Abstract
We investigated the prevalence, molecular epidemiology, and clinical significance of heterogeneous glycopeptide-intermediate Staphylococcus aureus (hGISA) isolates in 48 liver transplant recipients infected or colonized with methicillin-resistant S. aureus over a 5-year period. Strains were screened for hGISA on Mueller-Hinton agar containing 5 mg of teicoplanin per liter. Heterogeneous glycopeptide resistance was confirmed by the E-test method with a dense inoculum and a simplified method of population analysis. hGISA strains were found in 13 (27%) of the 48 patients studied. Eleven of the 13 strains shared a common multiresistant phenotype with homogeneous methicillin resistance and gentamicin resistance, and they were closely related according to the results of pulsed-field gel electrophoresis. Only 2 of the 13 patients infected or colonized with hGISA strains had previously received glycopeptide therapy. Most patients were successfully treated with vancomycin, but one patient who failed to respond to vancomycin subsequently died. These results suggest that the high prevalence of hGISA among our patients was due to the clonal spread of a multiresistant strain.
The first glycopeptide-intermediate Staphylococcus aureus (GISA) strain was found in Japan in 1997 (13). Since then, other cases of GISA infection have been reported in the United States, France, and Korea (7, 15, 21, 23, 26). These strains, for which vancomycin and teicoplanin MICs are >4 and <32 mg/liter, are usually isolated from patients who have severe underlying illnesses and who have previously been exposed to long-term vancomycin therapy. However, their frequency appears to be extremely low. Heterogeneous GISA strains (hGISA) were first described by Hiramatsu et al. (14) and are defined as strains for which vancomycin MICs are 1 to 4 mg/liter but which contain subpopulations that can grow on agar plates supplemented with 4 mg of vancomycin per liter. These strains may be the first step in the development of GISA strains (14). The reported prevalence of hGISA is highly variable, ranging from 0.6 to 65% by country and patient population (1, 4, 8, 10, 12, 14, 16, 20, 22, 28). The clinical significance of hGISA strains remains controversial. In some studies (1, 30) hGISA infections were found to be associated with a high rate of failure of vancomycin therapy, whereas in other studies (16) patients with hGISA infections were successfully treated with vancomycin. The clinical outcome may depend on the site of infection and other patient variables. Thus, heterogeneous resistance might be more clinically relevant in patients with a severe underlying condition, patients who have undergone surgery, or patients receiving immunosuppressive therapy, such as liver transplant (LT) recipients. Methicillin-resistant S. aureus (MRSA) is the leading cause of bacterial infection after an LT (2, 24, 25). No studies have looked at the impact of hGISA in LT recipients. In November 2002, an MRSA strain associated with a fatal infection in an LT recipient in our hospital who failed to respond favorably to vancomycin was identified as hGISA. This case initiated a retrospective study to investigate the prevalence, molecular epidemiology, and clinical significance of hGISA in LT recipients.
MATERIALS AND METHODS
Bacterial strains.
All MRSA strains isolated from LT recipients at Beaujon Hospital between January 1997 and January 2002 were included in the study. If MRSA isolates were recovered before and after glycopeptide treatment, both the pretreatment isolate and the posttreatment isolate were tested. S. aureus was identified by standard microbiological methods. Susceptibility to antibiotics was examined by the disk diffusion test on Mueller-Hinton agar according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (CASFM) (9, 18). MRSA strains were isolated from 48 (18.1%) of the 265 patients who benefited from LT during the study period. Methicillin-susceptible S. aureus was isolated from 49 of the 265 LT recipients. The most frequent sites from which MRSA was recovered were the lower respiratory tract (n = 17), a surgical wound (n = 15), blood (n = 14), and the abdominal cavity (n = 11). Several sites of infection were present in 13 patients. MRSA was isolated only from a nasal swab in eight patients.
MIC determination.
The MICs of vancomycin and teicoplanin were determined by the E-test method (AB Biodisk, Solna, Sweden) with an inoculum equivalent to a 0.5 McFarland standard on Mueller-Hinton agar according to the instructions of the manufacturer.
Screening for glycopeptide resistance.
Strains were screened for GISA and hGISA as recommended by CASFM (9). A suspension with a turbidity equivalent to a 2 McFarland standard (6 × 108 CFU/ml) was prepared, and 10 μl of the suspension was used to inoculate Mueller-Hinton agar containing 5 mg of teicoplanin per liter (prepared in-house). Reference strains S. aureus ATCC 25923 and Staphylococcus haemolyticus CIP 107204 were used as negative and positive controls, respectively. The plates were incubated at 37°C and examined after 24 and 48 h. If growth was not apparent within 48 h, the isolate was classified as glycopeptide-susceptible S. aureus (GSSA). Strains that grew on the screening agar plates were further analyzed by the E-test method and a simplified method of population analysis.
MICs were determined by the E-test as described previously for heterogeneous glycopeptide resistance detection (6, 22, 29) by using a heavy inoculum (2 McFarland units) and brain heart infusion (BHI) agar. The plates were incubated for 48 h at 37°C and examined for the presence of microcolonies indicative of heteroresistance, which were those isolates close to the edge of the growth inhibition ellipse. The interpretative criteria for hGISA categorization were vancomycin and teicoplanin MICs of ≥8 mg/liter or a teicoplanin MIC of ≥12 mg/liter (29).
Heterogeneous resistance was confirmed by a simplified method of population analysis as described previously (8, 14). The strains were cultured overnight in BHI broth, and 108 CFU was spread across the surface of BHI agar plates containing 4 mg of vancomycin per liter. The number of colonies was evaluated after 48 h of incubation at 37°C. The strain was considered heterogeneously resistant when it contained a subpopulation able to grow on this medium at a frequency of ∼10−6 (25 to 250 colonies).
Genotyping.
All isolates were typed by pulsed-field gel electrophoresis (PFGE) after DNA digestion with SmaI as described previously (2). PFGE banding patterns were compared by use of Gel Compar software (Applied Maths, Kortrijk, Belgium). The similarity between the isolates was determined by use of the Dice coefficient, and a dendrogram was constructed with clustering by the unweighted pair group method with arithmetic averages. Genotypes were numbered according to their position in the dendrogram.
Medical record review.
The patients' medical records were reviewed for pretransplantation and posttransplantation data, including demographic data, underlying disease, length of hospital stay, clinical and microbiological findings, antibiotic treatment, and outcome. Infections were determined according to the definitions of the Centers for Disease Control and Prevention.
During the study period, all candidates for LT were systematically screened for MRSA nasal carriage before surgery (2). Perioperative prophylaxis consisted of cefoxitin, which was used in association with vancomycin for patients identified as being previous MRSA nasal carriers. Immunosuppressive therapy consisted of azathioprine, corticosteroids, and cyclosporine or tacrolimus, as described previously (2).
Statistical analysis.
Data were entered into the Epi-Info 2000 program. Patients infected or colonized with hGISA and those infected or colonized with GSSA were compared by the chi-square test or Fisher's exact test for proportions and the Mann-Whitney test for means. The level of statistical significance was a P value of ≤0.05.
RESULTS
Detection of hGISA.
MRSA was isolated from 48 of the 265 LT recipients. The strains from 13 (27%) of the 48 patients grew on the glycopeptide screening agar plates within 48 h, resulting in a confluent growth or the formation of more than 10 colonies. These strains were also categorized as hGISA by the E-test method with a dense inoculum and BHI agar. The simplified method of population analysis confirmed that they contained a subpopulation able to grow on BHI agar containing 4 mg of vancomycin per liter, yielding 25 to 250 colonies after 48 h of incubation with an inoculum size of 108 CFU/ml. In contrast, the GSSA strains did not grow in the presence of 4 mg of vancomycin per liter. Thus, hGISA was isolated from 27% of LT recipients infected or colonized with MRSA and from 13 of 265 (4.9%) of all LT recipients between January 1997 and January 2002.
MIC determination.
The MIC ranges obtained by the standard E-test method for hGISA were 3 to 4 mg/liter for vancomycin and 4 to 8 mg/liter for teicoplanin. The MIC ranges for GSSA strains were 1 to 2 mg/liter for both vancomycin and teicoplanin. For 15 of the 48 patients, including 5 patients infected or colonized with hGISA, several isolates corresponding to pre- and posttherapy isolates were tested. The susceptibilities of the consecutive isolates were identical in all cases.
Molecular epidemiology of hGISA.
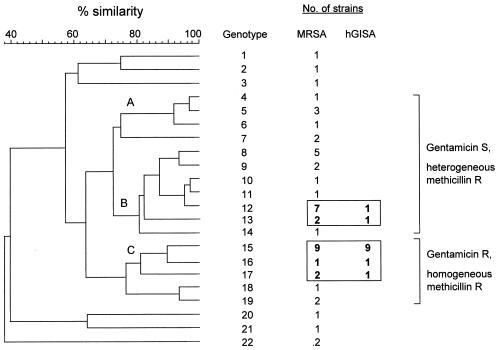
The dendrogram constructed from the PFGE results showed 22 banding patterns that differed by at least one band (Fig. 1). Three main clusters (clusters A, B, and C) were distinguished. A correlation was observed between the SmaI pattern and the antibiotic susceptibility phenotype. All of the strains in cluster C were characterized by homogeneous resistance to methicillin and resistance to gentamicin according to the results of the disk diffusion test. They were also resistant to amikacin, tobramycin, pefloxacin, erythromycin, clindamycin, rifampin, and, for most of the strains, tetracycline. This multiresistance phenotype was not found in any of the strains belonging to the other groups. Although a few gentamicin-resistant strains were found in other clusters, they differed from cluster C by their susceptibilities to erythromycin, clindamycin, or rifampin or combinations of these three antibiotics. Most strains in clusters A and B were characterized by heterogeneous resistance to methicillin and were resistant to amikacin, tobramycin, and pefloxacin but were susceptible to gentamicin, rifampin, and tetracycline.
FIG. 1.
Dendrogram constructed from the PFGE patterns of MRSA strains from 48 LT recipients. R, resistant; S, susceptible.
Eleven of the 15 strains belonging to cluster C were hGISA (Fig. 1). Nine of these 11 strains had identical banding patterns (genotype 15), and the patterns for the 2 others differed from the predominant pattern by only one band (genotype 16) and four bands (genotype 17), respectively. The two other hGISA strains had patterns completely distinct from those of the strains mentioned above and from one another, with the patterns differing by more than six bands. One of these strains was homogeneously resistant to methicillin, resistant to gentamicin, and susceptible to rifampin (genotype 13); the other one was heterogeneously resistant to methicillin and susceptible to gentamicin and rifampin (genotype 12).
Characteristics of patients.
The characteristics of the patients infected or colonized with MRSA are given in Table 1. Patients infected or colonized with hGISA and those infected or colonized with GSSA were similar in terms of age, sex, underlying disease, preoperative MRSA nasal carriage, and length of hospital stay before LT. Five (46.1%) of the 13 patients infected or colonized with hGISA had had an infection in the month preceding LT, whereas only 4 (11.4%) of the 35 patients infected or colonized with GSSA had had an infection (P = 0.05). The preoperative infection was due to MRSA in only one patient and to a nonstaphylococcal pathogen in the four other patients. Patients infected or colonized with hGISA were also more likely than patients infected or colonized with GSSA to have developed a nonstaphylococcal postoperative infection before the isolation of MRSA, although the difference was not statistically significant (P = 0.3). Only 2 of the 13 patients infected or colonized with hGISA had previously received glycopeptide therapy. In contrast, patients infected or colonized with hGISA had more frequently received a β-lactam agent in the 2 months preceding the acquisition of MRSA, before or after LT, than patients with GSSA (77% versus 40.7% [P = 0.03]).
TABLE 1.
Comparison of patients infected or colonized with hGISA and patients infected or colonized with GSSA
| Variable | Patients infected or colonized with:
|
P | |
|---|---|---|---|
| hGISA (n = 13) | GSSA (n = 35) | ||
| Mean age (yr [range]) | 48 (17-60) | 43 (16-60) | NSa |
| No. of males/no. of females | 10/3 | 25/10 | NS |
| No. (%) of patients with the following underlying liver disease: | NS | ||
| Alcoholic cirrhosis | 5 (38.5) | 13 (37.1) | |
| Posthepatitic cirrhosis | 2 (15.4) | 8 (22.9) | |
| Fulminant hepatitis | 2 (15.4) | 4 (11.4) | |
| Other | 4 (30.8) | 10 (28.6) | |
| No. of patients with MRSA nasal carriage before surgery | 2 (15.4) | 5 (14.7) | NS |
| Mean length of previous hospital stay (≤1 yr) (days) | 27 | 31 | NS |
| No. of patients with an infection before LT (≤1 month) | 6 (46.1) | 5 (15.6) | 0.05 |
| No. (%) of patients with the following postoperative complications: | |||
| Reoperated | 5 (38.5) | 13 (37.1) | NS |
| Renal deficiency | 5 (38.5) | 13 (37.1) | NS |
| Acute rejection | 3 (23.1) | 8 (22.9) | NS |
| Non-MRSA infection | 6 (46.1) | 9 (25.7) | NS |
| No. (%) of patients who previously received (≤2 mo) the following antibiotics: | |||
| β-Lactam agents | 10 (77) | 13 (40.6)b | 0.03 |
| Aminoglycosides | 1 (7.7) | 3 (9.4)b | NS |
| Fluoroquinolones | 5 (38.5) | 11 (34.4)b | NS |
| Glycopeptides | 2 (15.4) | 5 (14.3) | NS |
NS, nonsignificant.
Data were available for only 32 of the 35 patients.
Clinical significance.
Ten of the 13 patients from whom hGISA strains were isolated were infected, and only 3 were colonized. Among the patients infected or colonized with GSSA, 30 were infected and 5 were colonized. The times of onset, the sources of infection, and the rates of bacteremia were similar in the two groups (Table 2). The rate of MRSA-related mortality was not higher in patients infected with hGISA than in those infected with GSSA. The mean duration of glycopeptide treatment was also similar in the two groups. Details of the treatments and the clinical outcomes for the 10 hGISA-infected patients are presented in Table 3. One patient with a surgical wound infection was treated by debridement and did not receive systemic antibiotics. Seven patients were successfully treated with a glycopeptide, including six who were treated with vancomycin and one who was treated with teicoplanin, alone or in combination with a β-lactam agent. Another patient was initially treated with teicoplanin for catheter-related bacteremia. He did not respond favorably to therapy, and cultures of peripheral blood were still positive for hGISA 5 days after the initiation of teicoplanin and catheter removal. The median blood teicoplanin concentration was 10.2 mg/liter (range, 7.8 to 12.7 mg/liter). Teicoplanin was discontinued, and the patient was given a combination of vancomycin and cefpirome. Blood cultures were negative soon after this treatment was initiated, but defervescence was not achieved until an intra-abdominal abscess was found and drained 7 days later. In the last patient, hGISA infection was associated with a poor response to vancomycin and a fatal outcome. This patient developed fever and presented signs of shock 12 days postoperatively. Another operation was performed for intra-abdominal bleeding, and he was treated with piperacillin-tazobactam and amikacin. hGISA was isolated from blood cultures and intraabdominal specimens, and vancomycin was added to the regimen. Blood cultures became negative; but the patient's respiratory condition subsequently deteriorated, he developed tracheal bleeding, and hGISA was isolated from a bronchial distal aspirate at >106 CFU/ml. The mean trough blood vancomycin concentration was 15.3 mg/liter (range, 10.3 to 18.8 mg/liter). The patient died from shock and multiple organ failure 9 days after the initiation of vancomycin treatment.
TABLE 2.
MRSA infections and outcomes in 48 LT recipients
| Variable | Patients infected or colonized witha:
|
P | |
|---|---|---|---|
| hGISAb (n = 13) | GSSAc (n = 35) | ||
| Infection/colonization ratio | 10/3 | 30/5 | NSb |
| Time of onset (days) of MRSA infection | NS | ||
| 0-14 | 6/10 (60) | 19/30 (63) | |
| 14-30 | 3/10 (30) | 6/30 (20) | |
| >30 | 1/10 (10) | 5/30 (17) | |
| Site of infection | NS | ||
| Lower respiratory tract | 6/10 (60) | 11/30 (37) | |
| Abdomen | 4/10 (40) | 7/30 (23) | |
| Wound | 3/10 (30) | 12/30 (40) | |
| Intravascular device | 1/10 (10) | 5/30 (17) | |
| Other | 0 | 1/30 (3) | |
| Bacteremia | 3/10 (30) | 11/30 (37) | NS |
| Overall mortality | 1/13 (7.7) | 6/35 (17) | NS |
| MRSA-related mortality | 1/13 (7.7) | 2/35 (5.7) | NS |
| Mean duration (days [range]) of glycopeptide treatmentc | 18.5 (9-36) | 16 (7-56) | NS |
Data are number of patients to which the variable applies per total number of patients in the group (percent), unless otherwise indicated.
NS, nonsignificant.
Data were analyzed for surviving patients only.
TABLE 3.
Characteristics of patients with hGISA infection
| Age (yr)/sexa | Date of LT (day/mo/yr) | Liver disease | MRSA nasal carriage before LT | Previous antibiotic treatmentb | Time of onset after LT (days) | Site of infectionc | PFGE pattern | Treatment (duration [days])d | Clinical outcome |
|---|---|---|---|---|---|---|---|---|---|
| 60/M | 03/06/98 | Alcoholic cirrhosis | No | TAZ | 15 | LRTI | 15 | VAN (19) | Improved |
| 48/M | 08/06/98 | Hepatitis B and D viruses | No | AMX/CLAV | 25 | Wound | 15 | Surgery | Improved |
| 54/M | 20/10/98 | Alcoholic cirrhosis | Yes | None | 10 | Blood, peritonitis | 16 | VAN (18) | Improved |
| 49/F | 28/12/98 | Polykystosis | No | IMI, CIP | 21 | LRTI | 15 | TEI + CTX (9) | Improved |
| TEI (6) | Failure | ||||||||
| 52/M | 02/02/99 | Alcoholic cirrhosis | No | CTX, OFLO | 6 | KT, blood, LRTI, abdomen | 17 | VAN (21) + CPO (8) | Improved |
| 46/M | 06/06/99 | Fulminant hepatitis | No | OXA, OFLO, CAZ, TAZ | 60 | LRTI, wound | 15 | VAN (15) + CTX and surgery | Improved |
| 50/M | 31/04/01 | Hepatitis C virus | Yes | CIP, VAN, TAZ | 2 | LRTI | 15 | VAN (12) | Improved |
| 56/M | 16/07/01 | Alcoholic cirrhosis | No | FEP, AMIK | 14 | LRTI | 15 | VAN + FEP (14) | Improved |
| 59/M | 02/09/01 | Alcoholic cirrhosis | No | AMX/CLAV | 12 | Abdomen | 15 | VAN (36) and surgery | Improved |
| 58/M | 07/11/01 | Budd-Chiari syndrome | No | TAZ, CIP, TEI | 12 | Blood, LRTI, abdomen | 12 | VAN + TAZ (9) + AMIK and surgery | Death |
M, male; F, female.
Antibiotics received in the 2 months preceding MRSA infection. Abbreviations: AMIK, amikacin; AMX/CLAV, amoxicillin-clavulanate; CAZ, ceftazidime; CIP, ciprofloxacin; CTX, cefotaxime; FEP, cefepime; IMI, imipenem; OFLO, ofloxacin; OXA, oxacillin; TAZ, piperacillin-tazobactam; TEI, teicoplanin; VAN, vancomycin.
LRTI, lower respiratory tract infection; KT, catheter.
CPO, cefpirome. The other antibiotic abbreviations are as defined in footnote b.
DISCUSSION
Over the last few years there has been a lot of confusion about the laboratory methods used to detect heterogeneous glycopeptide resistance (11, 19). The Centers for Disease Control and Prevention recommends the use of BHI agar plates with 6 mg of vancomycin per liter for the detection of GISA, but hGISA strains fail to grow on this medium (27). CASFM recommends the screening of MRSA on Mueller-Hinton agar containing 5 mg of teicoplanin per liter, as the teicoplanin MICs are more elevated than the vancomycin MICs for hGISA and GISA (9, 22). In our study, we used three different screening methods to detect hGISA. All the strains shown to be positive by the CASFM agar screening method were subsequently shown to contain a subpopulation able to grow in the presence of 4 mg of vancomycin per liter by a simplified method of population analysis. They were also identified as hGISA by the E-test method with a dense inoculum and BHI agar, which is a reliable and sensitive method for the detection of heterogeneous glycopeptide resistance (29).
As reported previously (12, 22), the vancomycin and teicoplanin MICs for the hGISA strains were only moderately elevated compared with those for the GSSA strains. When consecutive isolates from a given patient were tested, strains with homogenous resistance were not selected after vancomycin therapy. This finding, consistent with those of another report (1), does not support the hypothesis that heterogeneous resistance is a preliminary step in the development of GISA under selective pressure with vancomycin.
The rate of hGISA among MRSA strains from our LT recipients was 27%. The results of screening consecutive nonselected MRSA isolates from Europe, South America, and Korea revealed low levels of prevalence, ranging from 0.5 to 5.8% (4, 10, 16, 20, 22, 28). Hiramatsu et al. (14) reported a highly variable prevalence in Japan, ranging from 1.3% in nonuniversity hospitals to 20% in one university hospital. Ariza et al. (1) reported a prevalence of 65% among MRSA isolates from orthopedic patients with surgical infections, most of whom had orthopedic implants. LT recipients represent a particular group of patients, characterized by the severity of their illness, the requirement for an intensive care unit stay, and the administration of immunosuppressive therapy after LT. Therefore, they may be a high-risk group for the acquisition of multiresistant strains of S. aureus, and the rate of hGISA in our study probably does not reflect the overall prevalence in a nonselected population.
Molecular typing by PFGE revealed that 11 of the 13 hGISA strains were highly related. Nine had identical banding patterns, and two further strains displayed very similar patterns. Moreover, these 11 hGISA strains exhibited a similar multidrug resistance phenotype, characterized by homogeneous resistance to methicillin and resistance to gentamicin. These findings suggest that these strains are closely related. Only 2 of the 13 hGISA strains had banding patterns and susceptibility phenotypes different from those of the predominant PFGE type. In contrast, most GSSA strains exhibited heterogeneous resistance to methicillin and were susceptible to gentamicin. This association between hGISA and this particular multidrug resistance phenotype has already been reported by others (12). Most of our patients infected or colonized with hGISA did not develop MRSA infection and did not receive glycopeptides before LT. This suggests that the high prevalence of hGISA in this study was due to the spread of a multiresistant strain rather than the acquisition of heterogeneous resistance by a preexisting endogenous strain following glycopeptide treatment. As the preoperative screening for MRSA nasal carriage was positive for only 2 of the 13 patients, the hGISA strain was probably acquired after LT by cross-contamination. Thus, as others have reported previously (12), this multiresistant hGISA strain may have been cryptically endemic in the LT unit and, possibly, in our hospital for several years.
At the beginning of the 1990s, most MRSA strains expressed homogeneous resistance to methicillin and were resistant to many antibiotics, including gentamicin. Since 1992, these multiresistant strains have been progressively replaced in French hospitals by new gentamicin-susceptible clones characterized by heterogeneous resistance to methicillin and susceptibility to various other antibiotics, such as rifampin (3, 5). The same trend has been observed in our hospital, and both types of strains were endemic during the study period. This changing epidemiology is probably related to the fitness and competitive growth advantage of new clones (3, 17). However, the selective pressure exerted by antibiotics, particularly β-lactam agents, may counterbalance the fitness advantage of gentamicin-susceptible MRSA, which expresses a more heterogeneous resistance to methicillin than gentamicin-resistant MRSA. This could explain why we found that LT recipients previously treated with a β-lactam agent were more likely to acquire an hGISA strain than a GSSA strain.
The clinical significance of hGISA remains controversial, and it is often difficult to determine the relative contributions of MRSA infection and the underlying conditions to the outcome (19). Despite the presence of a severe underlying disease and the use of immunosuppressive therapy, most of our LT recipients were successfully treated with glycopeptides. The MRSA-related mortality rate and duration of glycopeptide treatment did not differ significantly between hGISA-infected patients and GSSA-infected patients (Table 2). Two cases of glycopeptide therapeutic failures were observed in hGISA-infected patients. In the first patient, the serum teicoplanin level was insufficient, and there was an undrained purulent focus. Therefore, the failure of teicoplanin to eradicate the hGISA bacteremia could not be attributed to glycopeptide resistance in this patient. The other patient failed to respond favorably to vancomycin treatment, despite adequate levels in serum and the absence of undrained foci of infection, suggesting that glycopeptide resistance may have been involved in the therapeutic failure. However, this patient had a severe underlying condition and developed other postoperative complications such as intraabdominal and tracheal bleeding, and it is therefore difficult to know whether there is a direct association between the cause of death and heterogeneous glycopeptide resistance. Kim et al. (16) recently reported that most patients with hGISA infections can be successfully treated with vancomycin. Our results indicate that even in high-risk immunosuppressed patients, such as LT recipients, hGISA infection is not associated with an increased rate of glycopeptide treatment failure.
In conclusion, this study reports a high prevalence of hGISA in LT recipients, related to the dissemination of a multidrug-resistant strain expressing homogeneous resistance to methicillin. Most patients acquired hGISA after LT, and the risk of acquisition may have been increased by the previous administration of a β-lactam agent. Although glycopeptide resistance was potentially involved in the death of one patient who failed to respond to vancomycin, most patients infected or colonized with hGISA were successfully treated with vancomycin.
REFERENCES
- 1.Ariza, J., M. Pujol, J. Cabo, C. Pena, N. Fernandez, T. Linares, J. Ayats, and F. Gudiol. 1999. Vancomycin in surgical infections due to methicillin-resistant Staphylococcus aureus with heterogeneous resistance to vancomycin. Lancet 353:1587-1588. [DOI] [PubMed] [Google Scholar]
- 2.Bert, F., J.-O. Galdbart, V. Zarrouk, J. Le Mée, F. Durand, F. Mentré, J. Belghiti, N. Lambert, and B. Fantin. 2000. Association between nasal carriage of Staphylococcus aureus and infection in liver transplant recipients. Clin. Infect. Dis. 31:1295-1299. [DOI] [PubMed] [Google Scholar]
- 3.Bertrand, X., M. Thouverez, and D. Talon. 2000. Antibiotic susceptibility and genotypic characterization of methicillin-resistant Staphylococcus aureus strains in eastern France. J. Hosp. Infect. 46:280-287. [DOI] [PubMed] [Google Scholar]
- 4.Bierbaum, G., K. Fuchs, W. Lenz, and H.-G. Sahl. 1999. Presence of Staphylococcus aureus with reduced susceptibility to vancomycin in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 18:691-696. [DOI] [PubMed] [Google Scholar]
- 5.Blanc, D. S., P. Francioli, A. Le Coustumier, L. Gazagne, E. Lecaillon, P. Gueudet, F. Vandenesh, and J. Etienne. 2001. Reemergence of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus in France: a phylogenetic approach. J. Clin. Microbiol. 39:2287-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolmström, A., A. Karsson, and P. Wong. 1999. Micro-method conditions are optimal for detection of low level glycopeptide resistance in staphylococci. Clin. Microbiol. Infect. 5(Suppl. 3):113. [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1997. Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morb. Mortal. Wkly. Rep. 46:765-766. [PubMed] [Google Scholar]
- 8.Chesneau, O., A. Morvan, and N. El Solh. 2000. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 45:887-890. [DOI] [PubMed] [Google Scholar]
- 9.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2002. Communiqué 2001-2002. Société Française de Microbiologie, Paris, France. [Online.] http://www.sfm.asso.fr/.
- 10.Dos Santos Soares, M. J., M. C. Da Silva-Carvalho, B. T. Ferreira-Carvalho, and A. M. S. Figueiredo. 2000. Spread of methicillin-resistant Staphylococcus aureus belonging to the Brazilian epidemic clone in a general hospital and emergence of heterogeneous resistance to glycopeptide antibiotics among these isolates. J. Hosp. Infect. 44:301-308. [PubMed] [Google Scholar]
- 11.Fridkin, S. K. 2001. Vancomycin-intermediate and -resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin. Infect. Dis. 32:108-115. [DOI] [PubMed] [Google Scholar]
- 12.Guerin, F., A. Buu-Hoi, J. L. Mainardi, G. Kac, N. Colardelle, S. Vaupré, L. Gutmann, and I. Podglajen. 2000. Outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in a Parisian hospital. J. Clin. Microbiol. 38:2985-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 14.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 15.Kim, M. N., C. H. Pai, J. H. Woo, J. S. Ryu, and K. Hiramatsu. 2000. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38:3879-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, M. N., S. H. Hwang, Y. J. Pyo, H. M. Mun, and C. H. Pai. 2002. Clonal spread of Staphylococcus aureus heterogeneously resistant to vancomycin in a university hospital in Korea. J. Clin. Microbiol. 40:1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurent, F., H. Leličvre, M. Cornu, F. Vandenesch, G. Carret, J. Etienne, and J. P. Flandrois. 2001. Fitness and competitive growth advantage of new gentamicin-susceptible MRSA clones spreading in French hospitals. J. Antimicrob. Chemother. 47:277-283. [DOI] [PubMed] [Google Scholar]
- 18.Lecaillon, E., P. Gueudet, M. Wooton, T. R. Walsh, A. P. Macgowan, and M. E. Jones. 2002. Endemic heteroresistant glycopeptide intermediate Staphylococcus aureus (hGISA) comprising unrelated clonal types and not associated with vancomycin therapy. Pathol. Biol. 50:525-529. [DOI] [PubMed] [Google Scholar]
- 19.Liñares, J. 2001. The VISA/GISA problem: therapeutic implications. Clin. Microbiol. Infect. 7:S8-S15. [DOI] [PubMed] [Google Scholar]
- 20.Marchese, A., G. Balistreri, E. Tonoli, E. A. Debbia, and G. C. Schito. 2000. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J. Clin. Microbiol. 38:866-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ploy, M. C., C. Grélaud, C. Martin, L. de Lumley, and F. Denis. 1998. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet 351:1212. [DOI] [PubMed] [Google Scholar]
- 22.Reverdy, M. E., S. Jarraud, S. Bobin-Dubreux, E. Burel, P. Girardo, G. Lina, F. Vandenesch, and J. Etienne. 2001. Incidence of Staphylococcus aureus with reduced susceptibility to glycopeptides in two French hospitals. Clin. Microbiol. Infect. 7:267-272. [DOI] [PubMed] [Google Scholar]
- 23.Sieradzki, K., R. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 24.Singh, N., G. L. Paterson, F. Y. Chang, T. Gayowski, C. Squier, M. M. Wagener, and I. R. Marino. 2000. Methicillin-resistant Staphylococcus aureus: the other emerging resistant gram-positive coccus among liver transplant recipients. Clin. Infect. Dis. 30:322-327. [DOI] [PubMed] [Google Scholar]
- 25.Singh, N., T. Gayowski, J. D. Rihs, M. M. Wagener, and I. R. Marino. 2001. Evolving trends in multiple-antibiotic-resistant bacteria in liver transplant recipients: a longitudinal study of antimicrobial susceptibility patterns. Liver Transplant. 7:22-26. [DOI] [PubMed] [Google Scholar]
- 26.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, and W. R. Jarvis. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 27.Tenover, F. C. 2000. VRSA, VISA and GISA: the dilemma behind the name game. Clin. Microbiol. Newsl. 22:49-53. [Google Scholar]
- 28.Trakulsomboon, S., S. Danchaivijitr, Y. Rongrungruang, C. Driraputra, W. Susaemgrat, T. Ito, and K. Hiramatsu. 2001. First report of methicillin-resistant Staphylococcus aureus with reduced susceptibility to vancomycin in Thailand. J. Clin. Microbiol. 39:591-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh, T. R., A. Bolmström, A. Qwärnström, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong, S. S. Y., T. K. Ng, W.-C. Yam, D. N.-C. Tsang, P. C.-Y. Woo, S. K.-S. Fung, and K.-Y. Yuen. 2000. Bacteremia due to Staphylococcus aureus with reduced susceptibility to vancomycin. Diagn. Microbiol. Infect. Dis. 36:261-268. [DOI] [PubMed] [Google Scholar]