Abstract
The intimin gene eae, located within the locus of enterocyte effacement pathogenicity island, distinguishes enteropathogenic Escherichia coli (EPEC) and some Shiga toxin-producing E. coli (STEC) strains from all other pathotypes of diarrheagenic E. coli. EPEC is a leading cause of infantile diarrhea in developing countries, and intimin-positive STEC isolates are typically associated with life-threatening diseases such as hemolytic-uremic syndrome and hemorrhagic colitis. Here we describe the development of a PCR-restriction fragment length polymorphism (RFLP) assay that reliably differentiates all 11 known intimin types (α1, α2, β, γ, κ, ɛ, η, ι, λ, θ, and ζ) and three new intimin genes that show less than 95% nucleotide sequence identity with existing intimin types. We designated these new intimin genes Int-μ, Int-ν, and Int-ξ. The PCR-RFLP assay was used to screen 213 eae-positive E. coli isolates derived from ovine, bovine, and human sources comprising 60 serotypes. Of these, 82 were STEC isolates, 89 were stx-negative (stx−) and ehxA-positive (ehxA+) isolates, and 42 were stx− and ehxA-negative isolates. Int-β, the most commonly identified eae subtype (82 of 213 [38.5%] isolates), was associated with 21 serotypes, followed by Int-ζ (39 of 213 [18.3%] isolates; 11 serotypes), Int-θ (25 of 213 [11.7%] isolates; 15 serotypes), Int-γ (19 of 213 [8.9%] isolates; 9 serotypes), and Int-ɛ (21 of 213 [9.9%] isolates; 5 serotypes). Intimin subtypes α1, α2, κ, λ, ξ, μ, ν, and ι were infrequently identified; and Int-η was not detected. Phylogenetic analyses with the Phylip package of programs clustered the intimin subtypes into nine distinct families (α, β-ξ, γ, κ, ɛ-η-ν, ι-μ, λ, θ, and ζ). Our data confirm that ruminants are an important source of serologically and genetically diverse intimin-containing E. coli strains.
Enteropathogenic Escherichia coli (EPEC) and a subset of the Shiga toxin-producing E. coli (STEC) isolates known as the enterohemorrhagic E. coli (EHEC) represent two of the five pathotypes of the diarrheagenic E. coli recognized at present (32). EPEC and STEC isolates are commonly recovered from the feces of food-producing animals and pose threats to the health of humans and livestock (32). EPEC isolates are a leading cause of diarrhea, especially among infants in the developing world, and EHEC isolates are often recovered from patients with serious gastrointestinal and systemic diseases such as hemorrhagic colitis (HC) and hemolytic-uremic syndrome (HUS) (38). Unlike other diarrheagenic E. coli isolates, EPEC and many EHEC isolates share the ability to induce the formation of a characteristic histological feature known as an attaching-and-effacing (A/E) lesion on target epithelial cells. A/E lesions are characterized by localized destruction of brush border microvilli and the formation of polymerized actin pedestals beneath the intimately adhering bacteria (16). eae was the first gene to be associated with A/E activity and encodes the intimate bacterial adhesin known as intimin (22).
The expression of intimin among intimin-positive E. coli isolates is essential for colonization of the intestinal mucosa in newborn piglets, calves, adult cattle, and humans in in vitro organ cultures (8, 11, 39, 40). In addition to a role in A/E lesion formation, intimin may play a role in tissue tropism, since studies have shown that O157:H7 strains containing different intimin subtypes vary in their colonization patterns (39, 40). Intimin has also been reported to bind to β-integrins and perhaps another host cell receptor(s), and these interactions may affect tissue colonization patterns (9, 20, 39).
The origin of the locus of enterocyte effacement (LEE) is unknown. However, the A+T content of LEE (62%) compared with the average A+T content of the E. coli genome (49%) suggests that it was acquired by horizontal gene transfer (30). Although the N-terminal region of 670 amino acids of all intimin types is highly conserved, the C-terminal region of 280 amino acids, known as Int280, displays considerable sequence diversity and represents the surface-exposed, immunogenic region of the molecule that also contains Tir and epithelial cell binding activity (14). Twenty-three distinctive E. coli clones representing different pathotypes of the diarrheagenic E. coli have previously been identified by analysis of 20 housekeeping genes by multilocus enzyme electrophoresis (52). Distinct multilocus enzyme types and the conservation of flagellum antigens distinguished two phylogenetic groups within EPEC (EPEC 1 and EPEC 2) and EHEC (EHEC 1 and EHEC 2) (51, 52). Distinct serotypes and intimin subtypes were initially shown to associate within these groups. Serotypes O55:H6 and O127:H6 (Int-α) and serotypes O111:H2, O111:H−, O128:H2, and O45:H2 (Int-β) are representative of the EPEC 1 and EPEC 2 groups, respectively. The site of insertion of LEE in these clonal lineages disrupts the chromosomal gene selC (53). Similarly, in serotype O157:H7 (Int-γ) strains, representative of EHEC 1, and serotype O111:H8, O111:H11, O111:H−, O26:H11, and O111:H− (Int-β) strains, representative of EHEC 2, LEE is inserted in pheU (51-53).
Int-δ is usually expressed by EPEC strains belonging to serotype O86:H34 (1), and Int-ɛ is found among a range of human STEC serotypes (35). Some EHEC strains with serogroup O111 express a subset of Int-γ known as γ2 (51), whereas others have reported the presence of Int-β in serogroup O111 strains (1). Previous studies (44) have shown that strains with an O111:H8 serotype display a mosaic of divergent gene segments that alternatively cluster with intimin subtypes α, β, and γ and exhibit enough sequence divergence to warrant a new intimin subtype designation, identified as Int-θ. The Int-θ sequence is almost identical to that of Int-γ2 (35). Furthermore, Tarr and Whittam (44) showed that the eae gene from strains with serotype O111:H9 was more related to Int-ζ, which was described recently (23). The complexity of intimin subtypes identified among strains belonging to serotypes O111:H2, O111:H8, O111:H9, O111:H−, and O45:H2 suggests that there have been multiple acquisitions of LEE in the EHEC 2 and EPEC 2 clonal lineages (44).
Phylogenetic studies indicate that there are six distinct intimin families, designated α, β, γ, δ, ɛ, and θ (1, 35, 44). Recently, Zhang et al. (55) reported three novel intimin genes and designated them Int-η, Int-ι, and Int-κ. Moreover, sequences representing two intimin variants identified as ζ and λ have been deposited in GenBank (accession nos. AJ298279 and AF43953, respectively), but their phylogenetic relationships to the other intimin variants have not been studied in detail.
EPEC strains are defined as intimin-containing diarrheagenic E. coli strains that possess the ability to form A/E lesions on intestinal cells and that do not possess Shiga toxin genes (24). However, EPEC strains can be further classified as typical or atypical. Typical EPEC strains possess a virulence plasmid (EAF plasmid) that encodes genes for the bundle-forming pilus, which is required for localized adherence on cultured epithelial cells; atypical EPEC strains do not possess the EAF plasmid (24, 49). Typical and atypical EPEC strains (i) usually belong to different serotype clusters, (ii) differ in their adherence patterns on cultured epithelial cells, (iii) are typically found in different hosts (typical EPEC strains have only been recovered from humans), (iv) show different geographic distributions, and (v) display differences in their intimin types (49). More recently, eae-positive (eae+), EAF-negative, and ehxA-positive EPEC strains recovered from children with diarrhea have been described; but their intimin types could not be identified (49). Thus, typical and atypical EPEC strains appear to constitute two distinct groups of E. coli. Atypical EPEC strains appear to be more closely related to STEC strains in their serotype profiles, animal reservoirs, the toxins that they produce, and other genetic characteristics and as such are considered emerging pathogens (49).
Although typical EPEC strains have only been recovered from humans, ruminants are a recognized source of atypical EPEC strains (49). Furthermore, cattle and sheep are major reservoirs of STEC strains capable of causing severe infections in humans, such as HC and HUS. However, only a subset of the serotypes found in cattle and sheep has been reported to cause disease in humans, which usually occurs via fecal contamination through the food chain. Coupled with this is the recognition that non-O157 STEC strains, which usually contain eae, are more commonly recovered from patients with severe gastrointestinal and renal diseases in humans, especially in Australia, Sweden, South Africa, and several Latin American countries (12, 26, 28, 32, 36, 38, 50). Although a number of studies have determined the eae subtypes of non-O157 STEC strains isolated from humans, no extensive studies have been undertaken to determine the intimin subtypes from a serologically diverse collection of intimin-positive STEC and EPEC isolates recovered from cattle and sheep. The importance of subtyping of STEC virulence factors has already been shown to have clinical significance. Previous studies (5, 41) have demonstrated that STEC strains recovered from sheep usually possess stx1c and stx2d (Pierard) subtypes that are uncommonly associated with severe disease in humans (17, 54). A study to determine the eae subtypes in cattle and sheep STEC strains may also identify relationships between intimin subtypes, E. coli serotypes, and Shiga toxin subtypes that are more commonly associated with human infections; and such data may be clinically significant.
Present intimin subtyping methodologies are cumbersome, in that multiple assays are required to identify all known eae subtypes. A universal scheme capable of subtyping all known intimin subtypes has not previously been described. In the study described here we examined the distribution of intimin subtypes among a serologically diverse collection (60 serotypes) of 213 intimin-positive E. coli isolates derived from ruminant and human sources and developed an intimin typing scheme for all known intimin variants. Our strategy used a PCR assay to amplify 840 to 880 bp encoding the C-terminal 280 amino acids (Int280) of all known eae subtypes and restriction fragment length polymorphism (RFLP) analysis to differentiate them. Taking this approach, we discovered three new intimin types, identified here as Int-μ, Int-ν, and Int-ξ; determined their nucleotide sequences; and adapted our typing scheme to accommodate these three new subtypes. Comparison of the sequences of these genes with intimin sequences deposited in GenBank with the Phylip package of programs enabled us to examine the phylogenetic relationships among the members of the intimin gene family. Our intimin typing assay will have medical and veterinary clinical applications by facilitating the characterization of eae among E. coli isolates recovered from humans and animals experiencing a range of gastrointestinal and systemic conditions. These data will be useful in (i) gaining a better understanding of the relationship between the intimin subtype and the E. coli serotype for phylogenetic purposes, (ii) characterizing atypical EPEC strains from animal sources and understanding their role in human disease, and (iii) facilitating studies of the role of intimin in host specificity and tissue tropism.
MATERIALS AND METHODS
Bacterial strains.
Two hundred thirteen eae-positive E. coli isolates were used in this study. The Elizabeth Macarthur Agricultural Institute (Camden, New South Wales, Australia) provided 165 isolates. Of these, 86 were isolated from the feces of healthy sheep, 78 were isolated from the feces of healthy cattle, and 1 was isolated from a cow with diarrhea. The 165 isolates were recovered from feces collected from a geographically widespread area in two Australian states (New South Wales and Queensland) from 1997 to 2000. The 165 animal isolates were selected from a collection of over 2,000 STEC isolates on the basis of being eae+; more than one isolate from a single animal was avoided unless isolates from the same animal possessed different virulence factor profiles in the multiplex PCR (see below). Thirty-five human isolates were obtained from the Microbiological Diagnostic Unit (Melbourne, Australia). These consisted of 28 isolates from patients with HUS, bloody diarrhea, infantile diarrhea, infantile gastroenteritis, or diarrhea; 6 isolates from healthy babies; and 1 isolate from a human with an unknown history. Of these, 15 have been described previously and comprise 5 human diarrheal isolates of serotype O26:H− (33, 45); 1 isolate of serotype O26:H11 from a patient with diarrhea in Canada (34); 1 isolate of serotype O55:H6 from a patient with infantile diarrhea (45); 1 isolate of serotype O86:H− from a patient with infantile diarrhea in the United Kingdom (34); 2 isolates of serotype O111:H− from a patient with infantile diarrhea in the United Kingdom (25); 1 isolate of serotype O126:H2 from a patient with infantile diarrhea in the United Kingdom (46); 2 isolates of serotype O128:H2, 1 of which was from a healthy Australian human baby and the second of which was from a patient with infantile gastroenteritis in the United Kingdom (47); 1 isolate of serotype O128:H2 from a patient with infantile diarrhea in the United Kingdom (45), and 1 isolate of serotype O157:H− from a patient with HUS in Australia (3). Andre Burnens from the National Reference Laboratory for Food Borne Diseases (Bern, Switzerland) kindly provided 13 human isolates from patients with diarrhea or HUS (7). The source of the Swiss isolates has been described previously (41).
Intimin gene designation.
The nomenclature used to identify intimin subtypes is based on letters of the Greek alphabet (55). Zhang et al. (55) proposed a cutoff of less than 95% nucleotide sequence identity to define a new intimin allele, and this arbitrary value was applied in our study. We used Int-θ to describe intimin sequences that are almost identical to Int-γ2, since phylogenetic studies (44) suggested that this intimin subtype deserved a new intimin designation. Our analyses indicate that Int-κ (55) and Int-δ (2) possess 99.6% identical amino acids in the Int280 region. However, the complete nucleotide sequence for Int-δ is not available, and therefore, we use the designation Int-κ to describe this intimin subtype.
Detection of genes encoding intimin, Shiga toxins, and enterohemolysin by multiplex PCR.
DNA from all isolates was prepared and subjected to multiplex PCR for the detection of STEC virulence factors stx1, stx2, ehxA, and eae (37), except that the Instagene matrix (Bio-Rad, Richmond, Calif.) was used for the preparation of template DNA, as described previously (13). Amplified DNA fragments were resolved by gel electrophoresis with 2% (wt/vol) agarose. The gels were stained with ethidium bromide (5 μl/ml), visualized by UV illumination, and imaged with a GelDoc 1000 image analysis station (Bio-Rad).
Amplification and subtyping of the eae gene by PCR-RFLP analysis.
A single forward primer (primer EaeVF) and three reverse primers (primers EaeVR, EaeZetaVR, and EaeIotaVR) (Sigma Genosys, St. Louis, Mo.) (Table 1) were designed to amplify a 834- to 876-bp fragment (the fragment size varied, depending on the variant amplified) representing the 3′ variable regions (encoding the C-terminal Int280 amino acids) of all reported intimin variants. Instagene DNA preparations (5 μl) were each amplified in a reaction mixture that contained 10 mM Tris-HCl (pH 8.3), 10 mM KCl, 2 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 2 U of Taq DNA polymerase, and 50 pmol of each primer. The reaction volume was made up to 50 μl with distilled H2O. The thermal cycling conditions used to amplify this region of the intimin variants are shown in Table 1. A final extension cycle was performed at 72°C for 5 min. The amplified DNA fragments were resolved by agarose gel electrophoresis, as described above.
TABLE 1.
Primers used for amplifying and sequencing intimin subtypes
| Primer | Sequence (5′ to 3′) | Orientation | Product size (bp) | Intimin subtype amplified | No. of PCR cycles (cycling conditions) | Reference |
|---|---|---|---|---|---|---|
| EaeVF | AGYATTACTGAGATTAAG | Forward | All variants | This study | ||
| EaeVR | AAATTATTYTACACARAY | Reverse | 840-880a | 35 (94°C, 60 s; 41°C, 60 s; 72°C, 60 s) | This study | |
| EaeZetaVR | AGTTTATTTTACGCAAGT | Reverse | 840-880a | This study | ||
| EaeIotaVR | TTAAATTATTTTATGCAAAC | Reverse | 840-880a | This study | ||
| EaeUniVF | ACTCATTGTGGTGGAGC | Forward | 434b | All variants | 35 (94°C, 50 s; 52°C, 60 s; 72°C, 45 s) | This study |
| PatonR | CCACCTGCAGCAACAAGAGG | Reverse | 37 | |||
| Reid F | CTGAACGGCGATTACGCGAA | Forward | 917b | All variants | 30 (94°C, 60 s; 53°C, 120 s; 72°C, 180 s) | 43 |
| Reid R | CCAGACGATACGATCCAG | Reverse | ||||
| Gannon F | GTGGCGAATACTGGCGAGACT | Forward | 890b | All variants | 35 (94°C, 60 s; 58°C, 60 s; 72°C, 120 s) | 18 |
| Gannon R | CCCCATTCTTTTTCACCGTCG | Reverse | ||||
| EaeVRF1 | CACCTGGTCAGCAGA | Forward | 331b | All variants | 30 (94°C, 20 s; 62°C, 30 s; 72°C, 20 s) | This study |
| EaeVRR1 | ACCTCTGCCGTTCCAT | Reverse | This study | |||
| EaeVRF2 | AACAATGTACAGCTCACTAT | Forward | 543c | Int-μ | 30 (94°C, 30 s; 56°C, 40 s; 72°C, 30 s) | This study |
| EaeVRR2 | TACCGAGGCAAGACCATC | Reverse | This study | |||
| Eae64/4VRF1 | CGCAGTACGCAGAAGATT | Forward | 793d | Int-ν | 30 (94°C, 30 s; 60°C, 40 s; 72°C, 30 s) | This study |
| Eae64/4VRR1 | CCGAGCCAGATGTCAGTT | Reverse | This study | |||
| 365F1 | AACTTCCCTTTGAATACA | Forward | 298e | Int-λ | 30 (94°C, 30 s; 60°C, 30 s; 72°C, 30 s) | This study |
| EaeVRR1 | ACCTCTGCCGTTCCAT | Reverse | This study | |||
| 365F4 | TACGGCGGATAAGACT | Forward | 404f | Int-λ | 30 (94°C, 30 s; 54°C, 30 s; 72°C, 30 s) | This study |
| 365R4 | ACGTTACATCATAGCCC | Reverse | This study | |||
| 411F2 | CAGCTTACTATTACCGTTC | Forward | 471g | Int-ξ | 30 (94°C, 30 s; 54°C, 30 s; 72°C, 30 s) | This study |
| 411R2 | AGAGAAGGTCACTTTTTG | Reverse | This study | |||
| H41VRF2 | ATTACCGTTCTGTCGAAT | Forward | 439h | Int-α2 | 30 (94°C, 45 s; 55°C, 40 s; 72°C, 45 s) | This study |
| H41VRR1 | ATACCGGCTGACCATT | Reverse | This study |
Primers used to amplify and sequence the 3′ end of intimin variants; the size of the amplicon varied depending on the eae gene variant.
Primers used to sequence the conserved 5′ region of all intimin gene variants.
Primers used to sequence intimin μ; location within gene, 1618-2160 bp.
Primers used to sequence intimin ν; location within gene, 1399-2191 bp.
Primers used to sequence intimin λ; location within gene, 347-644 bp.
Primers used to sequence intimin λ; location within gene, 1689-2092 bp.
Primers used to sequence intimin ξ; location within gene, 1627-2097 bp.
Primers used to sequence intimin α2; location within gene, 1635-2074 bp.
Alignment of Int280 nucleotide sequences with the PileUp program (www.angis.org.au) enabled us to select restriction endonucleases which were predicted by computational analyses (Mapplot program; www.angis.org.au) to be capable of differentiating the known intimin subtypes. PCR products (10 μl) generated with the primer cocktail EaeVF, EaeVR, EaeZetaVR, and EaeIotaVR were incubated separately with 3 U of each of the restriction enzymes AluI, RsaI, and CfoI in the buffer provided by the manufacturer for a minimum of 4 h at 37°C. The restriction fragments were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Intimin subtypes were identified by comparing the restriction profile observed with the profile predicted by computational analyses (Table 2).
TABLE 2.
Predicted sizes of restriction fragments used for RFLP analysis of eae
| Intimin type | Expected fragment size(s) (bp) with the following restriction enzyme:
|
||
|---|---|---|---|
| AluI | RsaI | CfoI | |
| Int-α1 | 736, 113 | 725, 84,a 40a | 437, 412 |
| Int-α2 | 375, 334, 133, 7a | 590, 135, 84, 40a | 437, 412 |
| Int-β | 475, 374 | 528, 246, 75a | 618, 231 |
| Int-γ | 834 (uncut) | 432, 402 | 834 (uncut) |
| Int-ɛ | 384, 270, 190, 32a | 774, 102 | 345, 260, 195, 76a |
| Int-ζ | 605, 203, 38a | 345, 279, 135, 87a | 603, 225, 18a |
| Int-η | 384, 270, 190, 32a | 774, 102 | 421, 260, 195 |
| Int-θ | 527, 165, 110, 21,a 14a | 405, 354, 78a | 837 (uncut) |
| Int-ι | 602, 241 | 525, 318 | 843 (uncut) |
| Int-κ | 342, 214, 162, 131 | 231, 201, 159, 120, 84,a 54a | 624, 225 |
| Int-λ | 232, 214, 204, 151, 45a | 441, 318, 87a | 846 (uncut) |
| Int-μ | 824, 19a | 525, 318 | 843 (uncut) |
| Int-ν | 844, 32a | 774, 102 | 876 (uncut) |
| Int-ξ | 247, 214, 161, 148, 79a | 321, 318, 126, 84a | 313, 311, 225 |
The fragment was too small to visualize under the electrophoresis conditions used.
Subtyping of Shiga toxin genes.
The PCR primers, cycling conditions, and restriction endonucleases used for the amplification and RFLP analysis of stx2 and stx1 have been described previously (5, 6, 41).
DNA sequencing of eae genes.
The complete nucleotide sequences of the eae genes from three E. coli isolates of serotypes Ont:Hnt (where nt is nontypeable), O2-related:H19, and OR:H− (where R is rough) were determined since their respective RFLP profiles did not match those of any of the reported intimin variants. Sequencing of the eae gene from a human E. coli isolate of serotype O125:H6 possessing Int-α2 was also undertaken, since no prototype sequence of this subtype was available in public databases. The eae gene from a bovine E. coli isolate of serotype O2/74:H− was also sequenced. On the basis of RFLP and DNA sequence analyses, this isolate possessed Int-λ. However, the 5′ conserved region of Int-λ was not available in GenBank, so the whole intimin gene was sequenced. The strategy used to sequence these intimin genes involved the amplification of the 3′ region of the genes spanning nucleotides 1975 to nucleotides 2805 to 2847 (the fragment sizes varied, depending on the variant gene amplified), which encodes the C-terminal variable region known as Int280. The same primers (Table 1) were also used to sequence this region of the eae gene, and the remaining sequence was obtained by primer walking. To generate sequencing templates spanning the 5′ conserved regions of intimin subtypes, previously published primers (18, 37, 43) and primers generated by primer walking were used. PCR mixtures contained 100 to 500 ng of template DNA, 10 mM Tris-HCl (pH 8.3), 10 mM KCl, 2 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 2 U of Taq DNA polymerase, and 50 pmol of each primer; and the reaction volume was made up to 50 μl with distilled H2O. The primer sequences and the cycling conditions used to generate the sequencing templates are described in Table 1. PCR products were analyzed by agarose gel electrophoresis and purified with a QIAquick DNA purification kit (Qiagen). DNA sequencing reactions were performed with a Big Dye terminator cycle sequencing ready reaction DNA sequencing kit and electrophoresed with an ABI Prism 377 DNA sequencer (Perkin-Elmer, Santa Clara, Calif.). Compilation and analysis of DNA sequence data were performed with Auto Assembler software (Perkin-Elmer). Nucleotide and amino acid analyses were performed with programs accessed via the Australian National Genomic Information Service (www.angis.org.au).
Phylogenetic analysis.
The Clustal W program (48) was used to produce a multiple-sequence alignment of 46 inferred Int280 amino acid sequences, which included the 5 sequences determined in this study and 41 sequences retrieved from GenBank. Evolutionary gene trees were then estimated with the Phylip package of programs (http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html). Pairwise distances were calculated with the Protdist program, with the Dayhoff PAM (percent accepted mutation) matrix specified as the distance model. Int280 gene trees were then constructed by using the BIONJ program due to its superior performance compared with that of neighbor joining, particularly when substitution rates vary among lineages (19). Bootstrap analyses were subsequently performed (1,000 replicates) to assess the relative support for the nodes in the gene tree.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been submitted to GenBank and are available under accession numbers AF530553 to AF530557.
RESULTS
Prevalence of genes encoding Shiga toxins and enterohemolysin among eae+ E. coli strains by multiplex PCR.
The distributions of the stx genes and ehxA among the 213 isolates used in this study are shown in Table 3. All 213 isolates contained eae and belonged to 60 serotypes. Of these, 82 possessed stx genes and belonged to 25 serotypes, including O5:H−, O7:H−, O26:H−, O26:H11, O28:H−, O49:H−, O76:H7, O84:H−, O84:H2, O85:H49, O91:H21, O103:H2, O104:H11, O111:H−, O118:H16, O121:H19, O145:H−, O157:H−, O157:H7, O157:H21, OX3:H21, Ont:H−, Ont:Hnt, OR:H−, and OR:H31. Forty-eight of 82 (58.5%) isolates possessed stx1, 21 of 82 (25.6%) isolates possessed stx2, and 13 (15.9%) of the remaining 82 isolates contained both stx1 and stx2. Sixty-seven of these 82 (81.7%) isolates concomitantly possessed the enterohemolysin gene (ehxA). Of the remaining 131 isolates, 89 (67.9%) possessed ehxA but were stx negative (stx−) and 42 (32.1%) contained eae alone (stx−, ehxA negative [ehxA−]). Of 48 human isolates (23 serotypes), 21 (43.7%) contained only eae (putative EPEC isolates); 14 (29.1%) contained stx1, eae, and ehxA; 5 (10.4%) contained eae and stx1; 5 (10.4%) contained stx2 and eae; and 3 (6.2%) possessed stx2, eae, and ehxA. Although no human isolates simultaneously contained stx1, stx2, eae, and ehxA, 11 isolates of animal origin possessed all four genes.
TABLE 3.
Distributions of stx1, stx2, and ehxA among 213 eae-containing E. coli isolates
| Source | No. of animals or samples (%) | No. (%) of animals or samples containing E. coli isolates positive for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| stx1, eae | stx2, eae | stx1, stx2, eae | stx1, eae, ehxA | stx2, eae, ehxA | stx1, stx2, eae, ehxA | eae, ehxA | eae | ||
| Human | 48 (22.5) | 5 (10.4) | 5 (10.4) | 14 (29.1) | 3 (6.2) | 21 (43.7) | |||
| Bovine | 79 (37.1) | 1 (1.2) | 1 (1.2) | 18 (22.7) | 9 (11.3) | 6 (7.5) | 39 (49.3) | 5 (6.3) | |
| Ovine | 86 (40.4) | 2 (2.3) | 1 (1.1) | 11 (12.7) | 1 (1.1) | 5 (5.8) | 50 (58.1) | 16 (18.6) | |
| Total | 213 (100) | 5 (2.3) | 8 (3.7) | 2 (1.0) | 43 (20.2) | 13 (6.1) | 11 (5.2) | 89 (41.8) | 42 (19.7) |
Shiga toxin subtypes among eae+ E. coli strains.
Of 61 stx1-positive isolates, only 3 (4.9%) showed an RFLP profile indicative of the presence of stx1c (formerly stx1OX3). stx1c-positive (stx1c+) isolates belonged to serotypes O5:H− (one isolate) and O26:H− (two isolates). The stx1c+ O5:H− isolate was recovered from a patient with HUS in New Zealand, and the two stx1c+ O26:H− isolates (which also possessed stx1) were recovered from ovine feces. All three stx1c+ isolates possessed an Int-β intimin subtype.
Of 34 stx2-containing E. coli isolates, 16 (47%) possessed an stx2 subtype, 9 (26.5%) contained stx2vha, 3 (8.8%) contained stx2vhb, 2 (5.9%) possessed a combination of subtypes stx2 and stx2vhb, 1 (2.9%) contained a combination of subtypes stx2 and stx2vha, and 2 (5.9%) possessed the stx2d(pierard) subtype. One isolate of serotype O85:H49 was positive for a Shiga toxin 2 gene when it was isolated, but it appeared to have lost it upon storage. Isolates containing stx2 were serologically diverse and contained one of four intimin subtypes including Int-β (three isolates of serotypes O7:H− and O26:H− recovered from unrelated HUS patients and isolates of serotypes O26:H11 and OX3:H21 recovered from bovine feces), Int-γ (one isolate of serotype O145:H− from a patient with diarrhea), Int-ɛ (two isolates of serotype O121:H19, one from a patient with HUS and one from a patient with diarrhea), or Int-θ (five O111:H− isolates and an isolate of serotype O76:H7, all from bovine feces). Eight of the nine isolates containing stx2vha possessed Int-γ and belonged to the O157 serogroup; the remaining isolate was of serotype OR:H31. Similarly, all three isolates containing stx2vhb contained Int-γ and were of serotypes O145:H− (one isolate from a patient with HUS) and O157:H7 (two isolates from bovine feces). Isolates containing two stx2 subtypes belonged to serotypes O49:H− and OR:H− and possessed stx2 and stx2vhb (subtypes Int-ζ and Int-ɛ, respectively). A single isolate of serotype OR:H− possessed stx2 and stx2vha (Int-ζ). Only two isolates (serotypes O91:H21 and O104:H11, both from bovine feces) possessed stx2d (pierard) subtypes.
Development of a PCR-RFLP assay for subtyping of intimin genes.
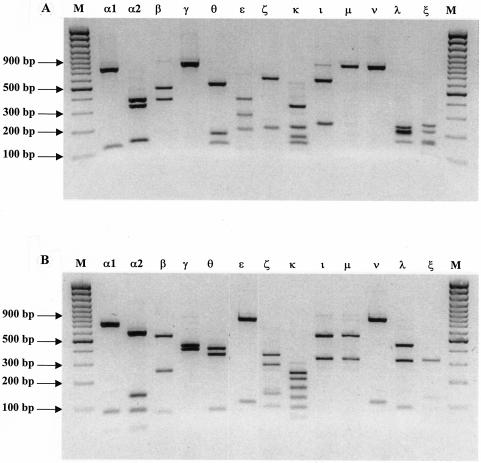
Primers were designed (Table 1) to amplify 840 to 880 bp of the 3′ ends of the genes for all known intimin subtypes. This region of eae encodes the C-terminal 280 amino acids and was used for RFLP analysis because it possesses the greatest degree of sequence variation between subtypes. An amplification product was generated for 10 known subtypes (α1, α2, β, γ, κ, ɛ, ι, λ, θ, and ζ) and 3 new subtypes designated Int-μ, Int-ν, and Int-ξ. Although we did not possess an E. coli isolate that contained the recently described Int-η subtype, sequence alignments indicate that our primer combination should be able to amplify an 876-bp fragment representing the 3′ end of the gene for this subtype. Computational analyses of aligned intimin gene sequences with the Mapplot program indicated that AluI, RsaI, and CfoI could potentially distinguish between all known intimin subtypes (Table 2). AluI differentiated 13 of 14 subtypes, although intimin subtypes γ, μ, and ν (Fig. 1) produced restriction fragments (824 to 844 bp) that could not be easily distinguished by agarose gel electrophoresis. RsaI distinguished 9 of 14 subtypes but failed to differentiate ι and μ (Fig. 1) and ɛ and ν (Fig. 1), and therefore, both enzymes were required to reliably differentiate 13 of the 14 intimin subtypes. CfoI was also used to subtype eae-containing strains because Int-ɛ and Int-η could not be differentiated with AluI and RsaI (Table 2). Representative RFLP profiles generated with these enzymes are depicted in Fig. 1A and B.
FIG. 1.
RFLP analysis of the 3′ 840 to 880 bp of all known intimin subtypes with AluI (A) and RsaI (B). Lanes: M, 100-bp-plus molecular weight marker; α1, O127:H− (human); α2, O125:H6 (human); β, O26:H11 (ovine); γ, O157:H− (ovine); θ, O111:H− (bovine); ɛ, O103:H3 (ovine); ζ, O84:H2 (ovine); κ, O37:H− (ovine); ι, Ont:H8 (ovine); μ, OR:H− (ovine); ν, O2-related:H19 (ovine); λ, O2/74:H− (bovine); and ξ, Ont:Hnt (bovine).
Int-β was the most common subtype identified among the E. coli isolates represented in this study (82 of 213 [38.5%] isolates) and was found to be associated with the greatest diversity of serotypes (n = 21) (Table 4). Int-ζ (39 of 213 [18.3%] isolates) was the second most common subtype and was represented by 11 serotypes (Table 4). Int-θ was identified as the third most common subtype (25 of 213 [11.7%] isolates; 15 serotypes), followed by Int-γ (22 of 213 [10.3%] isolates; 10 serotypes). Int-ɛ was identified in 21 isolates representing five serotypes. Int-λ (one isolate), Int-κ (seven isolates), and intimin ι (four isolates) were infrequently identified (Table 4); and Int-η was not identified. We did not observe PCR-RFLP profiles that indicated the presence of more than one intimin type in any one of the 213 E. coli isolates examined in this study, suggesting that only a single eae gene is present in each isolate. However, epidemiologically unrelated isolates with the same serotype or serogroup may possess different intimin subtypes (see below).
TABLE 4.
Associations between serotype, virulence profile, and intimin subtype
| Int-type | Serotype
|
|||
|---|---|---|---|---|
| eae+, stx+ | eae+, stx+, ehxA+ | eae+, ehxA+ | eae+ | |
| Int-α1 | O85:H49a | O127:H− | ||
| O85:H49a | ||||
| O55:H6 | ||||
| Int-α2 | O125:H6 | |||
| Int-β | O26:H− | O5:H−a | O5:H11a | O15:H− |
| O26:H11 | O7:H−a | O26:H11 | O15:H2a | |
| OX3:H21a | O26:H− | O109:H−a | O26:H− | |
| O26:H11 | O145:H−a | O88:H25a | ||
| O104:H11a | O157:H11a | O111:H− | ||
| O118:H16 | Ont:H− | O126:H2a | ||
| OR:H− | Ont:H11 | O128:H2a | ||
| Ont:Hnt | ||||
| Int-γ | O157:H− | O28:H−a | O28:H−a | |
| O157:H− | O28:HRa | |||
| O157:H7 | O98:H−a | |||
| O157:H21a | O172:H1a | |||
| OR:H31a | OR:Hnta | |||
| Int-ɛ | O102:H19 | O103:H2 | O15:H2a | |
| O103:H2 | OR:H− | O166:H49a | ||
| Int-ζ | O49:H− | O5:H11 | O28:H31 | |
| O84:H− | O84:H2 | O156:H1 | ||
| O91:H21 | Ont:H25 | Ont:H25 | ||
| Ont:H− | OR:H2 | |||
| OR:H− | ||||
| Int-θ | O111:H− | O76:H7 | O3:H−a | O3:H8a |
| O111:H− | O5:H11a | O153:H11/21a | ||
| O35:H25a | O156-related:H8a | |||
| O76:H7a | Ont:H7a | |||
| O84:H25a | Ont:H8a | |||
| O103:H2a | ||||
| O172:H25a | ||||
| Ont:H−a | ||||
| Ont:H25a | ||||
| Int-ι | Ont:H8 | |||
| Int-κ | O49:H− | O37:H− | ||
| OR:H− | O86:H− | |||
| Ont:HR | ||||
| Int-λ | O2/74:H− | |||
| O145:H− | ||||
| Int-μ | OR:H− | |||
| Int-ν | O2-related:H19 | |||
| Int-ξ | Ont:Hnt | |||
Not previously reported to possess these intimin types.
Distribution of intimin subtypes among E. coli isolates from cattle, sheep, and humans.
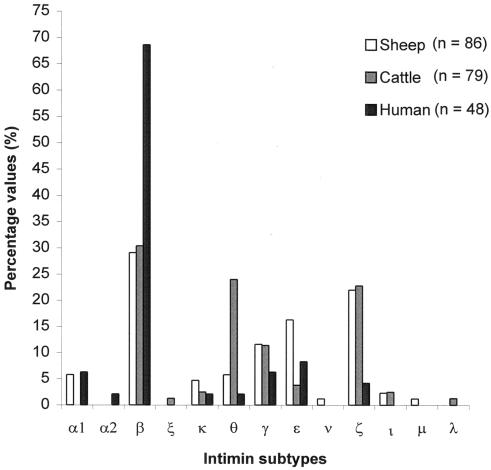
Figure 2 shows the distribution and relative frequencies of the intimin subtypes among cattle, sheep, and humans. E. coli isolates possessing, most intimin types were isolated from both ruminant species and humans, with the following exceptions: Int-α1 (no cattle isolates), Int-α2 (no ruminant isolates), Int-μ (no cattle or human isolates), Int-ξ (no sheep or human isolates), Int-λ (no sheep or human isolates), and Int-ν (no cattle or human isolates). E. coli isolates possessing Int-ι were also not identified among the human isolates (Fig. 2). Interestingly, we did not identify an E. coli isolate possessing Int-α2 among the 165 intimin-positive isolates recovered from both ruminant species. Thus, isolates with Int-α2 may be representative of typical EPEC, as the only isolate that we identified with this intimin subtype possessed a O125:H6 serotype and did not possess the stx or the ehxA gene. Several of these intimin subtypes (Int-μ, Int-ν, and Int-ξ) are reported here for the first time and may be rare. However, it is premature to hypothesize that these intimin species associate exclusively with a particular host.
FIG. 2.
Frequency and host distribution of intimin subtypes in 213 E. coli isolates from sheep, cattle, and humans. n, total number of E. coli isolates examined in each group.
Relationship among intimin subtype, virulence gene profile, and E. coli serotype.
The relationship among the intimin subtype, virulence gene profile, and E. coli serotype is shown in Table 4. The 82 stx-positive (stx+) E. coli isolates examined in this study possessed 6 intimin subtypes and were represented by 26 serotypes. Of 131 non-stx+ E. coli isolates, 89 (67.9%) possessed ehxA and comprised 27 serotypes and 7 intimin types. The remaining 42 isolates (which possessed eae alone) belonged to 25 serotypes and 11 intimin types. Several of the 42 eae+, stx−, and ehxA− isolates, including isolates of serotypes O26:H− (Int-β), O55:H6 (Int-α1), O86:H− (Int-κ), O111:H− (Int-β), O126:H2 (Int-β), O127:H− (Int-α1), and O128:H2 (Int-β), were isolated prior to 1955 (25, 33, 34, 45-47); and the majority of these were from patients with infantile diarrhea. Thus, these isolates probably represent typical EPEC isolates, although further studies are needed to confirm this.
In many instances, E. coli isolates belonging to a particular serotype often possessed the same intimin subtype, irrespective of the host species from which it was recovered. In some instances the same intimin subtype was identified within a serogroup. This was most strikingly exemplified by isolates belonging to the O26 serogroup (O26:H− and O26:H11), which all possessed Int-β, irrespective of the host source. However, isolates belonging to a particular serogroup but displaying different H (flagellum) types more commonly possessed different intimin subtypes. For example, isolates of serogroup O84 possessed Int-ζ (O84:H−, O84:H2) Int-θ (O84:H25), and Int-α1 (O84:H49). Importantly, epidemiologically unrelated isolates belonging to serotypes O5:H11, O15:H2, O49:H−, O103:H2, O111:H−, and O145:H− possessed different intimin subtypes, suggesting that isolates of these serotypes represent different clonal lineages. It should be noted that isolates possessing the same serotype but different virulence gene attributes (e.g., O49:H−, O111:H−, and O145:H−) may belong to the EHEC or the EPEC lineage and thus are not expected to possess identical intimin types. Furthermore, isolates belonging to the Ont and OR serogroups probably represent phylogenetically diverse clones, and new antisera and molecular methods are required to distinguish serotypes and/or clonal lineages within these two serogroups before any valid conclusions may be drawn.
Sequence and phylogenetic analysis of novel eae genes.
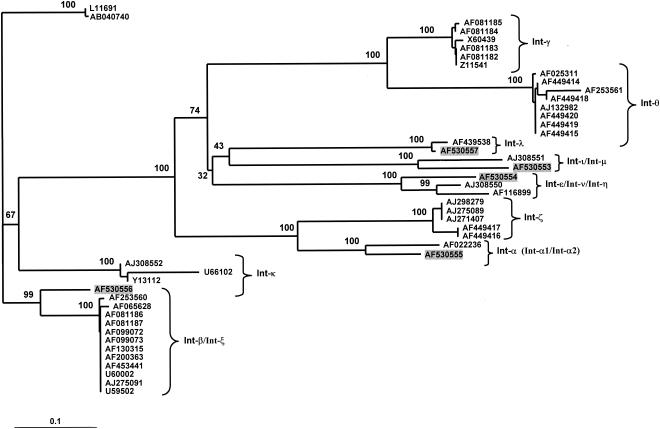
The RFLP profiles of the eae genes from three isolates of serotypes Ont:Hnt, O2-related:H19, and OR:H− did not match any pattern predicted with the intimin gene sequences deposited in GenBank, suggesting that these isolates possess novel intimin subtypes. DNA sequence analysis of the eae genes from these strains was performed by using a panel of primers published previously and by primer walking (Table 1). Alignment of the Int280 sequences deduced from these three genes showed considerable sequence divergence with known intimin subtypes. The predicted amino acid sequence of the intimin gene characterized from an ovine isolate (VR45) of serotype OR:H− showed 89.1% nucleotide sequence identity with Int-ι (GenBank accession no. AJ308551). Phylogenetic analysis confirmed the close relationship of these two intimin subtypes (Fig. 3).
FIG. 3.
Neighbor-joining gene tree based on the C-terminal Int280 amino acids of different intimin subtypes (amino acids 658 to 938) showing the resolutions of various intimin families. The numbers after the branches are the accession numbers in GenBank for the intimin sequences. The highlighted accession numbers represent the sequences determined in this study. The numbers at the nodes correspond to bootstrap proportions. The scale bar indicates the number of amino acid replacements per site.
The predicted amino acid sequence of the intimin gene from an ovine isolate (VR64/4) of serotype O2-related:H19 showed 91.7% nucleotide sequence identity with the Int-ɛ sequence. In addition, it also showed 93.1% nucleotide sequence identity with the Int-η sequence. Phylogenetic analysis resolved these three intimin subtypes as being the closest relatives. The predicted intimin sequence from a bovine isolate (KB411) with serotype Ont:Hnt revealed 90.9% nucleotide sequence identity with the Int-β sequence. Phylogenetic analysis confirmed the association of the sequence of the intimin from Ont:Hnt with the Int-β sequence (Fig. 3). Intimin sequences from ovine isolates of serotypes OR:H− and O2 related:H19 and the bovine isolate of serotype Ont:Hnt showed less than 95% nucleotide sequence identity with the sequences of the intimin subtypes in public databases. According to the arbitrary cutoff values described by Zhang et al. (55), this level of sequence divergence is enough to warrant new intimin gene designations. We propose that they be identified as Int-μ, Int-ν, and Int-ζ, respectively, in accordance with the accepted nomenclature (55).
The Int280 sequence from an E. coli isolate (KB365) of serotype O2/74:H− showed 98.8% nucleotide sequence identity with the Int-λ sequence (GenBank accession no. AF439538). RFLP analysis showed that the sequence of the intimin gene from O2/74:H− was indistinguishable from that of Int-λ. Since only a partial (Int280) sequence for Int-λ exists in GenBank, we were unable to accurately determine the degree of sequence identity between the two intimin sequences. However, phylogenetic analysis strongly supports the affiliation of the Eae protein sequence from serotype O2/74:H− with Int-λ (Fig. 3).
DISCUSSION
The intimin typing scheme described here is capable of distinguishing between all 11 previously described intimin variants (α1, α2, β, γ, κ, ɛ, η, ι, λ, θ, and ζ) and another three new intimin types identified here as μ, ν, and ξ. The application of this typing scheme to 213 E. coli strains representing 60 different serotypes is the most comprehensive intimin subtyping study described so far. Although humans, cattle, and sheep harbor eae+ E. coli strains possessing a range of intimin types, we observed that some intimin types were preferentially associated with specific flagellum types. Int-β was the most common subtype identified in this study, and 78 of 82 Int-β-positive isolates (95.1%) possessed H−, H2, or H11 flagellar antigens. Int-κ was identified in only seven isolates comprising five serotypes (O37:H−, O49:H−, O86:H−, Ont:H−, and OR:H−), none of which expressed a flagellar antigen. Only a limited number of E. coli serotypes (O55:H6, O125:H6, O127:H−, O127:H6, O157:H−, and O157:H45) have been reported to possess Int-α (referred to as Int-α1 in this study), and the H6 flagellar type predominates (35). Twenty-five isolates with Int-θ belonged to 15 serotypes and possessed a range of flagellar antigens, including H−, H2, H7, H8, H11, and H25. Of these 25, 19 (76%) possessed the H−, H7, and H25 flagellar types. Int-ζ and Int-γ were associated with a range of flagellar antigens (Int-ζ flagellar types comprised H−, H1, H2, H11, H21, H25, and H31; and Int-γ flagellar types comprised H−, H1, H7, H21, H31, HR, and Hnt). However, 31 of 39 (79.5%) Int-ζ-positive isolates possessed the H− and H25 flagellar antigens, and 12 of 19 (63.2%) Int-γ-positive isolates possessed the H− flagellar type. Of 21 isolates with Int-ɛ, 19 (90.5%) possessed the H2 and H19 flagellar types. Further studies with a larger collection of eae+ isolates from different geographic locations are required to confirm these flagellar antigen-intimin subtype associations.
Phylogenetic analysis of 46 Int280 sequences (including 5 from this study) confirmed (100% bootstrap support; Fig. 3) the previous division of the intimin family into the six subtypes α, β, γ, κ, ɛ, and θ (1, 35, 44). In addition, our analysis supported the validity of the newly designated Int-ζ subtype (23) and showed that it is most closely related to Int-α. Similarly, the newly designated ι (55) and λ groups were resolved as distinct groups (Fig. 3), but their relationship to other intimin types was less clear. Our analysis indicates that the λ, ι-μ, and ɛ-η-ν groups of intimins are most closely related to each other rather than to any other intimin subtype (32% bootstrap support), but the relationships among these three groups of subtypes could not be determined with confidence. The sequence data suggest that the λ subtype is most closely related to the ι-μ subtype group, with ɛ, η, and ν being more remotely related to the other two groups, but bootstrap support for this is unconvincing (43%). The branches with low levels of support (41 and 47% bootstrap proportions) are very short, suggesting that these intimin subtypes diverged over a short period of time. Furthermore, the branches leading to the ɛ-η-ν, ι-μ, and λ groups are very long, indicating that each of these groups is quite divergent from the other groups.
Although the Int-μ, Int-ν, and Int-ξ subtypes showed considerable sequence diversity in the C-terminal Int280 region, each retained amino acid residues considered essential for interactions with Tir (15, 29, 42). Two cysteine residues which form the disulfide bond required for epithelial cell binding activity (15) and four tryptophan residues (W117/776, W136/795, W222/881, and W240/899) (the positions are numbered according to the Int280α sequence and the complete intimin α sequence) which reside within the receptor-binding superdomain of intimin (2) were conserved. W240/899, which is located on a conserved loop on the D3 domain, is important in A/E lesion formation and intimin-Tir interactions (2), and its replacement with alanine (W240/899A) in site-directed mutagenesis studies generated a phenotype in which intimin could no longer bind to Tir or induce A/E lesions on HEp-2 cells or colonic hyperplasia in vivo (42). Similarly, the phenotype associated with W136/795A showed an intimin-Tir interaction but no A/E lesion formation, and this tryptophan residue is believed to play a central role in maintaining the integrity of the D2 and D3 superdomain (42). The remaining two tryptophan residues are postulated to play roles in Tir-independent host-receptor interactions (42). The preservation of these tryptophan residues among 14 different intimin subtypes supports the hypothesis that these residues are essential for the biological function(s) of intimin.
According to Trabulsi et al. (49), typical EPEC strains produce virulence factors encoded by LEE and plasmid EAF, although some strains possess genes for a cytolethal distending toxin and the enteroaggregative heat-stable toxin (EAST 1). In contrast, atypical EPEC strains are more heterogeneous in their virulence characteristics, commonly possessing EAST 1, ehxA, aggregative adherence, and the afimbrial adhesin (49). Of 131 eae+, stx− isolates examined in our study, 89 (67.9%) possessed ehxA and none were recovered from humans. Furthermore, none of the 48 human isolates examined in this study were stx−, eae+, and ehxA+. Since typical EPEC strains have not been recovered from animal reservoirs, these isolates probably represent atypical EPEC isolates. However, we cannot discount the possibility that some of these isolates were STEC isolates that have lost stx genes. Human isolates with the virulence profile eae+ stx−, and ehx− possess a diverse range of 11 intimin types. We are not sure if isolates with rarer intimin types (ι, μ, ξ, and ν) that we isolated from cattle and sheep (serotypes Ont:H8, Ont:Hnt, OR:H−, and O2-related:H19) play a role in gastrointestinal disease in humans. The isolates of serotypes O26:H− (Int-β), O55:H6 (Int-α1), O86:H− (Int-κ), O111:H− (Int-β), O126:H2 (Int-β), O127:H− (Int-α1), and O128:H2, (Int-β) isolated prior to 1955 are mostly from patients with infantile diarrhea and may represent typical EPEC isolates. Further studies are required to examine the adherence patterns of eae+ and stx− isolates on tissue culture cells and to determine if these isolates possess genes for EAST 1, bundle-forming pilus, afimbrial adhesin, and cytolethal distending toxin (49).
Of the 213 intimin-containing E. coli isolates tested, 82 possessed at least one Shiga toxin gene (putative EHEC isolates), and 73 of these displayed non-O157:H− and non-O157:H7 serotypes. Although epidemiological studies strongly implicate E. coli O157:H7 and O157:H− strains as serious disease-causing agents, the role of most non-O157 STEC strains remains obscure. Serogroup O157 STEC isolates are not the predominant cause of HC and HUS in some countries, including Australia (12), Sweden (50), South Africa (32), Chile (32), and Argentina (28). Furthermore, recent studies suggest that non-O157 STEC strains are responsible for up to 30% of cases of these diseases in the United States (26, 31, 36). We characterized 21 non-O157 STEC serotypes among isolates from humans in this study. Eleven of these serotypes contained Int-β; two contained Int-ζ, Int-α1, and Int-ɛ; and only a single serotype each possessed Int-γ, Int-θ, Int-α2, and Int-κ. Ten of these serotypes (O5:H−, O7:H−, O26:H−, O26:H11, O118:H16, and OX3:H21 [all Int-β]; O76:H7 and O111:H− [Int-θ]; O145:H− [Int-γ]; and O121:H19 [Int-ɛ]) have been recovered from patients with HC and HUS; and most of these belong to the EHEC 2 clonal cluster (52). Collectively, our data show that the vast majority of serologically diverse eae+ STEC isolates typically possess Shiga toxin gene subtypes that comprise the stx2, stx2vha, and stx2vhb subtypes (or combinations of these genes) and/or the non-stx1c subtype. Furthermore, 67 of 82 (81.7%) stx+ and eae+ E. coli isolates also possessed ehxA. These virulence gene patterns are commonly associated with STEC serotypes recovered from patients with bloody diarrhea, HC, and HUS (17) and are increasingly being isolated from patients with diarrhea.
Studies clearly show that different STEC serotypes preferentially colonize healthy cattle, sheep, and swine and that only a small percentage (approximately 10%) of these isolates possess intimin (4, 5, 6, 10, 21, 27, 41). Although intimin probably plays an important role in tissue tropism, little is known about the effects of intimin sequence variation on cell adherence and host range. In this study we show that most intimin variants are equally distributed among E. coli isolates recovered from both sheep and cattle, which strongly suggests that intimin plays little, if any, role in the ability of different serotypes to preferentially colonize ruminant hosts. The STEC serotypes commonly recovered from outbreaks of HUS and HC (O157:H−, O157:H7, O111:H−, O111:H2, O26:H11, and others) typically possess the Int-γ, Int-β, or Int-θ subtype. However, STEC isolates of serotypes O5:H−, O7:H−, O157:H21, O26:H−, O28:H−, O76:H7, O103:H2, O104:H11, O111:H−, O118:H16, O145:H−, OX3:H21, Ont:H−, Ont:Hnt, and OR:H− also possess these intimin subtypes; and many of these have been isolated from sporadic cases of HUS or related afflictions. These data suggest that intimin subtype may be a good diagnostic indicator of the potential of STEC isolates derived from ruminant sources to cause disease in humans. Furthermore, the observation that the vast majority of eae+ STEC isolates recovered from ruminants possess stx2, stx2vha, or stx2vhb or combinations of these genes and/or stx1 (non-stx1c subtype) genes that are commonly associated with EHEC strengthens this hypothesis. Although the available evidence suggests that the stx2 subtype plays a key role in the severity of disease induced by EHEC (17, 54), the intimin subtyping assay reported here should facilitate studies to further our understanding of the associations among serotype, eae and stx subtype, and the phylogenetic relationships between various STEC serotypes.
Acknowledgments
We acknowledge the financial support of Meat and Livestock Australia for parts of this study. V.R. is the recipient of a University of Wollongong Overseas Postgraduate Research Scholarship and University of Wollongong Postgraduate Award. K.B. is the recipient of an Australian Postgraduate Scholarship.
We thank Alexander Kuzevski for skillful technical assistance.
REFERENCES
- 1.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batchelor, M., S. Prasannan, S. Daniell, S. Reece, I. Connerton, G. Bloomberg, G. Dougan, G. Frankel, and S. Matthews. 2000. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J. 19:2452-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettelheim, K. A., M. Whipp, S. P. Djordjevic, and V. Ramachandran. 2002. First isolation outside Europe of sorbitol fermenting verocytotoxigenic Escherichia coli (VTEC) belonging to O group O157. J. Med. Microbiol. 51:1-2. [DOI] [PubMed] [Google Scholar]
- 4.Beutin, L., D. Geier, H. Steinruck, S. Zimmermann, and F. Scheutz. 1993. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J. Clin. Microbiol. 31:2483-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brett, K. N., V. Ramachandran, M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2003. stx1c is the most common Shiga toxin 1 subtype in Shiga toxin-producing Escherichia coli (STEC) excreted by sheep, but not isolates from cattle. J. Clin. Microbiol. 41:926-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brett, K. N., M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2003. Bovine non-O157 Shiga toxin 2-containing Escherichia coli isolates commonly possess stx2EDL933 and/or stx2vhb subtypes. J. Clin. Microbiol. 41:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnens, A. P., P. Boss, F. Orskov, I. Orskov, U. B. Schaad, F. Muller, R. Heinzle, and J. Nicolet. 1992. Occurrence and phenotypic properties of verotoxin producing Escherichia coli in sporadic cases of gastroenteritis. Eur. J. Clin. Microbiol. Infect. Dis. 11:631-634. [DOI] [PubMed] [Google Scholar]
- 8.Dean-Nystrom, E. A., B. T. Bosworth, H. W. Moon, and A. D. O'Brien. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect. Immun. 66:4560-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deibel, C., P. Dersch, and F. Ebel. 2001. Intimin from Shiga toxin-producing Escherichia coli and its isolated C-terminal domain exhibit different binding properties for Tir and a eukaryotic surface receptor. Int. J. Med. Microbiol. 290:683-691. [DOI] [PubMed] [Google Scholar]
- 10.Djordjevic, S. P., M. A. Hornitzky, G. Bailey, P. Gill, B. Vanselow, K. Walker, and K. A. Bettelheim. 2001. Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian slaughter-age sheep. J. Clin. Microbiol. 39:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., S. Tzipori, M. L. McKee, A. D. O'Brien, J. Alroy, and J. B. Kaper. 1993. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J. Clin. Investig. 92:1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, E. J., R. M. Robins-Browne, E. V. O'Loughlin, V. Bennett-Wood, J. Bourke, P. Henning, G. G. Hogg, J. Knight, H. Powell, and D. Redmond. 2001. Nationwide study of hemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagan, P. K., M. A. Hornitzky, K. A. Bettelheim, and S. P. Djordjevic. 1999. Detection of Shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl. Environ. Microbiol. 65:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frankel, G., D. C. Candy, P. Everest, and G. Dougan. 1994. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect. Immun. 62:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel, G., D. C. Candy, E. Fabiani, J. Adu-Bobie, S. Gil, M. Novakova, A. D. Phillips, and G. Dougan. 1995. Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli. Infect. Immun. 63:4323-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 18.Gannon, V. P., S. D'Souza, T. Graham, R. K. King, K. Rahn, and S. Read. 1997. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J. Clin. Microbiol. 35:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gascuel, O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 20.Hartland, E. L., M. Batchelor, R. M. Delahay, C. Hale, S. Matthews, G. Dougan, S. Knutton, I. Connerton, and G. Frankel. 1999. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32:151-158. [DOI] [PubMed] [Google Scholar]
- 21.Hornitzky, M. A., B. A. Vanselow, K. Walker, K. A. Bettelheim, B. Corney, P. Gill, G. Bailey, and S. P. Djordjevic. 2002. Virulence properties and serotypes of Shiga toxin-produsing Escherichia coli from healthy Australian cattle. Appl. Environ. Microbiol. 68:6439-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jores, J., K. Zehmke, J. Eichberg, L. Rumer, and L. Wieler. 2003. Description of a novel intimin variant (type ζ) in the bovine O84:NM verotoxin-producing Escherichia coli strain 537/89 and the diagnostic value of intimin typing. Exp. Biol. Med. 228:370-376. [DOI] [PubMed] [Google Scholar]
- 24.Kaper, J. B. 1996. Defining EPEC. Rev. Microbiol. 27:130-133. [Google Scholar]
- 25.Kauffmann, F., and A. Dupont. 1950. Escherichia coli strains from infantile epidemic gastroenteritis. Acta Pathol. Microbiol. Scand. 27:552-564. [DOI] [PubMed] [Google Scholar]
- 26.Kehl, S. C. 2002. Role of the laboratory in the diagnosis of enterohemorrhagic Escherichia coli infections. J. Clin. Microbiol. 40:2711-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch, C., S. Hertwig, R. Lurz, B. Appel, and L. Beutin. 2001. Isolation of a lysogenic bacteriophage carrying the stx1OX3 gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39:3992-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez, E. L., M. M. Contrini, and M. F. De Rosa. 1998. Epidemiology of Shiga toxin-producing Escherichia coli in South America, p. 30-37. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 29.Luo, Y., E. A. Frey, R. A. Pfuetzner, A. L. Creagh, D. G. Knoechel, C. A. Haynes, B. B. Finlay, and N. C. Strynadka. 2000. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405:1073-1077. [DOI] [PubMed] [Google Scholar]
- 30.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 31.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ørskov, F. 1951. On the occurrence of Escherichia coli belonging to O-group 26 in cases of infantile diarrhea and white scours. Acta Pathol. Microbiol. Scand. 29:373-378. [PubMed] [Google Scholar]
- 34.Ørskov, F. 1954. Studies on a number of Escherichia coli strains belonging to O group 86. Acta Pathol. Microbiol. Scand. 35:179-186. [DOI] [PubMed] [Google Scholar]
- 35.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, C. H., H. J. Kim, and D. L. Dixon. 2002. Importance of testing stool specimens for Shiga toxins. J. Clin. Microbiol. 40:3542-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 181:1496-1500. [DOI] [PubMed] [Google Scholar]
- 40.Phillips, A. D., S. Navabpour, S. Hicks, G. Dougan, T. Wallis, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer's patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramachandran, V., M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2001. The common ovine Shiga toxin 2-containing Escherichia coli serotypes and human isolates of the same serotypes possess a Stx2d toxin type. J. Clin. Microbiol. 39:1932-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reece, S., C. P. Simmons, R. J. Fitzhenry, M. Batchelor, C. Hale, S. Matthews, A. D. Phillips, G. Dougan, and G. Frankel. 2002. Mutagenesis of conserved tryptophan residues within the receptor-binding domain of intimin: influence on binding activity and virulence. Microbiology 148:657-665. [DOI] [PubMed] [Google Scholar]
- 43.Reid, S. D., D. J. Betting, and T. S. Whittam. 1999. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J. Clin. Microbiol. 37:2719-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarr, C. L., and T. S. Whittam. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J. Bacteriol. 184:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, J., and K. A. Bettelheim. 1966. The action of chloroform-killed suspensions of enteropathogenic Escherichia coli in ligated rabbit-gut segments. J. Gen. Microbiol. 42:309-313. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, J., and R. E. Chanter. 1952. The isolation of serological types of bacteri in two residential nurseries and their relation to infantile gastroenteritis. J. Pathol. Bacteriol. 64:715-727. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, J., and R. E. Chanter. 1955. Escherichia coli O128 causing gastroenteritis of infants. J. Clin. Pathol. 8:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trabulsi, L. R., R. Keller, and T. A. Tardelli Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welinder-Olsson, C., M. Badenfors, T. Cheasty, E. Kjellin, and B. Kaijser. 2002. Genetic profiling of enterohemorrhagic Escherichia coli strains in relation to clonality and clinical signs of infection. J. Clin. Microbiol. 40:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whittam, T. S., and E. A. McGraw. 1996. Clonal analysis of EPEC serogroups. Rev. Microbiol. 27:7-16. [Google Scholar]
- 52.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wieler, L. H., T. K. McDaniel, T. S. Whittam, and J. B. Kaper. 1997. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol. Lett. 156:49-53. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, W., M. Bielaszewska, T. Kuczius, and H. Karch. 2002. Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, W. L., B. Köhler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]