Abstract
Antiviral drugs, most notably the neuraminidase inhibitors, are an important component of control strategies aimed to prevent or limit any future influenza pandemic. The potential large-scale use of antiviral drugs brings with it the danger of drug resistance evolution. A number of recent studies have shown that the emergence of drug-resistant influenza could undermine the usefulness of antiviral drugs for the control of an epidemic or pandemic outbreak. While these studies have provided important insights, the inherently stochastic nature of resistance generation and spread, as well as the potential for ongoing evolution of the resistant strain have not been fully addressed. Here, we study a stochastic model of drug resistance emergence and consecutive evolution of the resistant strain in response to antiviral control during an influenza pandemic. We find that taking into consideration the ongoing evolution of the resistant strain does not increase the probability of resistance emergence, however it increases the total number of infecteds if a resistant outbreak occurs. Our study further shows that taking stochasticity into account leads to results that can differ from deterministic models. Specifically, we find that rapid and strong control can not only contain a drug sensitive outbreak, it can also prevent a resistant outbreak from occurring. We find that the best control strategy is early intervention heavily based on prophylaxis at a level that leads to outbreak containment. If containment is not possible, mitigation works best at intermediate levels of antiviral control. Finally, we show that the results are not very sensitive to the way resistance generation is modeled.
Introduction
It is almost certain that sooner or later, a new influenza A virus will emerge against which humans have little or no immunity and that is able to spread through human populations and potentially cause a pandemic (7, 47). In the face of this threat, researchers have been studying control strategies that might prevent or mitigate such a pandemic (11–13, 16, 32, 33). Most proposed intervention strategies rely to some extent on the use of antivirals, most notably the neuraminidase inhibitors (15, 36). Unfortunately, the strong selection pressure exerted by the extensive use of drugs often leads to the evolution of drug resistance (8, 27, 30). Most situations encountered so far in the realm of antibiotic resistance involve time-scales on the order of years before a large fraction of hosts harbors a resistant strain (28, 30). However, the high mutation rate of viruses can lead to a much more rapid evolution of resistance. One premier example is the evolution of resistance that occurs in HIV during treatment with a single drug (6). Since influenza is also a relatively fast evolving virus with a high mutation rate (37, 38), it is possible that drug resistance can become a problem during the course of a single pandemic outbreak.
A number of modeling studies investigated the possible impact of resistance emergence and spread during an influenza outbreak (1, 9, 14, 29, 35, 39, 41, 49). While these studies have provided important insights, a few aspects remain to be fully addressed. Most importantly, the majority of studies are based on deterministic models. This ignores the stochastic nature of the rare events that lead to initial resistance generation and spread. while a few recent studies were based on stochastic models (9, 49), these studies only considered outbreaks in small populations (less than 103 – 104 individuals). Further, these studies did not consider continued evolution of the resistant strain. While resistance usually carries a fitness cost, the resistant mutants can undergo further evolution, acquiring so called compensatory mutations that restore their fitness while retaining the resistant phenotype (3, 34). The result can be a strain that is at the same time drug resistant and has a fitness close to – and in the worst case even higher than – the original drug-sensitive strain. Limited in vitro evidence suggests that compensatory mutations might occur for neuraminidase inhibitor resistant influenza (52). Only one study considered compensatory mutations for influenza drug resistance (35). However, this study is based on a deterministic framework, and due to the rarity of these compensatory mutation events, a stochastic framework is more appropriate (22).
Here, we study a stochastic model of neuraminidase inhibitor resistance emergence and consecutive evolution of the resistant strain in response to antiviral control during an influenza pandemic in a large population. Our study shows that taking stochasticity into account leads to results that can differ from deterministic models. Specifically, we find that rapid and strong control can contain not only a drug sensitive outbreak but also prevent a resistant outbreak from occurring. We find that the best control strategy to prevent resistance emergence and reduce the total number of infecteds is early intervention heavily based on prophylaxis at a level that leads to outbreak containment. If containment is not possible, mitigation works best at intermediate levels of control. Taking into consideration the ongoing evolution of the resistant strain does not increase the probability of resistance emergence, however it increases the total number of infecteds if a large resistant outbreak occurs. We also show that the results are largely insensitive with respect to the detailed implementation of the resistance generation process.
The Model
We model the outbreak using a stochastic, compartmental, SIR-type model. A schematic flow diagram of the model is shown in Figure 1, Table 1 gives the transitions and their propensities which fully specify the model, while Table 2 summarizes the variables and parameters of the system.
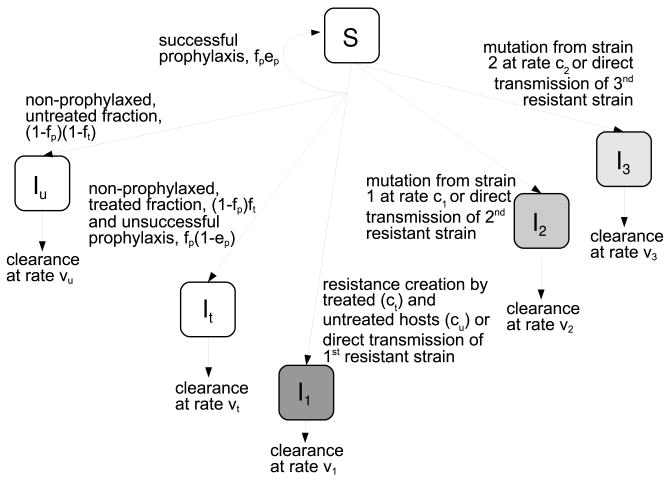
Fig. 1.
Schematic of the compartmental model describing the infection dynamics. The compartments are susceptibles, S, persons infected with the drug sensitive strain that are untreated, Iu, persons infected with the drug sensitive strain that receive treatment, It, and persons infected with the first, second and third resistant strain, I1, I2 and I3. The first resistant strain is the one initially generated, ongoing evolution leads to further mutations that increase fitness of the resistant strain, resulting in I2 and subsequently in I3. Table 1 show the possible transitions and their propensities, Table 2 summarizes the model parameters. Further details are given in the text.
Table 1.
Possible transitions and their propensities (the propensity multiplied with the time step gives the probability that a given event occurs).
| transitions | propensity |
|---|---|
| S → S − 1, Iu → Iu + 1 | (1 − fp)(1 − ft)(βu(1 − cu)Iu + βt(1 − ct)It)S |
| S → S − 1, It → It + 1 | (fp(1 − ep) + (1 − fp)ft)(βu(1 − cu)Iu + βt(1 − ct)It)S |
| S → S − 1, I1 → I1 + 1 | (βucuIu + βtctIt)S + β1(1 − c1)I1S |
| S → S − 1, I2 → I2 + 1 | β1c1I1S + β2(1 − c2)I2S |
| S → S − 1, I3 → I3 + 1 | β2c2I2S + β3I3S |
| Iu → Iu − 1 | νuIu |
| It → It − 1 | νtIt |
| I1 → I1 − 1 | ν1I1 |
| I2 → I2 − 1 | ν2I2 |
| I3 → I3 − 1 | ν3I3 |
Table 2.
Model parameters. Values are specific for neuraminidase inhibitor treatment and resistance emergence in influenza A.
| symbol | meaning | values | comment |
|---|---|---|---|
| fp | fraction of uninfecteds receiving prophylaxis | 0 – 1 | varied |
| ft | fraction of infecteds receiving treatment | 0 – 1 | varied |
| ep | efficacy of prophylaxis | 0.8 | based on AVES in (50), AVESd in (19) |
| νu | clearance rate (1/mean duration of infection) of untreated infected hosts | 1/4.8d−1 | based on (5) |
| νt | clearance rate of treated infected hosts | 1/3.4d−1 | reduction of infectious period by ≈ 30 %, based on (42, 48) |
| ν1, ν2, ν3 | clearance rate of resistant infected hosts | 1/4.8d−1 | assumption that resistant strain leads to same duration of infection as sensitive strain |
| ct | probability of resistance generation for treated hosts | 10−3 | based on Fig. 4A in (20), assuming treatment at day one for the more realistic (IR) model |
| cu | probability of resistance generation for untreated hosts | 10−5 | similar to value for ineffective (late) treatment in (20), Fig. 4A, IR model |
| c1, c2 | probability of resistance generation of compensatory mutants | 10−3 | see text |
| Ru | reproductive number of susceptible strain (in the absence of treatment) | 2.0 | (44, 45) |
| Rt | reproductive number of susceptible strain (in the presence of treatment) | 0.68 | based on (50) |
| R1, R2, R3 | reproductive numbers of resistant strains | 1.5, 1.75, 2.0 | assumed |
| βu, βt β1, β2, β3 | transmission parameters | calculated as Riνi/N0 | |
| N0 | population size | 3 × 108 | U.S. population |
We consider a pandemic outbreak in the United States. We assume that for a novel, pandemic strain, no immunity exists, the whole population is susceptible (S). Susceptible hosts receive prophylaxis with a uniform probability, or phrased differently, a fraction fp of susceptible hosts receive prophylaxis, which has an efficacy of ep. If prophylaxis fails, hosts become infected. We assume that all infected persons will become ill and show symptoms, we ignore the possibility of asymptomatic infections. Infecteds are divided into five different compartments. A fraction ft of hosts infected with the drug sensitive strain receive antiviral treatment (It), while the remainder of the hosts infected with the sensitive strain do not (Iu). Following Lipsitch et al. (29), we assume that failed prophylaxis leads to a course of infection comparable to a treated host. Additionally, three compartments for resistant mutants are considered, (I1, I2 and I3). We assume that treatment has no effect on the resistant strains. All infected hosts leave the infected stage after some time, either through recovery or death. The rates of “clearing” the infection by either means are listed in Table 2.
There are several important differences that distinguish our model from previous ones. Most previous models include a conversion rate from wild-type infected to resistant infected hosts. This assumes that once a host converts to a resistant one, the infection “starts over”. In contrast, we assume here that resistance can emerge during treatment, and with probability ct cause new infections that are dominated by the resistant strain. We believe that this way of implementing resistance generation is more realistic. Additionally, we allow resistance to arise and spread with a small probability, cu, in untreated patients. Values for ct and cu are chosen based on estimates we obtained in a previous study (20). Specifically, we chose the value for ct as obtained from the immune response model with treatment occurring one day after infection, while the value for cu was chosen slightly lower than that obtained for the no treatment case (see Figure 4 in (20)). Also note that in our model, prophylaxis has no direct effect on the generation of resistant infecteds. Instead, prophylaxis influences resistance generation through the fact that failed prophylaxis places infecteds into the treated class (which is more likely to give rise to resistance than the untreated class).
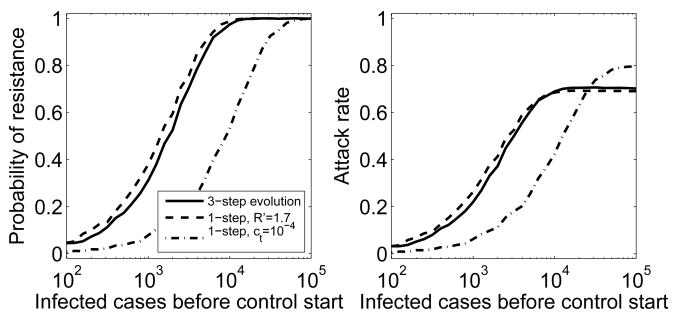
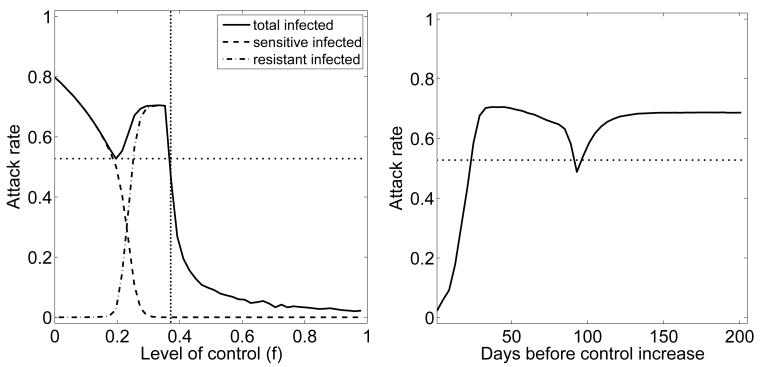
Fig. 4.
Different evolutionary trajectories of the resistant strain. Control is strong enough to contain the sensitive outbreak (Rf = 0.8), as previously shown in Figure 3. Left: Probability of resistance emergence for the 3-step evolutionary trajectory previously shown in Figure 3 (solid line), a 1-step process with ct = 10−3 as before and R′ = 1.7 (dashed line), and a 1-step process with c′ = 10−4 leading to a strain with fitness R′ = 2 (dash-dotted). Right: Average attack rate. Attack rate here and in the following figures is the average over all stochastic simulations, independent of the occurrence of a resistant outbreak.
Our model includes the evolution of the resistant strain. While back-mutations to the fitter, susceptible strain are possible, it is often more likely that instead of reversion to the original, drug sensitive genotype, the resistant mutant undergoes further, so called compensatory mutations (22, 26, 34). These mutations reduce the fitness cost that comes with resistance, while at the same time retaining the resistant mutation. The result can be a strain that is at the same time resistant and has a fitness similar to the initial, sensitive strain. Evolution of compensatory mutants could occur along a single linear pathway or there could be multiple routes with multiple possibilities for compensatory mutations to increase fitness. For illustrative purposes, we choose a simple, linear pathway with 3 levels of fitness for the resistant strain. While the positive selection pressure experienced by the fitter, compensated mutants could be less strong compared to the selection pressure induced by drug treatment, there is no data for estimates of the rate at which compensated mutants arise and spread. As a conservative estimate, we assume that resistant mutants with increased fitness are generated at the same rate as the initial generation of resistance during treatment, i.e. we choose c1 = c2 = ct.
Since the creation of resistance is a rare event, stochasticity is important. Therefore, we use a stochastic model. The model is a variation of a discrete time, Monte-Carlo simulation, often referred to as the Gillespie algorithm (17). The Gillespie algorithm produces exact trajectories of the stochastic process. Since a straightforward implementation of the Gillespie algorithm would be computationally too expensive for the population size we consider, we instead use a recently introduced hybrid stochastic solver known as partitioned leaping algorithm (23). The algorithm uses the exact Gillespie method for low numbers and reaction rates, i.e. when stochasticity is important, but switches automatically to computationally more efficient methods using Poisson, Langevin and deterministic approximations when appropriate (23). This leads to a significant reduction in execution time, while still essentially retaining the “exactness” of the Gillespie algorithm. The simulations are implemented in Fortran 90, the code is available from the authors upon request.
Results
Antiviral control affects the drug sensitive strain by reducing its fitness, defined in our setting as the average number of secondary infections caused by an infected host, the reproductive number (2, 10, 25). The reproductive number of the sensitive strain is given by the largest eigenvalue of the matrix M = FV−1 (43), where
and
From this one finds that the reproductive number in the presence of control is
| (1) |
Note that we ignored the negligible contributions of resistance generation, i.e. we set ci ≈ 0. The reproductive numbers for untreated and treated hosts Ru and Rt, as well as the other parameters are given in Table 2. If Rf < 1, on average less than one new host gets infected with the sensitive strain and therefore the outbreak will die down. For Rf > 1, containment of the outbreak is likely to fail. However, treatment or prophylaxis will still reduce the number of hosts infected with the sensitive strain. Note that the antiviral has no effect on the fitness of the resistant strains.
For any infectious disease outbreak, there will be a time lag between the occurrence of the first infection, the recognition of the outbreak as such, and implementation of control measures. Since the probability that resistance is generated depends on the number of infected hosts, rapid containment of the outbreak will reduce the probability that a resistant mutant is generated and spreads. In the following sections, we study how resistance emergence depends on the number of infecteds before control starts. We consider this for different scenarios by varying the evolutionary pathway of the resistant strain and the type (prophylaxis versus treatment) and strength of the antiviral control.
Compensatory mutations do not change the probability of resistance emergence, but increase the number of infecteds in large outbreaks
We start by considering how ongoing evolution of the resistant strain can influence the probability of resistance generation and the size of a pandemic outbreak. For the first scenario, we assume that antiviral control is not strong enough to control the drug sensitive strain, that is we have Rf > 1. The control effort leads to only a mitigation in outbreak size. Since in this situation, a large number of infections occur, resistance is always generated. In the presence of compensatory mutations, the resistant strain can evolve to higher fitness and therefore contribute to a larger outbreak, increasing the attack rate by ≈ 20% (Fig. 2). Including ongoing evolution also increases the variance in the outbreak size, which is expected since stochastic effects are most important when a new resistant strain is created, which happens three times for the scenario with ongoing evolution, versus only one time in the absence of further evolution. Since the dynamics is nonlinear, it is not expected that the mean of the stochastic simulations agrees with the result found from the equivalent deterministic model. However, as Fig. 2 shows, there is relatively good agreement. If control starts early, almost all infections are caused by the resistant strain(s). If control starts later, the sensitive strain causes a significant outbreak before the resistant strains emerge and cause their own outbreaks. These multiple smaller outbreaks cause less infections compared to one large outbreak, leading to the observed decline in overall attack rate for late control. This phenomenon has been noted previously (22, 29) and we will return to it in a later section.
Fig. 2.
Attack rate in the absence and presence of compensatory mutations. Control starts after the indicated number of infections have occurred, with treatment and prophylaxis chosen at equal levels, (ft = fp). Control can only mitigate the outbreak (Rf = 1.2). Attack rate is defined as the total number of infecteds divided by the population size. Boxplots are results from 2000 stochastic simulations, lines show results from the equivalent deterministic model. The black boxes and solid line are results in the absence of ongoing evolution through compensatory mutations, the gray boxes and dashed line show results in the presence of ongoing evolution. The dotted line shows the attack rate in the absence of control. The resistant strains have R1 = 1.5, R2 = 1.75 and R3 = 2.0, for the case with compensatory mutations, c1 = c2 = ct.
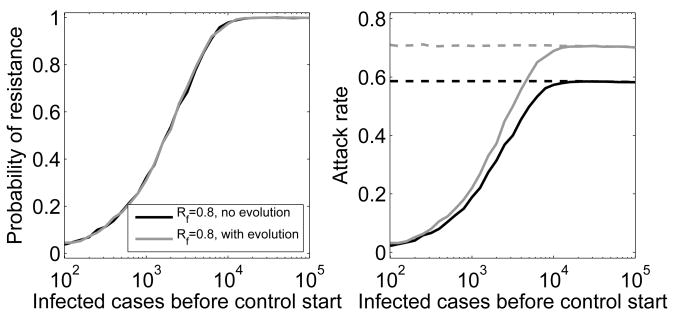
In the next scenario, we assume that the control effort is strong enough to lead to Rf < 1, i.e. the outbreak caused by the sensitive strain can be contained. If containment occurs before the resistant strain has emerged, no major epidemic occurs. If, on the other hand, the resistant strain is generated and starts to spread, antiviral control efforts become ineffective and a large epidemic caused by the resistant strain occurs. We find that including evolution of the resistant strain barely changes the probability of resistance emergence (Fig. 3). However, as was the case in the mitigation scenario above, the increase in fitness of the resistant strain due to compensatory mutations leads to an increase in attack rate by as much as ≈ 20%.
Fig. 3.
Probability of resistance emergence and attack rate in the absence and presence of compensatory mutations. Control is strong enough to contain the sensitive outbreak (Rf = 0.8). Left: Probability of resistance emergence in the absence (black) and presence (gray) of compensatory mutations. Resistance is considered to have emerged if at least 5% of the total number of infections that have occurred during the outbreak are resistant infecteds (a higher/lower percentage leads to a right/left shift of the curves). Right: Solid lines show attack rate averaged over all 2000 stochastic simulations, dashed lines show attack rate averaged only over those simulations where a (resistant) outbreak occurred. The variance in attack rate for the stochastic simulations when an outbreak occurs is very small, we therefore only plot the mean instead of showing boxplots. These mean values agree closely with the deterministic results (not plotted). Rest as explained previously.
Evolution through compensatory mutations can be mapped onto a one-step process
While continued evolution of a resistant strain is without doubt going to occur, the details of the evolutionary process can not be predicted. Above, we assumed that fitness increases in equal steps, from R1 = 1.5 over R2 = 1.75 to R3 = 2.0, the original fitness of the sensitive strain. We further assumed that the probability of these events happening was the same as the probability of emergence during treatment, i.e. ct = c1 = c2. However, other scenarios are equally likely. The increase in fitness could for instance occur in unequal steps or the probabilities for these events could differ. While it is impossible to explore all these scenarios (see (22) for some more details), it is worth investigating if and how the evolutionary trajectory can be mapped onto a simple process where a resistant strain emerges and does not undergo further compensatory mutations. Two mappings might be expected to be possible. First, a 3-step trajectory with fitness levels R1 = 1.5, R2 = 1.75 and R3 = 2.0 could be equivalent to a single step to a resistant strain with some different fitness R′. Alternatively, the three probabilities ct, c1 and c2, might be mapped into a single probability c′, directly leading to the final strain. As Figure 4 shows, while it is indeed possible to find a 1-step process with resistant fitness R′ that produces a result similar to that of the 3-step process, it is not possible to map the jump probabilities into a single one. The reason for this latter finding is is that the main “bottleneck” in the process is caused by the initial generation of resistance. Once the resistant strain has been generated, it starts to spread and quickly reaches levels at which the generation of fitter strains is almost certain. Therefore, the initial rate of resistance generation is crucial in determining the probability of resistance emergence, and therefore the average attack rate. Changing this rate to a different value, c′, does not lead to dynamics that resembles a process with three transition rates.
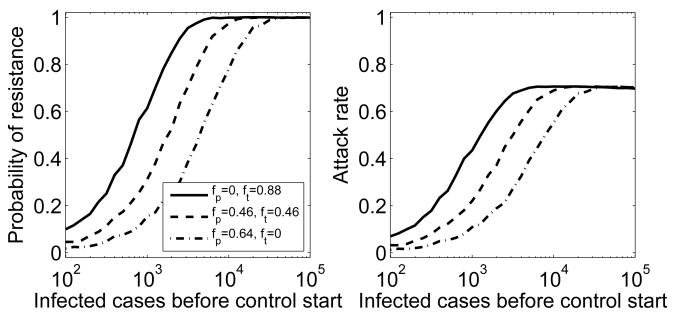
Early control based on strong prophylaxis is the best control strategy to prevent resistance emergence
While treatment with neuraminidase inhibitors will be important to reduce morbidity and mortality of individuals, epidemiological control can be achieved with treatment or prophylaxis. While resistance is more likely to emerge during treatment, the fact that prophylaxed individuals are only susceptible to the resistant strain leads to strong selective pressure for resistance (29). Nevertheless, we find that if control is strong enough to contain the outbreak caused by the sensitive strain, prophylaxis fares better in preventing resistance emergence and therefore reducing the attack rate (Fig. 5). Stronger control measures (i.e. a further reduction in Rf) contain the sensitive outbreak faster, thereby further reducing the probability that resistance emerges. This leads to a shift of the curves in Figure 5 towards the right (not shown). Equivalently, resistance emergence becomes more likely and the curves shift to the left if control is less strong and containment takes longer. Note that again, the distribution underlying the average for the attack rate is bimodal, consisting of a fraction of simulations for which no resistant outbreak occurred, and another fraction (given by the probability of resistance emergence) for which resistant outbreaks occur. Changing the level of control does not change the size of a resistant outbreak once it occurs, it only changes the probability of such a resistant outbreak to occur.
Fig. 5.
Probability of resistance emergence and attack rate for different control strategies. Control starts after the indicated number of infections have occurred. It leads to Rf = 0.8, which is strong enough to contain the outbreak caused by the sensitive strain. Control strategies are: only treatment (solid line), equal levels of prophylaxis and treatment (dashed line) and only prophylaxis (dash-dotted line). Evolution through compensatory mutations is included.
Optimal mitigation occurs at intermediate control strength
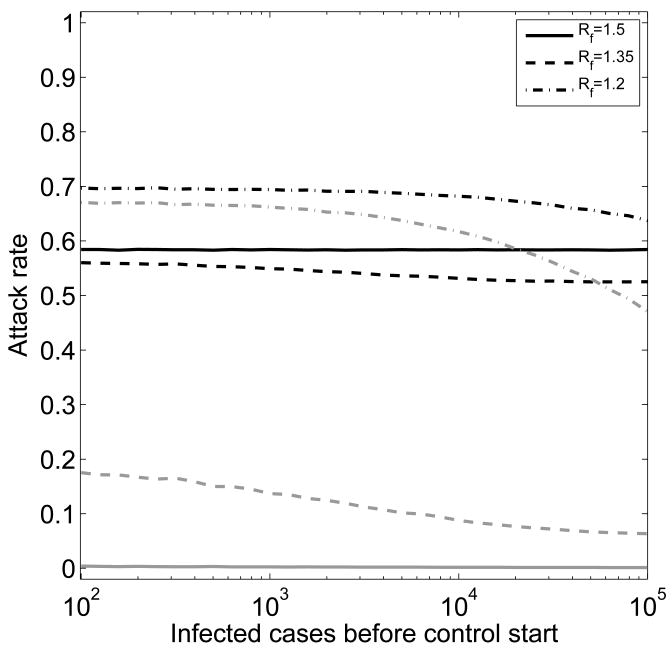
If control can not contain the outbreak, but instead can only mitigate its strength, resistance is very likely to be generated. However, if control does not bring the fitness of the sensitive strain below that of the initially generated resistant mutant, the sensitive strain will dominate. The resistant strain will cause few infections, not enough to have a significant chance of generating further mutants with fitness levels above that of the sensitive strain. The pandemic is almost certain to end before resistance can emerge (i.e. account for more than 5% percent of infecteds) and virtually all infections are caused by the sensitive strain (Fig. 6, Rf = 1.5). Increasing control measures to a level where Rf < R1 leads to a decrease of sensitive infecteds, but now resistance will emerge and contribute to the attack rate (Fig. 6, Rf = 1.35). Once control measures are strong enough to sufficiently suppress the sensitive strain, the resistant strain will dominate and lead to a large resistant outbreak, which in turn leads to an overall increase in infecteds (Fig. 6, Rf = 1.2). Therefore, if it is not possible to contain the outbreak, an intermediate level of control is optimal. We will discuss this point in more detail in the next section. In contrast to the containment scenario (Rf < 1), for the mitigation scenario (Rf > 1) the type of control (prophylaxis or treatment) has almost no impact on the overall attack rate (not shown).
Fig. 6.
Average attack rate for different levels of control. Treatment and prophylaxis are chosen equal (f = fp+ft). Control starts after the indicated number of infections have occurred. Control can only reduce, not contain the outbreak caused by the sensitive strain. The solid, dashed and dash-dotted lines show results for Rf = 1.5, Rf = 1.35 and Rf = 1.2. Black lines are total attack rate, gray lines indicate fraction of cases caused by the resistant strains. Evolution through compensatory mutations is included as previously described.
Optimal treatment strategies differ between stochastic and deterministic models
Above results suggest, and previous studies have shown, that if there are two outbreaks, one caused by the drug sensitive strain and one by a drug resistant strain, an intermediate level of antiviral control can lead to a minimum in the total number of infecteds (29, 35). This can be explained by one of our previous studies, where we showed that the minimization of an “overshoot” – defined as the excess infections that occur during the waning phase of an outbreak – will lead to an optimal control strategy for multiple outbreaks, such as a drug sensitive outbreak followed by a drug resistant one (21). Essentially, two small outbreaks, sensitive and resistant, lead to less overshoot and therefore a smaller overall number in infecteds compared to one large outbreak (21, 29). These studies are based on a deterministic modeling framework, for which resistance is always generated. We decided to see if these proposed strategies are still optimal when stochasticity is taken into account. Figure 7 shows that for Rf > 1 (area left of the dotted vertical line in Fig. 7), there is indeed an intermediate level of control which minimizes the attack rate, in agreement with the results obtained for the mitigation scenario above. However, if control can be implemented such that Rf < 1, Figure 7 suggests that more control is better, since it can reduce the probability of resistance generation (area right of the dotted vertical line in Fig. 7). This is in contrast to results obtained using a deterministic framework (35), for which resistance is always generated and causes a second outbreak. In such a deterministic scenario, high levels of antiviral use lead to rapid generation of resistance and an increased overall attack rate, with an intermediate control level producing the lowest attack rate. The stochastic framework suggests that rapid and strong control that might lead to quick containment of the outbreak is best.
Fig. 7.
Attack rate for varying levels of control. Control starts after 500 infected cases have occurred, treatment and prophylaxis are used equally (f = ft = fp). Left: control strength is varied from 0 to 1 and kept constant throughout the outbreak. The vertical dotted line indicates the level of control for which Rf = 1. Right: Low level of control (f = 0.1) for the indicated number of days, followed by a switch to strong control (f = 0.9). The horizontal dotted line indicates the minimum attack rate obtained for the constant treatment schedule. Evolution through compensatory mutations is included.
It was also shown that for a deterministic model, a strategy of initial low control, followed by a sudden increase in control strength once enough sensitive infecteds are depleted, could perform better compared to a strategy that is based on a constant level of treatment (35). However, little initial control is more likely to lead to generation of a secondary resistant outbreak, while rapid and strong control might contain not only the sensitive outbreak, but also prevent resistance generation. Figure 7 confirms this. We plot attack rate for a situation where treatment starts at f = 0.1 and increases to f = 0.9 at the indicated time. The figure shows that a scenario at which the switch to stronger control occurs at around 90 days leads to a local minimum in the attack rate, again due to minimization of the overshoot (21). However, rapid switch to strong control leads to the largest reduction in attack rate. This suggests that using a “start low, then increase” control strategy as suggested in (35) might be suitable if a secondary resistant outbreak is unavoidable. However, if control can reduce the number of infecteds enough to prevent generation and spread of resistance, then one should implement strong control measures at high levels as soon as possible.
Details of modeling resistance emergence lead to small differences in results
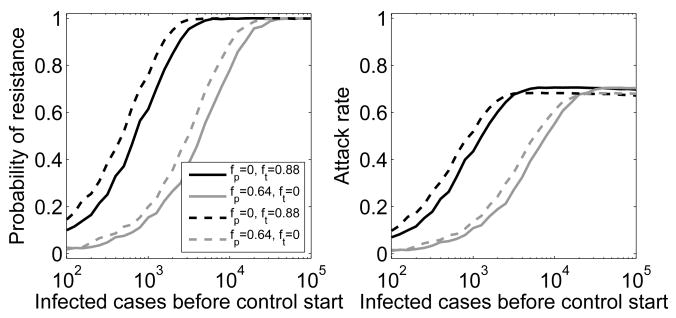
Our study implements the process of resistance generation in a way that differs from previous studies. Most previous models assume that a fraction of treated hosts exit the class of sensitive infecteds and enter the class of resistant infecteds, thereby essentially “starting over”. In contrast, we assume that resistance can emerge during treatment (and at much lower levels in the absence of treatment), and with a certain probability cause new infections that are dominated by the resistant strain. We believe that our implementation of resistance generation is more realistic. To see if the different implementations of the resistance generation process are important, we compared our model to previous ones. Specifically, we assumed – as done in previous models – that as infected hosts leave their compartment, a small fraction, c′i, enter a new resistant infected compartment (i.e. treated and untreated hosts enter the resistant compartment I1, the first resistant strain enters compartment I2, etc.). It is not clear what the rates for c′i should be – especially since we would argue that modeling resistance generation in this way does not correspond directly to a biologically realistic mechanism. To allow some comparison, we assume here that the fractions c′i are equal to the ci. Figure 8 shows that using the two different ways of implementing resistance generation leads to small differences but overall close agreement. We find the same for other types and levels of control (not shown).
Fig. 8.
Probability of resistance emergence and attack rate for different implementation of resistance generation. Control based on treatment only (black) or prophylaxis only (gray) for the model with resistance generation as used here (solid) and as used in previous studies (dashed). See text for details on the two model implementations. Evolution through compensatory mutations is included.
Discussion
Several conclusions can be drawn from our study. First, we find that the ongoing evolution of the resistant strain can contribute significantly to an increase in outbreak size. The fitness of the resistant mutants is not known. If the initially generated resistant mutant spreads poorly (i.e. R1 < Rf), it could take very long before compensatory mutations are created that improve the fitness to a level where the strain can spread widely (4, 22). While there was initial hope that strains resistant to the neuraminidase inhibitors have strongly reduced fitness, recent data suggest that at least some resistant mutants spread almost as well as the wild-type (24, 40, 51). Therefore, assuming that resistant strains with fitness value similar to the ones we choose here will emerge is (unfortunately) reasonable. For the (assumed but plausible) scenario where the rates of compensatory mutation are as high as those of resistance generation during treatment and the fully compensated resistant strain has a fitness the same as the drug sensitive strain, we find that the number of infecteds can increase by as much as 20% owing to the evolution of the resistant strain.
Second, our results show that if it is possible to quickly contain an outbreak caused by a drug sensitive strain, it might also be possible to prevent resistance generation and an outbreak by the resistant strain. For that to occur, it is crucial to start control early and at high levels. Additionally, our results suggest that prophylaxis is the better control strategy to prevent resistance emergence. However, prophylaxis of a fraction of the total population will likely require many more doses of antivirals and is more problematic logistically, compared with treatment of infecteds. This could be prevented by using targeted prophylaxis (32). In any case, additional factors will likely influence the question of treatment versus prophylaxis. If a pandemic strain with a high level of virulence were to spread, treatment might be crucial to reduce mortality and could take precedence over prophylaxis.
Third, we find – in agreement with earlier studies (29, 35) – that if containment is not possible and outbreak mitigation is the best possible outcome, intermediate levels of control minimize the number of infecteds, owing to a reduction in overshoot caused by two smaller outbreaks (a sensitive and a resistant one) compared to one large outbreak (either a sensitive or a resistant one) (21).
Fourth, we find that details in which resistance generation is implemented in the model do not significantly affect the results. This is reassuring, as it suggests a some robustness of the results obtained by different models.
The inclusion of stochasticity and the consideration of evolution of the resistant virus gives a somewhat more realistic model compared to most previous ones that were used to study generation and spread of neuraminidase inhibitor resistance in influenza. Nevertheless, we still made a number of simplifying assumptions. Our model assumes a homogeneous population. Based on results by others, we expect that heterogeneity will likely change the detailed dynamics of the outbreak, but the overall qualitative results will probably not change (29). We also assume that every infected case is symptomatic. If asymptomatic cases do not spread the virus, then including those into our model simply reduces the reproductive numbers and therefore makes a given level of control more effective. If, however, asymptomatic cases spread, and at the same time are not detected (i.e. do not receive treatment), it could undermine treatment based control strategies. This would argue further for the importance of prophylaxis as the better control strategy from an epidemiological standpoint. Implicit in our model formulation is the assumption that infectious periods are exponentially distributed. It has been shown that the assumption that infectious periods are exponentially distributed can lead to different results in for instance parameter estimation and dynamical details, compared to models that assume gamma-distributed infection periods (31, 46). One way our results could be affected is that an exponential distribution leads to a few hosts with unrealistically long infection times. These hosts could potentially impact the probability that resistance is generated, especially in the multi-step process including compensatory mutations. Based on our experience using models with both exponential and gamma-distributed infection times, we believe that a gamma-distributed model would not affect the qualitative results. However, we have not formally tested this for the scenarios studied here, and it might merit further investigation.
If an outbreak were to occur, treatment or prophylaxis will not be random and uniform as we implemented it in our simple model, but instead public health authorities will likely use a combination of targeted antiviral prophylaxis, contact tracing, preferential treatment of certain groups, etc. Therefore, to carefully assess intervention methods that take into account drug availability, as well as details in drug delivery, e.g. at what day post infection people start taking the drug, for how long they continue to do so, and how that affects transmission, requires more detailed, agent-based models (12, 13, 16, 18, 33). Such models that include resistance into agent based models are in development (Neil Ferguson, personal communication).
To summarize, our results suggest that if we are able to detect an outbreak early and intervene quickly, it might be possible to not only control a sensitive outbreak, but also to prevent the emergence and spread of resistance. If on the other hand intervention is not quick enough, or control measures are not able to stop the outbreak, then the emergence of resistance is very likely. Therefore, while antivirals will certainly be an important component of pandemic control, we should not rely on them too much. Instead, a comprehensive approach based on good surveillance and rapid response with first-line control mechanisms such as antivirals and behavior changes such as social distancing measures, as well as a concerned effort to rapidly produce a potent vaccine, will be the best answer to an influenza pandemic (18). In fact, such a multi pronged approach seems the most promising approach against most future, novel emerging pathogens.
Acknowledgments
We acknowledge support from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alexander ME, Bowman CS, Feng Z, Gardam M, Moghadas SM, Rst G, Wu J, Yan P. Emergence of drug resistance: implications for antiviral control of pandemic influenza. Proc Biol Sci. 2007 Jul;274(1619):1675–1684. doi: 10.1098/rspb.2007.0422. URL http://dx.doi.org/10.1098/rspb.2007.0422. [DOI] [PMC free article] [PubMed]
- 2.Anderson RM, May RM. Infectious Diseases of Humans - Dynamics and Control. Oxford Science Publications; Oxford: 1991. [Google Scholar]
- 3.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Current Opinion in Microbiology. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 4.Antia R, Regoes R, Koella J, Bergstrom C. The role of evolution in the emergence of infectious diseases. Nature. 2003;426(6967):658–61. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrat F, Vergu E, Ferguson NM, Lemaitre M, Cauchemez S, Leach S, Valleron AJ. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008 Apr;167(7):775–785. doi: 10.1093/aje/kwm375. URL http://dx.doi.org/10.1093/aje/kwm375. [DOI] [PubMed]
- 6.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004 Mar;350(10):1023–1035. doi: 10.1056/NEJMra025195. URL http://dx.doi.org/10.1056/NEJM2ra025195. [DOI] [PubMed]
- 7.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. URL http://dx.doi.org/10.1146/annurev.med.51.1.407. [DOI] [PubMed]
- 8.Dancer SJ. How antibiotics can make us sick: the less obvious adverse effects of antimicrobial chemotherapy. Lancet Infectious Diseases. 2004;4:611–619. doi: 10.1016/S1473-3099(04)01145-4. [DOI] [PubMed] [Google Scholar]
- 9.Debarre F, Bonhoeffer S, Regoes RR. The effect of population structure on the emergence of drug resistance during influenza pandemics. J R Soc Interface. 2007 Oct;4(16):893–906. doi: 10.1098/rsif.2007.1126. URL http://dx.doi.org/10.1098/rsif.2007.1126. [DOI] [PMC free article] [PubMed]
- 10.Diekmann O, Heesterbeek JAP, Metz JAJ. On the definition and the computation of the basic reproductive ratio R0 in models for infectious diseases in heterogeneous populations. Journal of Mathematical Biology. 1990;28:365–382. doi: 10.1007/BF00178324. [DOI] [PubMed] [Google Scholar]
- 11.Duerr HP, Brockmann SO, Piechotowski I, Schwehm M, Eichner M. Influenza pandemic intervention planning using influsim: pharmaceutical and non- pharmaceutical interventions. BMC Infect Dis. 2007;7:76. doi: 10.1186/1471-2334-7-76. URL http://dx.doi.org/10.1186/1471-2334-7-76. [DOI] [PMC free article] [PubMed]
- 12.Ferguson NM, Cummings DAT, Cauchemez S, Fraser C, Riley S, Meeyai A, Iamsirithaworn S, Burke DS. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005 Sep;437(7056):209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson NM, Cummings DAT, Fraser C, Cajka JC, Cooley PC, Burke DS. Strategies for mitigating an influenza pandemic. Nature. 2006 Jul;442(7101):448–452. doi: 10.1038/nature04795. URL http://dx.doi.org/10.1038/nature04795. [DOI] [PMC free article] [PubMed]
- 14.Ferguson NM, Mallett S, Jackson H, Roberts N, Ward P. A population-dynamic model for evaluating the potential spread of drug-resistant influenza virus infections during community-based use of antivirals. J Antimicrob Chemother. 2003 Apr;51(4):977–990. doi: 10.1093/jac/dkg136. URL http://dx.doi.org/10.1093/jac/dkg136. [DOI] [PubMed]
- 15.Gani R, Hughes H, Fleming D, Griffin T, Medlock J, Leach S. Potential Impact of Antiviral Drug Use during Influenza Pandemic. Emerg Infect Dis. 2005;11(9):1355–62. doi: 10.3201/eid1109.041344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germann TC, Kadau K, Longini IM, Macken CA. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A. 2006 Apr;103(15):5935–5940. doi: 10.1073/pnas.0601266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillespie DT. Exact Stochastic Simulation of Coupled Chemical Reactions. The Journal of Physical Chemistry. 1977;81(25):2340–2361. [Google Scholar]
- 18.Halloran ME, Ferguson NM, Eubank S, Longini IM, Cummings DAT, Lewis B, Xu S, Fraser C, Vullikanti A, Germann TC, Wagener D, Beckman R, Kadau K, Barrett C, Macken CA, Burke DS, Cooley P. Modeling targeted layered containment of an influenza pandemic in the United States. Proc Natl Acad Sci U S A. 2008 Mar;105(12):4639–4644. doi: 10.1073/pnas.0706849105. URL http://dx.doi.org/10.1073/pnas.0706849105. [DOI] [PMC free article] [PubMed]
- 19.Halloran ME, Hayden FG, Yang Y, Longini IM, Monto AS. Antiviral effects on influenza viral transmission and pathogenicity: observations from household-based trials. Am J Epidemiol. 2007 Jan;165(2):212–221. doi: 10.1093/aje/kwj362. URL http://dx.doi.org/10.1093/aje/kwj362. [DOI] [PubMed]
- 20.Handel A, Longini IM, Antia R. Neuraminidase Inhibitor Resistance in Influenza: Assessing the Danger of Its Generation and Spread. PLoS Comput Biol. 2007 Dec;3(12):e240. doi: 10.1371/journal.pcbi.0030240. URL http://dx.doi.org/10.1371/journal.pcbi.0030240. [DOI] [PMC free article] [PubMed]
- 21.Handel A, Longini IM, Antia R. What is the best control strategy for multiple infectious disease outbreaks? Proc Biol Sci. 2007 Mar;274(1611):833–837. doi: 10.1098/rspb.2006.0015. URL http://dx.doi.org/10.1098/rspb.2006.0015. [DOI] [PMC free article] [PubMed]
- 22.Handel A, Regoes RR, Antia R. The role of compensatory mutations in the emergence of drug resistance. PLoS Comput Biol. 2006 Oct;2(10):e137. doi: 10.1371/journal.pcbi.0020137. URL http://dx.doi.org/10.1371/journal.pcbi.0020137. [DOI] [PMC free article] [PubMed]
- 23.Harris LA, Clancy P. A “partitioned leaping” approach for multiscale modeling of chemical reaction dynamics. J Chem Phys. 2006 Oct;125(14):144107. doi: 10.1063/1.2354085. URL http://dx.doi.org/10.1063/1.2354085. [DOI] [PubMed]
- 24.Herlocher ML, Truscon R, Elias S, Yen HL, Roberts NA, Ohmit SE, Monto AS. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J Infect Dis. 2004 Nov;190(9):1627–1630. doi: 10.1086/424572. URL http://dx.doi.org/10.1086/424572. [DOI] [PubMed]
- 25.Hethcote HW. The Mathematics of Infectious Diseases. SIAM Review. 2000;42:599–653. [Google Scholar]
- 26.Levin B, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154(3):985–97. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nature Medicine. 2004;10(12):S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 28.Lipsitch M. The rise and fall of antimicrobial resistance. Trends in Microbiology. 2001;9(9):438–444. doi: 10.1016/s0966-842x(01)02130-8. [DOI] [PubMed] [Google Scholar]
- 29.Lipsitch M, Cohen T, Murray M, Levin BR. Antiviral Resistance and the Control of Pandemic Influenza. PLoS Med. 2007 Jan;4(1):e15. doi: 10.1371/journal.pmed.0040015. URL http://dx.doi.org/10.1371/journal.pmed.0040015. [DOI] [PMC free article] [PubMed]
- 30.Livermore DM. Minimising antibiotic resistance. Lancet Infect Dis. 2005 Jul;5(7):450–459. doi: 10.1016/S1473-3099(05)70166-3. URL http://dx.doi.org/10.1016/S1473-3099(05)70166-3. [DOI] [PubMed]
- 31.Lloyd AL. Destabilization of epidemic models with the inclusion of realistic distributions of infectious periods. Proceedings of the Royal Society London B; 2001. pp. 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longini IM, Halloran ME, Nizam A, Yang Y. Containing pandemic influenza with antiviral agents. American Journal Of Epidemiology. 2004 Apr;159(7):623–633. doi: 10.1093/aje/kwh092. [DOI] [PubMed] [Google Scholar]
- 33.Longini IM, Nizam A, Xu S, Ungchusak K, Hanshaoworakul W, Cummings DAT, Halloran ME. Containing pandemic influenza at the source. Science. 2005 Aug;309(5737):1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 34.Maisnier-Patin S, Andersson DI. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol. 2004 Jun;155:360–369. doi: 10.1016/j.resmic.2004.01.019. URL http://dx.doi.org/10.1016/j.resmic.2004.01.019. [DOI] [PubMed]
- 35.Moghadas SM, Bowman CS, Rst G, Wu J. Population-wide emergence of antiviral resistance during pandemic influenza. PLoS ONE. 2008;3(3):e1839. doi: 10.1371/journal.pone.0001839. URL http://dx.doi.org/10.1371/journal.pone.0001839. [DOI] [PMC free article] [PubMed]
- 36.Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med. 2005 Sep;353(13):1363–1373. doi: 10.1056/NEJMra050740. URL http://dx.doi.org/10.1056/NEJMra050740. [DOI] [PubMed]
- 37.Nobusawa E, Sato K. Comparison of the mutation rates of human influenza A and B viruses. J Virol. 2006 Apr;80(7):3675–3678. doi: 10.1128/JVI.80.7.3675-3678.2006. URL http://dx.doi.org/10.1128/JVI.80.7.3675-3678.2006. [DOI] [PMC free article] [PubMed]
- 38.Parvin JD, Moscona A, Pan WT, Leider JM, Palese P. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol. 1986 Aug;59(2):377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Regoes RR, Bonhoeffer S. Emergence of drug-resistant influenza virus: population dynamical considerations. Science. 2006 Apr;312(5772):389–391. doi: 10.1126/science.1122947. [DOI] [PubMed] [Google Scholar]
- 40.Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Butler EN, Wallis TR, Klimov AI, Gubareva LV. Surveillance for neuraminidase inhibitor resistance among human influenza a and b viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother. 2008 Sep;52(9):3284–3292. doi: 10.1128/AAC.00555-08. URL http://dx.doi.org/10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed]
- 41.Stilianakis NI, Perelson AS, Hayden FG. Emergence of drug resistance during an influenza epidemic: insights from a mathematical model. J Infect Dis. 1998 Apr;177(4):863–873. doi: 10.1086/515246. [DOI] [PubMed] [Google Scholar]
- 42.Treanor JJ, Hayden FG, Vrooman PS, Barbarash R, Bettis R, Riff D, Singh S, Kinnersley N, Ward P, Mills RG. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. US Oral Neuraminidase Study Group. JAMA. 2000 Feb;283(8):1016–1024. doi: 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- 43.van den Driessche P, Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math Biosci. 2002;180:29–48. doi: 10.1016/s0025-5564(02)00108-6. [DOI] [PubMed] [Google Scholar]
- 44.Viboud C, Tam T, Fleming D, Handel A, Miller MA, Simonsen L. Transmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic. Vaccine. 2006 Nov;24(4446):6701–6707. doi: 10.1016/j.vaccine.2006.05.067. URL http://dx.doi.org/10.1016/j.vaccine.2006.05.067. [DOI] [PubMed]
- 45.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proceedings of the Royal Society B; Feb, 2007. pp. 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wearing HJ, Rohani P, Keeling MJ. Appropriate models for the management of infectious diseases. PLoS Med. 2005 Jul;2(7):e174. doi: 10.1371/journal.pmed.0020174. URL http://dx.doi.org/10.1371/journal.pmed.0020174. [DOI] [PMC free article] [PubMed]
- 47.Webster RG. The importance of animal influenza for human disease. Vaccine. 2002;20:S16–S20. doi: 10.1016/s0264-410x(02)00123-8. [DOI] [PubMed] [Google Scholar]
- 48.Whitley RJ, Hayden FG, Reisinger KS, Young N, Dutkowski R, Ipe D, Mills RG, Ward P. Oral oseltamivir treatment of influenza in children. Pediatr Infect Dis J. 2001 Feb;20(2):127–133. doi: 10.1097/00006454-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 49.Xu Y, Allen LJS, Perelson AS. Stochastic model of an influenza epidemic with drug resistance. J Theor Biol. 2007 Sep;248(1):179–193. doi: 10.1016/j.jtbi.2007.05.009. URL http://dx.doi.org/10.1016/j.jtbi.2007.05.009. [DOI] [PMC free article] [PubMed]
- 50.Yang Y, Longini IM, Halloran ME. Design and evaluation of prophylactic interventions using infectious disease incidence data from close contact groups. Journal Of The Royal Statistical Society Series C-Applied Statistics. 2006;55:317–330. doi: 10.1111/j.1467-9876.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yen HL, Herlocher LM, Hoffmann E, Matrosovich MN, Monto AS, Webster RG, Govorkova EA. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob Agents Chemother. 2005 Oct;49(10):4075–4084. doi: 10.1128/AAC.49.10.4075-4084.2005. URL http://dx.doi.org/10.1128/AAC.49.10.4075-4084.2005. [DOI] [PMC free article] [PubMed]
- 52.Yen HL, Hoffmann E, Taylor G, Scholtissek C, Monto AS, Webster RG, Govorkova EA. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J Virol. 2006 Sep;80(17):8787–8795. doi: 10.1128/JVI.00477-06. URL http://dx.doi.org/10.1128/JVI.00477-06. [DOI] [PMC free article] [PubMed]