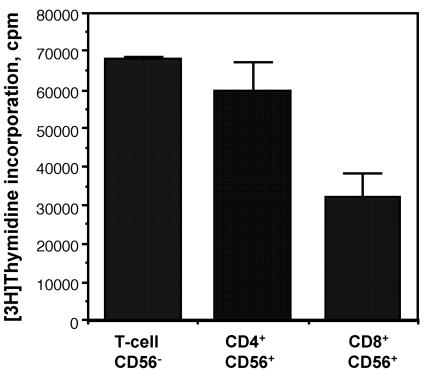
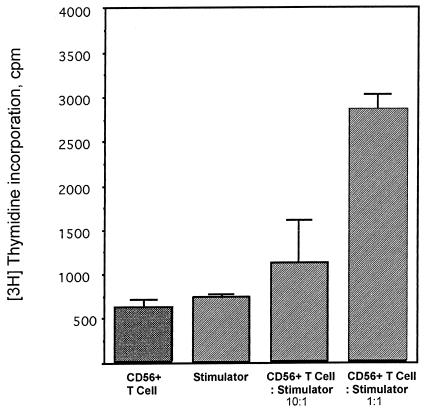
Because the frequencies of coreceptor expression suggested that up to 20% of CD56+ T cells expressed CD4 (Fig. 1), a molecule functioning as a ligand for MHC class II, we examined the responding CD56+-T-cell subsets in more detail. The CD56+ T cells were separated into CD8+ and CD4+ populations and cultured with the superantigen SEB. The predominant cell types responding to SEB were CD4+ (Fig. 6). These results indicate that a portion of CD4+ T cells coexpressing NK markers recognized ligands presented by HLA-DR molecules. Because CD56+ T cells responded to bacterial superantigens in association with HLA-DR, we next examined whether these cells would respond to HLA-DR as an alloantigen. Results from cocultures indicated that CD56+ T cells proliferated in a dose-dependent manner in response to irradiated MNC from HLA-disparate donors (Fig. 7). These results suggested that CD56+ T cells recognized processed antigen presented by HLA-DR and that this alloresponse was at least partially dependent on CD4+ cells.
FIG. 6.
CD4+ cells are the predominant SEB-responding cells in CD56+ T cells. CD56− T (mixed CD4+ and CD8+) cells and CD4+ or CD8+ CD56+ T cells (2 × 105) were cultured (37°C, 5% CO2) with SEB (0.36 nM) and gamma-irradiated (1,500 rads) autologous PBMC (2 × 105 cells/well). After 60 h of culture, cells were pulsed with [3H]thymidine (1 μCi/well) for 10 h. Cells were harvested after a total of 70 h of culture, and cellular proliferation was assessed by measuring incorporated radioactivity. The data represent mean counts per minute ± standard errors of the means of triplicate wells. Results presented in the figure were representative of three experiments.
FIG. 7.
Contribution of HLA-DR and CD4 to the alloresponse of CD56+ T cells. CD56+ T cells (2 × 105) were cultured with gamma-irradiated PBMC (2 × 104 and 2 × 105) from an HLA-DR-mismatched donor for 6 days (37°C, 5% CO2) and pulsed for 10 h with [3H]thymidine (1 μCi/well). Proliferation was assessed by incorporation of radioactivity. Results are from a representative experiment (total of three experiments).
CD4+CD56+ T cells recognized processed tetanus toxoid presented by HLA-DR.
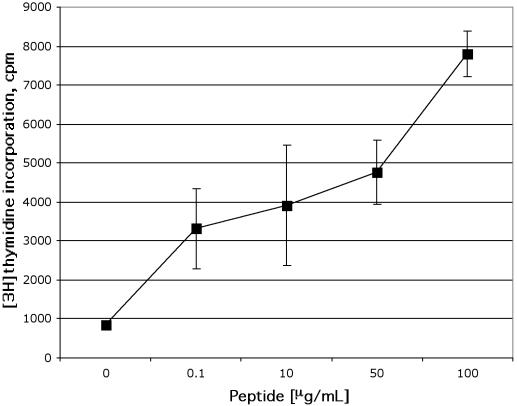
We next examined MHC class II-dependent recognition of processed exogenous antigens by CD56+ T cells. To confirm that direct recognition of ligands presented by DR molecules was a result of CD4+CD56+ T cells and not copurified cell types, we isolated clones from human PBMC activated with the recombinant C fragment of tetanus toxin. These clones were maintained and propagated by restimulation with a synthetic tetanus toxin peptide presented by irradiated autologous MNC and IL-2 supplements. The results presented in Fig. 8 from clone G5 with the CD4+ CD56+ CD8− CD81+ CD28low phenotype demonstrate that this T-cell clone was activated in a dose-dependent manner upon recognition of the HLA-DR binding peptide derived from tetanus toxin. These results indicate that CD4+ T cells can stably express CD56 and retain peptide specificity.
FIG. 8.
CD4+CD56+ NKT cells activated by tetanus toxoid-derived peptide. The CD4+CD56+ NKT cell clone G5 was generated from purified CD3+CD56+ NKT cells of human PBMC stimulated with tetanus toxin ex vivo. CD4+CD56+ NKT cells were activated by peptide derived from tetanus toxoid. The G5 clone was cultured with autologous irradiated PBMC (2 × 105) and peptide (LYNGLKFIIKRY) derived from the C fragment of tetanus toxin for 6 days. Cells were pulsed for an additional 10 h with [3H]thymidine (1 μCi/well), and cellular proliferation was measured by incorporation of radioactivity. The T-cell proliferation of the control culture (T cells + APC without peptide) was <1,000 cpm. The data are presented as mean counts per minute ± standard errors of the means of triplicate wells. Results shown in the figure were from one representative experiment of three.
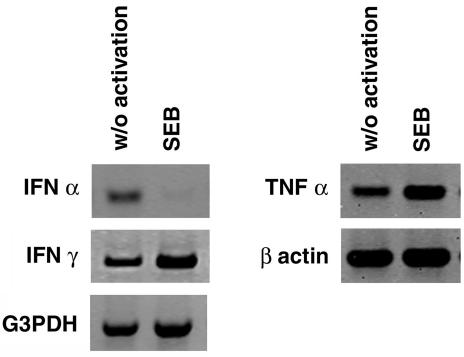
CD56+ T cells directly activated with SEB produced IFN-γ and TNF-α but not IFN-α.
Our earlier results indicated that CD56+-T-cell subsets were activated by superantigens in the presence of APC and suggested that superantigen binding to HLA-DR was necessary for optimal CD56+-T-cell activation. To examine further the requirement for HLA-DR-dependent activation by an accessory cell, CD56+ T cells were treated directly with SEB with or without APC. The results shown in Fig. 9 and Table 1 suggest that direct SEB treatment induced TNF-α and IFN-γ production. Significantly, autopresentation of SEB resulted in complete down-regulation of IFN-α expression (Fig. 9). Although IFN-α expression also occurred in cocultures of CD56+ T cells and APC, it was not possible to distinguish the cellular source of expression (data not shown). The detection of Th1 cytokines in activated culture agrees with previous observations that CD56+ T cells responded suboptimally to superantigens by autopresentation.
FIG. 9.
Cytokine production by CD56+ T cells activated directly with SEB. CD56+ NKT cells were cultured with (100 ng/ml) or without (w/o) SEB in the absence of APC. After 16 h of culture, supernatants were collected for cytokine analysis (Table 1) and cells were used to make RNA for reverse transcription-PCR analysis for measuring transcriptional activation of cytokine genes. The data shown are from a representative experiment (total of three experiments). G3PDH, glyceraldehyde-3-phosphate dehydrogenase.
TABLE 1.
Cytokine production by CD56+ T cells activated with SEB
| Treatment | Amt of cytokine detected (pg/ml)a
|
||
|---|---|---|---|
| IFN-α | TNF-α | IFN-γ | |
| Expt 1 | |||
| None | 31.2 | 81 | 40 |
| SEB | 31.2 | 668 | 658 |
| Expt 2 | |||
| None | 31.2 | 30 | 21 |
| SEB | 31.2 | 1,000 | 1,000 |
Culture supernatants were collected 16 h after treatment with (100 ng/ml) or without SEB. Cytokines were measured by enzyme-linked immunosorbent assay, and the results were from equal numbers of cells.
DISCUSSION
Subsets of T cells expressing NK markers and biased TCR-recognizing antigens, usually restricted by CD1 proteins, appear to fill specialty roles in regulating immune responses. However, the physiological significance of the more abundant T cells expressing NK markers and a normal distribution of TCR is less certain. Though representing a relatively minor cell population in peripheral blood, CD56 expression is characteristic for most T cells of the liver (13). Recent reports, primarily from mouse studies, indicated an association of NK+ T cells with viral infections. As demonstrated by data obtained with antigenic peptides and a panel of bacterial superantigens, our results show that responses of a CD4+ subset of CD56+ T cells are attributable to recognition of MHC class II ligands. A recent report also demonstrated that both CD56+ and CD57+ T cells are activated by the superantigen SEA through an IL-12-dependent pathway (1). These data, combined with results from previous studies showing that CD56+ T cells lysed both HLA class I- and II-bearing target cells (22, 32), suggest that common T-cell functions can be ascribed to these MNC subsets. Yet some CD56+ T cells possess additional features clearly not found in prototypical NK or T cells, suggesting a phenotype intermediate between these two cell lineages. Expression of the NK receptor KIR is initiated after TCR rearrangement (30). However, it is unknown whether the same expression hierarchy holds for CD56. A small, but significant, number of CD56+ T cells become dendritic cells in response to incubation with granulocyte-macrophage colony-stimulating factor (25). Receptors for granulocyte-macrophage colony-stimulating factor were reported to be absent following commitment to the lymphocyte cell lineage from precursor cells in the mouse (17), although equivalent human data are not available. In addition, CD56+ T cells spontaneously lyse U937 but not K562 tumor cell targets, whereas true NK cells have the opposite pattern of cytolysis (25). While acquisition of certain features, such as cytolysis of non-NK target cells, accompanies expression by T cells, a direct functional role for CD56 expression remains obscure. As one distinctive feature, cytokine responses differ from those of resting CD56− T cells. Treatment with IL-15 produced a 10-fold expansion of CD56+ T cells from peripheral blood and increased cytolytic activity towards leukemic cell targets (9). Increased IL-15 expression also correlated with a reduced rate of human liver allograft rejection (9). In contrast, Lyngstrand et al. observed that IL-2 expansion of peptide-specific T-cell lines resulted in an increased number of cells expressing CD56, whereas IL-15 treatment reduced the yield of CD56+ T cells (20).
Our observation that CD56+ T cells recognize MHC class II-restricted antigens suggests an association with adaptive immunity to infectious diseases. A selective expansion of cytotoxic T lymphocytes with a CD4+ CD56+ surface phenotype was previously noted following infection with hepatitis B virus (3). It was also reported that CD56+ T cells were increased in the blood of patients infected with the malarial parasites Plasmodium falciparum or Plasmodium vivax during the early phase of the infection (33) or with other microbial infections (15). A more recent report indicated infiltration of CD4+CD56+ T cells into the nasopharynx following herpes simplex virus infection, which the authors postulated to be a florid antiviral immune response (28). Additional studies will be necessary to determine the physiological role of CD56 expression during T-lymphocyte development and immune responses. In the clinic, it may be useful to monitor fluctuations in the numbers of these T-cell subsets found in peripheral blood following vaccinations or infections.
Acknowledgments
We thank Sarah L. Norris for assistance with statistical analyses and the excellent facilities of the Department of Blood Transfusion, National Institutes of Health, for the processing of MNC.
REFERENCES
- 1.Ami, K., T. Ohkawa, Y. Koike, K. Sato, Y. Habu, T. Iwai, S. Seki, and H. Hiraide. 2002. Activation of human T cells with NK cell markers by staphylococcal enterotoxin A via IL-12 but not with IL-18. Clin. Exp. Immunol. 128:453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assarson, A., T. Kambayashi, J. K. Sandberg, S. Hong, M. Taniguchi, L. V. Kaer, H.-G. Ljunggren, and B. J. Chambers. 2000. CD8+ T cells rapidly acquire NK1.1 and NK cell-associated molecules upon stimulation in vitro and in vivo. J. Immunol. 165:3673-3679. [DOI] [PubMed] [Google Scholar]
- 3.Barnaba, V., A. Franco, M. Proli, R. Benvenuto, G. D. Petrillo, V. L. Burgio, I. Santillo, C. Balsano, M. S. Bonavita, G. Cappelli, V. Colizzi, G. Cutrona, and M. Ferrarini. 1994. Selective expansion of cytotoxic T lymphocytes with a CD4+CD56+ surface phenotype and a T helper type 1 profile of cytokine secretion in the liver of patients chronically infected with hepatitis B virus. J. Immunol. 152:3074-3087. [PubMed] [Google Scholar]
- 4.Bavari, S., and R. G. Ulrich. 1995. Staphylococcal enterotoxin A and toxic shock syndrome toxin compete with CD4 for human major histocompatibility complex class II binding. Infect. Immun. 6:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendelac, A. 1995. Mouse NK1+ T cells. Curr. Opin. Immunol. 7:367-374. [DOI] [PubMed] [Google Scholar]
- 6.Borrego, F., M. Ulbrecht, E. H. Weiss, J. E. Coligan, and A. G. Brooks. 1998. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J. Exp. Med. 187:813-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braud, V. M., S. Allan, C. A. O'Callaghan, K. Soderstrom, A. D'Andrea, G. S. Ogg, S. Lazetic, N. T. Young, J. I. Bell, J. H. Phillps, L. L. Lanier, and A. J. McMicheal. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2 A, B and C. Nature 391:795-799. [DOI] [PubMed] [Google Scholar]
- 8.Capone, M., M. Troesch, G. Eberl, B. Hausmann, E. Palmer, and H. R. MacDonald. 2001. A critical role for the T cell receptor α-chain connecting peptide domain in positive selection of CD1-independent NKT cells. Eur. J. Immunol. 31:1867-1875. [DOI] [PubMed] [Google Scholar]
- 9.Cookson, S., D. G. Doherty, S. Todryk, P. Gibbs, B. Portmann, J. O'Grady, M. Rela, N. Heaton, and S. Norris. 2003. Hepatic expression of IL-15 mRNA is associated with liver graft acceptance. Transpl. Immunol. 11:39-48. [DOI] [PubMed] [Google Scholar]
- 10.Cui, J., T. Shin, T. Kuwano, H. Sato, E. Kanno, and M. Taniguchi. 1997. Requirement for Vα14 NKT cells in IL-12-mediated rejection of tumors. Science 278:1623-1626. [DOI] [PubMed] [Google Scholar]
- 11.Dang, Y., and K. D. Heyborne. 2001. Regulation of uterine NKT cells by a fetal class I molecule other than CD1. J. Immunol. 166:3641-3644. [DOI] [PubMed] [Google Scholar]
- 12.Dellabona, P., E. Padovan, G. Casorati, M. Brockhaus, and A. Lanzavecchia. 1994. An invariant V alpha24-J alpha Q/V beta11 T cell receptor is expressed in all individuals by clonally expanded CD4−8− T cells. J. Exp. Med. 180:1171-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doherty, D. G., S. Norris, L. Madrigal-Estebas, G. McEntee, O. Traynor, J. E. Hegarty, and C. O'Farrelly. 1999. The human liver contains multiple populations of NK cells, T cells, and CD3+CD56+ natural T cells with distinct cytotoxic activities and Th1, Th2, and Th0 cytokine secretion patterns. J. Immunol. 163:2314-2321. [PubMed] [Google Scholar]
- 14.Emoto, M., Y. Emoto, I. B. Buchwalow, and S. Kaufmann. 1999. Induction of IFN-γ producing CD4+ natural killer T cells by Mycobacterium bovis bacillus Calmette Guerin. Eur. J. Immunol. 29:650-659. [DOI] [PubMed] [Google Scholar]
- 15.Jason, J., I. Buchanan, L. K. Archibald, O. C. Nwanyanwu, M. Bell, T. A. Green, A. Eick, A. Han, D. Razsi, P. N. Kazembe, H. Dobbie, M. Midathada, and W. R. Jarvis. 2000. Natural T, γδ, and NK cells in mycobacteial, Salmonella, and human immunodeficiency virus infections. J. Infect. Dis. 182:474-481. [DOI] [PubMed] [Google Scholar]
- 16.Kambayashi, T., E. Assarsson, J. Michaelsson, P. Berglund, A. D. Diehl, B. J. Chambers, and H.-G. Ljunggren. 2000. Emergence of CD8+ T cells expressing NK cell receptors in influenza A virus-infected mice. J. Immunol. 165:4964-4969. [DOI] [PubMed] [Google Scholar]
- 17.Kondo, M., D. C. Scherer, T. Miyamoto, A. G. King, K. Akashi, K. Sugamura, and I. L. Weissman. 2000. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature 407:383-386. [DOI] [PubMed] [Google Scholar]
- 18.Lantz, O., and A. Bendelac. 1994. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class-specific CD4+ and CD4−CD8− T cells in mice and humans. J. Exp. Med. 180:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, N., M. Llano, M. Carretero, A. Ishitani, F. Navaro, M. Lopez-Botet, and D. E. Geraghty. 1998. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 95:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyngstrand, S. T., P. A. Wurtzen, N. Odum, M. H. Nissen, and C. Ropke. 2002. IL-15 induces unspecific effector functions in human peptide-specific CD8+ T-cell cultures. Scand. J. Immunol. 56:602-610. [DOI] [PubMed] [Google Scholar]
- 21.Makino, Y., R. Kanno, T. Ito, K. Higashino, and M. Taniguchi. 1995. Predominant expression of invariant Vα14+ TCRα chain in NK1.1+ T cell populations. Int. Immunol. 7:1157-1161. [DOI] [PubMed] [Google Scholar]
- 22.Pittet, M. J., D. E. Speiser, D. Valmori, J.-C. Cerottini, and P. Romero. 2000. Cytolytic effector function in human circulating CD8+ T cells closely correlates with CD56 surface expression. J. Immunol. 164:1148-1152. [DOI] [PubMed] [Google Scholar]
- 23.Porcelli, S., A. D. Gerdes, A. Fertig, and S. P. Balk. 1996. Human T cells expressing an invariant Vα24-J.αQTCRα are CD4− and heterogeneous with respect to TCRβ expression. Hum. Immunol. 48:63-67. [DOI] [PubMed] [Google Scholar]
- 24.Saikh, K. U., P. Brandler, J. Sesno, and R. G. Ulrich. 1998. Are DNA based vaccines useful for protection against secreted bacterial toxin? Tetanus toxin as a test case. Vaccine 16:1029-1038. [DOI] [PubMed] [Google Scholar]
- 25.Saikh, K. U., T. Kissner, and R. G. Ulrich. 2002. Regulation of HLA-DR and co-stimulatory molecule expression on natural killer T cells by granulocyte-macrophage colony-stimulating factor. Immunology 106:363-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slifka, M. K., R. P. Pagarigan, and J. L. Witton. 2000. NK markers are expressed on a high percentage of virus-specific CD8+ and CD4+ T cells. J. Immunol. 164:2009-2015. [DOI] [PubMed] [Google Scholar]
- 27.Stiles, B. G., S. Bavari, T. Krakauer, and R. G. Ulrich. 1993. Toxicity of staphylococcal enterotoxins potentiated by lipopolysaccharide: major histocompatibility complex class II molecule dependency and cytokine release. Infect. Immun. 61:5333-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taddesse-Heath, L., J. I. Feldman, G. A. Fahle, S. H. Fischer, L. Sorbara, M. Raffeld, and E. S. Jaffe. 2003. Florid CD4+, CD56+ T-cell infiltrate associated with herpes simplex infection simulating nasal NK-/T-cell lymphoma. Mod. Pathol. 16:166-172. [DOI] [PubMed] [Google Scholar]
- 29.Takii, Y., S. Hashimoto, T. Iiai, H. Watanabe, K. Hatakeyama, and T. Abo. 1994. Increase in the proportion of granulated CD56+ T cells in patients with malignancy. Clin. Exp. Immunol. 97:522-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhrberg, M., N. M. Valiante, N. T. Young, L. L. Lanier, J. H. Philipps, and P. Parham. 2001. The repertoire of killer cell Ig-like receptor and CD94:NKG2A receptors in T cells: clones sharing identical αβ TCR rearrangements express highly diverse killer Ig-like receptor patterns. J. Immunol. 166:3923-3932. [DOI] [PubMed] [Google Scholar]
- 31.Ulrich, R. G., M. A. Olson, and S. Bavari. 1998. Development of engineered vaccines effective against structurally related bacterial superantigens. Vaccine 16:1857-1864. [DOI] [PubMed] [Google Scholar]
- 32.Vergelli, M., H. Le, J. M. V. Noort, S. Dhib-jalbut, H. McFarland, and R. Martin. 1996. A novel population of CD4+CD56+ myelin-reactive T cells lyses target cells expressing CD56/neural cell adhesion molecule. J. Immunol. 157:679-688. [PubMed] [Google Scholar]
- 33.Watanabe, H., A. Weerasinghe, C. Miyaji, C. H. Sekikawa, S. Toyabe, M. K. Mannor, S. R. Morshed, R. C. Halder, J. Kobayashi, H. Toma, Y. Sato, K. Iwai, H. Matsuoka, and T. Abo. 2003. Expansion of unconventional T cells with natural killer markers in malaria patients. Parasitol. Int. 52:61-70. [DOI] [PubMed] [Google Scholar]