Abstract
A duplex real-time SYBR Green LightCycler PCR (LC-PCR) assay with DNA extraction using the QIAamp DNA Stool Mini kit was evaluated with regard to detection of 8 of 17 species of food- or waterborne pathogens in five stool specimens in 2 h or less. The protocol used the same LC-PCR with 20 pairs of specific primers. The products formed were identified based on a melting point temperature (Tm) curve analysis. The 17 species of food- or waterborne pathogens examined were enteroinvasive Escherichia coli, enteropathogenic E. coli, enterohemorrhagic E. coli, enterotoxigenic E. coli, enteroaggregative E. coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Yersinia pseudotuberculosis, Campylobacter jejuni, Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, Aeromonas spp., Staphylococcus aureus, Clostridium perfringens, and Bacillus cereus. No interference with the LC-PCR assay was observed when stool specimens were artificially inoculated with each bacterial species. The detection levels were approximately 105 food- or waterborne pathogenic bacteria per g of stool. The protocol for processing stool specimens for less than 104 food- or waterborne pathogenic bacteria per g of stool requires an overnight enrichment step to achieve adequate sensitivity. However, the rapid amplification and reliable detection of specific genes of greater than 105 food- or waterborne pathogenic bacteria per g in samples should facilitate the diagnosis and management of food- or waterborne outbreaks.
The traditional cultural techniques for the direct isolation and identification of food- or waterborne pathogens from stool specimens in food poisoning outbreaks are time-consuming and laborious. Efforts have been made in diagnostic laboratories to reduce the time required for identification of food- or waterborne pathogens. Real-time PCR is currently used for the diagnosis of infectious agents, and there are few reports of the application of real-time PCR for the direct detection of Escherichia coli strain O157:H7 (11) and Plesiomonas shigelloides (20) in stool specimens. However, traditional PCR is often used for the detection of infectious agents from stool specimens and food samples, and there are a number of previous reports of PCR use for detection of food- or waterborne pathogens, including E. coli (6, 14, 12, 27, 37, 38, 40), Salmonella spp. (26), Shigella spp. (34), pathogenic Yersinia spp. (18, 30), Campylobacter jejuni (4, 19, 25, 39), Vibrio cholerae (21, 22), Vibrio parahaemolyticus (13, 24), Vibrio vulnificus (38), Aeromonas spp. (16), Staphylococcus aureus (2), Clostridium perfringens (15), and Bacillus cereus (38). The published PCR protocols used for detection of food- or waterborne pathogens differ and include the use of different PCR cycler machines, annealing temperatures, and buffer systems. For routine laboratory analysis, the development of streamlined PCR methods would be ideal. In addition, not all of the published PCR methods are so sensitive or specific. Therefore, careful choice of PCR primers, modification of some existing PCR primers, and development of new PCR methods for detection and identification of a wide range of food- or waterborne pathogens are all necessary for the development of a standardized PCR protocol (38). Moreover, interference and inhibitors should be considered when using PCR to detect food- or waterborne pathogens in stool specimens. In a recent study, the efficiency of the QIAamp DNA Stool Mini kit (Qiagen, Hilden, Germany) for extraction of PCR-quality DNA from liquid stool specimens was checked (4). The method's efficiency rating of 86%, based on positive PCR signals for the target, was investigated previously (9). Moreover, traditional PCR methods require amplification in a thermocycler and product separation by gel electrophoresis, both of which are time-consuming and laborious. However, products of PCR can also be detected by using fluorescent probes or a DNA binding dye, such as SYBR Green. The non-sequence-specific SYBR Green assay is less expensive than the fluorescent probe-based assays utilizing Taqman probes or molecular beacons (14). Real-time PCR assays can be automated and are sensitive and rapid. They can also quantify PCR products with greater reproducibility while eliminating the need for post-PCR processing, thus preventing carryover contamination (14). Our study is a preliminary evaluation of a duplex real-time SYBR Green fluorescent PCR assay carried out with a LightCycler (LC) to directly detect food- or waterborne pathogens in DNA extracted from stool specimens. We used the newly available QIAamp DNA Stool Mini kit (Qiagen). We identified the products formed by using a Tm curve analysis.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study and their sources are listed in Table 1. Aerobic bacteria, except for Yersinia spp., Vibrio spp., and Aeromonas spp., were cultured overnight at 37°C in tryptic soy broth yeast extract medium (TSBYE). Yersinia spp. isolates were cultured in TSBYE at 28°C for 48 h. Vibrio spp. and Aeromonas spp. isolates were cultured overnight at 37°C in alkali peptone water (pH 8.3). The TSBYE contained 30 g of tryptic soy broth (Difco Laboratories, Detroit, Mich.), 6 g of yeast extract (Difco), and 1,000 ml of water. The alkali peptone water contained 5 g of bact-peptone (Difco), 10 g of NaCl, and 1,000 ml of water. The Clostridium perfringens strains were cultured overnight at 37°C in TSBYE using anaerobic incubation (GasPack; BBL) overnight at 37°C. The Campylobacter jejuni strains were cultured at 37°C for 48 h in Preston medium using microaerobic incubation (GasPack; BBL).
TABLE 1.
Bacterial strains assayed by LC-PCR
| Bacterial strain (gene) | Source | PCR result with each primer sete:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| virA | EAE | JMS1 | JMS2 | LT | STa | EAST | aggR | invA | yadA | JL | rtx C | CT | Tdh | Trh | VV | AHC | SA | BC | GAP | ||
| Escherichia coli EIEC O124:HNM (virA) | EA32a | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| E. coli EPEC O55 (eaeA) | EC-2736b | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| E. coli EPEC O153 (eaeA and astA) | EC-2649b | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | ||
| E. coli EHEC O26:H11 (Stx1) | SE-02005 | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| E. coli EHEC O157:H7 (Stx2) | SE020025 | − | + | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| E. coli EHEC O157:H7 (Stx1 and Stx2) | SE-02027 | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| E. coli ETEC O148 (LT, ST, and astA) | EC-3515b | − | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| E. coli ETEC O169 (ST and astA) | EC-4725b | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| E. coli EAEC O111 (aggR and astA) | EC-4131b | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − |
| Shigella sonnei | I00031 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Shigella flexneri | I00032 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Salmonella enteritidis | Sal-2339 | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| Salmonella enterica serovar Typhimurium | Sal-2304 | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| Y. enterocolitica O3/B4 | Pa241 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| Y. pseudotuberculosis O4b | SP988 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| Campylobacter jejuni | SC 009 | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| V. cholerae O1 | ATCC 14035 | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| V. cholerae O139 | NIID63-93c | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − |
| V. cholerae O22 | NIID169-68c | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| V. cholerae non-O1 | SVP84 | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| V. parahaemolyticus O3:K6 (tdh) | SVP02 | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| V. parahaemolyticus O3:K6 (trh) | NIIDK4c | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | |
| V. vulnificus | SVV1526 | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + | − | − | − | − |
| A. hydrophila O1 | ATCC 7966 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − |
| A. trota O4 | NIID2315-80 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − |
| Staphylococcus aureus | SS 05 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − |
| B. cereus | SB 11 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| Clostridium perfringens | H2d | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
Strain kindly donated by K. Sugiyama, Shizuoka Prefectural Institute of Public Health, Shizuoka, Japan.
J. Yatsuyanagi, Akita Prefectural Institute of Public Health, Akita, Japan.
E. Arakawa, Natural Institute of Infectious Disease (Tokyo, Japan).
S. Kaneko, Tokyo Metropolitan Institute of Public Health.
+, positive result; −, negative result. See Table 2 for primer sets.
Viable counts were obtained by culturing each dilution (10 μl) overnight at 37°C on tryptic soy agar (TSA) plates for aerobic bacteria and TSA plates containing 3% NaCl for Vibrio spp. Yersinia spp. strains were cultured at 28°C for 48 h. The Clostridium perfringens strains were cultured on TSA overnight at 37°C using anaerobic conditions. The Campylobacter jejuni strains were cultured at 37°C for 48 h on Skirrow agar plates under microaerobic conditions.
Primers used in duplex LC-PCR.
The 20 LC-PCR primer pairs used in this study for the detection of E. coli (enteroinvasive E. coli [EIEC], enteropathogenic E. coli [EPEC], enterohemorrhagic E. coli [EHEC], enterotoxigenic E. coli [ETEC], and enteroaggregative E. coli [EAEC]), Salmonella spp., Shigella spp., Yersinia enterocolitica, Yersinia pseudotuberculosis, Campylobacter jejuni, V. cholerae, V. parahaemolyticus, V. vulnificus, Aeromonas spp., S. aureus, Clostridium perfringens, and B. cereus are listed in Table 2. The primer set of yadA667-F and yadA851-R for the detection of Y. enterocolitica and Y. pseudotuberculosis was constructed from the yadA gene on the plasmid present in virulent Yersinia spp. (33). Other primer pairs were those used in earlier publications (see references listed in Table 2). All oligonucleotide primers were synthesized by Invitrogen (Yokohama, Japan) and were of unpurified grade.
TABLE 2.
LC-PCR primers and the LC-PCR sensitivities
| Species and subgroups | Target gene (description) | PCR primer and sequence (5′-3′) | Product size (bp) | Tma | Sensitivityb
|
Reference | |
|---|---|---|---|---|---|---|---|
| Direct LC-PCR | LC-PCR after enrichment | ||||||
| Escherichia coli | |||||||
| EPEC and EHEC | eaeA (E. coli attaching and effacing) | EAE-a, ATGCTTAGTGCTGGTTTAGG | 248 | 78.2 ± 0.2 | 3 | 3.3 | 37 |
| EAE-b, GCCTTCATCATTTCGCTTTC | |||||||
| EHEC | stx1 (Shiga toxin) | JMS1F, GTCACAGTAACAAACCGTAACA | 95 | 81.4 ± 1.0 | 4.1 | 2.6 | 14 |
| JMS1R, TCGTTGACTACTTCTTATCTGGA | |||||||
| stx2 | JMS2F, CGACCCCTCTTGAACATA | 108 | 81.4 ± 0.3 | 4 | 2 | 14 | |
| JMS2R, GATAGACATCAAGCCCTCGT | |||||||
| ETEC (LT) | LT (heat-labile enterotoxin) | LT-1, AGCAGGTTTCCCACCGGATCACCA | 132 | 81.4 ± 0.3 | 4.3 | 2.3 | 12 |
| LT-2, GTGCTCAGATTCTGGGTCTC | |||||||
| ETEC (ST) | ST (heat-stable enterotoxin) | STa-F, GCTAATGTTGGCAATTTTTATTTCTGTA | 190 | 76.6 ± 0.2 | 4.5 | 3.5 | 6 |
| STa-R, AGGATTACAACAAAGTTCACAGCAGTAA | |||||||
| EAEC | astA (EAECheat-stable enterotoxin) | EAST-1S, GCCATCAACACAGTATATCC | 106 | 84.9 ± 0.6 | 3.6 | 2 | 40 |
| EAST-1AS, GAGTGACGGCTTTGTAGTCC | |||||||
| aggR (transcriptional activator for EAEC aggregative adherence fimbria I expression) | aggRks1, GTATACACAAAAGAAGGAAGC | 254 | 78.9 ± 0.2 | 4.6 | 2 | 27 | |
| aggRkas2, ACAGAATCGTCAGCATCAGC | |||||||
| EIEC | virA (virulence of plasmid) | virA-F, CTGCATTCTGGCAATCTCTTCACA | 215 | 81.4 ± 0.3 | 3.3 | 6 | 34 |
| virA-R, TGATGAGCTAACTTCGTAAGCCCTCC | |||||||
| Shigella spp. | virA | 4.8 | 6 | ||||
| Salmonella spp. | invA (invasion) | invA 139, GTGAAATTATCGCCACGTTCGGGCAA | 284 | 85.8 ± 0.4 | 5 | 1.3 | 26 |
| invA 141, TCATCGCACCGTCAAAGGAACC | |||||||
| Yersinia enterocolitica | yadA (Yersinia adhesion) | yadA667-F, TGTTCTCATCTCCATATGC | 203 | 85.5 ± 0.8 | 4.5 | 2.9 | This study |
| yadA851-R2, TCCTTTCGCTGCTTCAGCA | |||||||
| Y. pseudotuberculosis | yadA | >203 | 84.3 ± 0.3 | 5.7 | 1.6 | ||
| Campylobacter jejuni | gyrA (DNA gyrase: a primary target of the fluoroquinolone antibiotics) | JL238, TGGGTGCTGTTATAGGTCGT | 192 | 80.3 ± 0.3 | 3 | 2 | 39 |
| JL239, GCTCATGAGAAAGTTTACTC | |||||||
| Vibrio cholerae | RTX toxin (repeat in toxin) | rtxC-F, CGACGAAGATCATTGACGAC | 265 | 85.8 ± 0.5 | 5 | 2 | 3 |
| rtxC-R, CATCGTCGTTATGTGGTTGC | |||||||
| CT (cholera toxin) | CT-F, ACAGAGTGAGTACTTTGACC | 308 | 80.4 ± 0.3 | 4.8 | 2.3 | 22 | |
| CT-R, ATACCATCCATATATTTGGGAG | |||||||
| V. parahaemolyticus | tdh (thermostable direct hemolysin) | Tdh199-F, GGTACTAAATGGCTGACATC | 251 | 80.4 ± 0.4 | 4.7 | 2 | 24 |
| Tdh199-R, CCACTACCACTCTCATATGC | |||||||
| trh (tdh-related hemolysin) | Trh250-F, GGCTCAAAATGGTTAAGCG | 250 | 79.6 ± 0.1 | 4.7 | 2 | 24 | |
| Trh251-R, CATTTCCGCTCTCATATGC | |||||||
| V. vulnificus | Cytotoxin hemolysin | VV-1, CTCACTGGGGCAGTGGCT | 383 | 84.4 ± 0.3 | 5 | 1.9 | 38 |
| VV-2, CCAGCCGTTAACCGAACCA | |||||||
| Aeromonas hydrophila | Cytolytic enterotoxin | AHCF1, GAGAAGGTGACCACCAAGAACA | 232 | 89.0 ± 0.5 | 4.7 | 4 | 16 |
| AHCR1, AACTGACATCGGCCTTGAACTC | |||||||
| Clostridium perfringens | Enterotoxin | GAP11, GGTTCATTAATTGAAACTGGTG | 154 | 76.0 ± 0.6 | 3.9 | 3.8 | 15 |
| GAP12, AACGCCAATCATATAAATTACAGC | |||||||
| Staphylococcus aureus | Nuclease | SA-1, GCGATTGATGGTGATACGGTT | 276 | 79.0 ± 0.3 | 4.5 | 6.9 | 2 |
| SA-2, CAAGCCTTGACGAACTAAAGC | |||||||
| Bacillus cereus | Hemolysin | BC-1, CTGTAGCGAATCGTACGTATC | 185 | 79.8 ± 0.9 | 3.9 | 8 | 38 |
| BC-2, TACTGCTCCAGCCACATTAC | |||||||
Values represent means ± standard deviations of 10 to 18 tests.
Sensitivity values represent the minimum numbers of cells necessary for the assay to yield positive LC-PCR results. Values are given as number of cells log10 per gram.
LC-PCR with broth cultures.
One milliliter of broth culture was centrifuged at 12,000 × g for 3 min. The pellet was then washed in 1 ml of distilled water, centrifuged, and suspended into 1 ml of distilled water. Each 200 μl of suspension, containing 107 to 109 food- or waterborne bacterial cells, was treated with the QIAamp DNA Stool Mini kit (Qiagen) according to the manufacturer's instructions. However, we opted to incubate the specimens in lysis buffer at 95°C rather than at 70°C (4, 20). All runs included one negative DNA control consisting of PCR-grade water and a DNA sample extracted from a pathogenic bacterium. For LC-PCR, we used glass capillary tubes and LC FastStart SYBR Green kits (Roche Diagnostics, Laval, Quebec, Canada) and the LC instrument as described by the manufacturer. Each reaction tube contained 2 μl of LC Master SYBR Green I, 1.6 μl of 25 mM MgCl2 (final concentration, 3 mM), 12.4 μl of PCR-grade H2O, 2 μl of a 10 mM concentration of primer set, and 2 μl of template DNA in a 20-μl PCR mixture. The cycling profile for the assay consisted of 95°C for 10 min, which was followed by 32 to 42 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 10 s, and elongation at 72°C for 15 s. Fluorescence signals were measured once in each cycle at the end of the extension step. After PCR amplification, Tm curve analysis was performed. The LC-PCR products were cooled to 65°C and then heated to 95°C at a rate of 0.2°C per s. The obtained fluorescence signals were continuously monitored to confirm amplification specificity during 1 h of analyzing time. The Tm peaks of the products were calculated for 10 or more assays on each sample and were based on the initial fluorescence curve (F/T) by plotting the negative derivative of fluorescence over temperature versus temperature (−dF/dT versus T).
Duplex LC-PCR with stool samples.
Stool samples (1 g) were weighed aseptically, placed into sterile tubes, and homogenized with 9 ml of distilled water. Two hundred microliters of stool suspension was then treated with the QIAamp DNA Stool Mini kit (Qiagen), and duplex LC-PCR was amplified as described above. In artificial stool contamination experiments, each 20-μl bacterial suspension (including 107 to 109 CFU per ml) was inoculated into 180 μl of a 10-fold suspension of pathogenic bacterium-free stool sample, which was then treated with QIAamp DNA Stool Mini kits. For direct duplex LC-PCR with stool samples, each reaction tube contained 2 μl of LC Master SYBR Green I, 1.6 μl of 25 mM MgCl2, 10.4 μl of PCR-grade H2O, 2 μl of a 10 mM concentration from each of two primer sets, and 2 μl of template DNA in a 20-μl PCR mixture. Two primer sets, each having different Tm curves based on the illness characteristics and incubation period (Fig. 1), were selected for the detection of bacteria. To determine the detection limit in the stool specimens of the assay, a serial 10-fold dilution of the above DNA samples in PCR-grade water was amplified with LC-PCR. To quantify target bacteria in stool specimens in subsequent experiments, DNA samples extracted from the stool samples were artificially inoculated with the target bacteria and were used for the formation of a standard curve. Two microliters of a serial 10-fold dilution of DNA in PCR-grade water was prepared and analyzed under the conditions specified above.
FIG. 1.
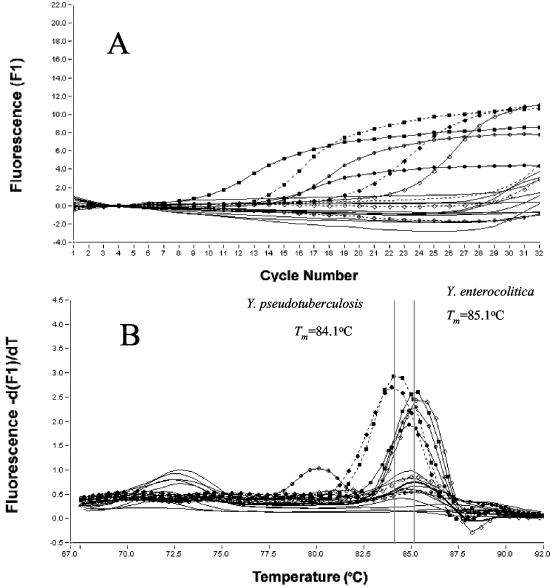
Amplification (A) of yadA in LC-PCR assays and melting curve analysis (B) on amplification products of Y. enterocolitica, Y. pseudotuberculosis, and 11 nonpathogenic Yersinia strains. - - - - - - - - , water (negative control); —⋄—, Y. enterocolitica O3/B4; —•—, Y. enterocolitica O5,27/B2; —○—, Y. enterocolitica O8/B1B; —▪—, Y. enterocolitica O9/B2; - - - - ⋄- - - - , Y. enterocolitica O3/B4 plasmid negative; - - - - •- - - - , Y. pseudotuberculosis O3; - - - - ○- - - - , Y. pseudotuberculosis O3 plasmid negative; - - - - ▪- - - - , Y. pseudotuberculosis O4a; ____, 11 nonpathogenic strains of Yersinia spp.
LC-PCR with stool samples after enrichment.
Cotton swabs of stool samples (0.1 g) were suspended in enrichment broth medium (10 ml) for each food- or waterborne bacterium, as described above, except for cultures of aerobic bacteria in buffered peptone water (BPW; BBL). In artificial stool contamination experiments, 10 μl of a serial 10-fold dilution of food- or waterborne pathogenic bacterial cells was added to 10 ml from a stool suspension. One milliliter of enrichment culture was centrifuged at 12,000 × g for 3 min. The pellet was then washed in 1 ml of distilled physiological saline, centrifuged, and suspended in 500 μl of distilled water. Each 200-μl suspension was treated with QIAamp DNA Stool Mini kits and amplified by LC-PCR as described above.
Application of duplex LC-PCR for instances of waterborne outbreak.
The use of duplex LC-PCR was experimentally tested in a waterborne outbreak which occurred in 23 out of 33 elementary school children (age range, 9 to 12 years) and three teachers. The outbreak happened during a school excursion from drinking water at a waterfall in a mountainous area of the town Yokota in the Shimane Prefecture on 4 October 2002. In 26 patients, acute gastrointestinal symptoms, such as abdominal pain (92%), diarrhea (81%), vomiting (23%), nausea (15%), and fever (23%), occurred 6 to 78 h after drinking water from the waterfall. To investigate the possibility of infection by acute gastrointestinal organisms, 11 stool specimens were collected from symptomatic patients with diarrhea and abdominal pain, and 20 liters of water from the waterfall was collected on 10 October 2002. Eleven other stool specimens were collected from symptomatic patients on 11 October 2002. Seven stool diarrhea specimens were selected from the 11 stool specimens brought into our laboratory at 7:00 p.m. on 10 October 2002. They were screened by using LC-PCR within 3 h. The next morning, the remaining four normal or hard stool specimens were tested with LC-PCR. The specimens were diluted 10-fold with distilled water. Within 1 h, the template DNA samples from 200 μl of these stool suspensions were directly extracted using QIAamp DNA Stool Mini kits. In anticipation of a positive reaction, the duplex real-time LC-PCR was performed within 1 h with 30 glass capillary tubes on the LC instrument (procedure performed in one run). Three groups of two primer sets were used for the detection of the following gastrointestinal infectious organisms: Shigella spp. and Campylobacter jejuni (primer sets of virA-F and virA-R and JL238 and JL239), Salmonella spp. and ETEC of heat-labile enterotoxin (LT) positive (primer sets of invA 139 and invA 141 and LT-1 and LT-2), and EHEC (primer sets of JMS1F and JMS1R and JMS2F and JMS2R). Next, the single LC-PCR, with a primer pair of EAE-a and EAE-b for detection of EPEC and EHEC was done within 1 h. This was done because at that time this duplex real-time LC-PCR assay was still being developed. Simultaneously, a conventional PCR assay was done using seven primer sets for the detection of gastrointestinal infectious organisms such as ETEC of heat-stable enterotoxin positive, EAEC (aggR and astA genes), Y. enterocolitica, Y. pseudotuberculosis, V. cholerae, V. parahaemolyticus and Clostridium perfringens. The results of the conventional PCR assay were confirmed by LC-PCR after the development of this duplex real-time LC-PCR assay. Then, stool specimens (0.1 g) and a water sample passed through a filter (0.45-μm pore size) were cultured in 10 ml of BPW overnight at 37°C. Next, DNA extracted from the enrichments was assayed with conventional PCR. The primer pairs used for detection of the EPEC eaeA gene and the EAEC astA gene showed positive reactions in the above PCR assays. The stool specimens of 15 patients (patients 8 to 22 in Table 3) collected on 10 and 11 October were examined by using the same methods described earlier on the 7 patients. Based on the PCR assays, PCR-positive samples and these enrichments were spread onto Chromocult agar (Merck) plates after HCl treatment with 0.25 M HCl-0.5% NaCl solution for 30 s (8). They were then incubated overnight at 37°C. Next, 100 blue colonies on the plates were examined with conventional PCR analysis for the presence of an eaeA gene- or the astA gene-positive organism. Finally, PCR-positive bacteria were identified and serotyped as described previously (8). Figure 2 shows fluorescent amplification curves and Tm analysis of real-time SYBR Green LC-PCR products.
TABLE 3.
Detection and isolation of EPEC and EAEC with LC-PCR and conventional PCR analysis and culture analysis of 22 stool samples and a water sample in a waterborne outbreak
| Sampling dates (yr/mo/day) | Patient no. or contaminated water | Results with detection methodd:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| EPEC (eaeA gene positive)
|
astA gene-positive E. coli
|
||||||||
| Direct analysis
|
After enrichment
|
Direct analysis
|
After enrichment
|
||||||
| CFU/g by LC-PCRa | Isolation (CFU/g) | Conventional PCR | Serotype of isolates | CFU/g by LC-PCRa | Isolation | Conventional PCR | Serotype of isolates | ||
| 2002/10/10 | 1 | −*b | − | − | − | − | − | ||
| 2 | −* | − | − | − | − | − | |||
| 3 | −* | − | − | − | − | − | |||
| 4 | *2 × 107 | 2 × 107 | + | O125 | 2 × 108 | + | OUT | ||
| 5 | −* | − | − | − | − | − | |||
| 6 | −* | − | − | − | − | − | |||
| 7 | −* | − | − | − | − | − | |||
| 8 | − | − | − | − | − | − | |||
| 9 | − | − | − | − | − | + | O1 | ||
| 10 | − | − | − | − | − | − | |||
| 11 | − | − | − | − | − | − | |||
| Contaminated water | NTc | NT | + | O166, OUT | NT | NT | + | O27, OUT | |
| 2002/10/11 | 12 | − | NT | − | − | NT | − | ||
| 13 | 2 × 106 | NT | + | O166, OUT | 3 × 105 | NT | + | ||
| 14 | − | NT | + | OUT | − | NT | − | ||
| 15 | − | NT | − | − | NT | − | |||
| 16 | − | NT | − | − | NT | − | |||
| 17 | − | NT | − | 4 × 106 | NT | + | OUT | ||
| 18 | 2 × 106 | NT | + | OUT | − | NT | − | ||
| 19 | − | NT | − | − | NT | − | |||
| 20 | 3 × 105 | NT | + | OUT | − | NT | − | ||
| 21 | − | NT | − | − | NT | − | |||
| 22 | − | NT | − | − | NT | − | |||
The CFU/gram of feces was counted by LC-PCR quantitative analysis.
Seven stool samples were examined first by using duplex LC-PCR on 10 October 2002.
NT, not tested.
+, positive result; −, negative result. OUT, untypeable.
FIG. 2.
Fluorescent amplification curves and melting curve analysis of real-time SYBR Green LC-PCR products using the EAE primer set (A) and EAST primer set (B) from 22 human stool samples in a waterborne outbreak-associated EPEC and astA-positive E. coli isolate. Panel A shows detection of EPEC with the EAE primer set: —○—, water (negative control); , EPEC (EC-2736); - - - - ♦- - - - , patient no. 4; - - - - ○- - - - , patient no. 13; - - - - ⋄- - - - , patient no. 18; - - - - x- - - - , patient no. 20; __, 18 other patients. Panel B shows detection of astA gene-positive E. coli with the EAST primer set: - - - - ○- - - - , water (negative control); , EAEC (EC-4131); - - - - ♦- - - - , patient no. 4; - - - - ○- - - - , patient no. 13; - - - - ▪- - - - , patient no. 17; __, 19 other patients.
Application of LC-PCR to swine cecal contents for the detection of pathogenic Yersinia organisms.
After a bacteriological culture analysis from a survey of pathogenic Yersinia from slaughtered pigs on 9 and 11 December 2002, 21 Yersinia-positive and two negative samples were selected from 200 cecal samples stored at −80°C for 3 weeks. Yersinia strains, including Y. enterocolitica serotype O3 biotype 3 VP− (O3/B3 VP−), O3/B4, O5,27/B2, and Y. pseudotuberculosis serotype O3, were isolated from 13 samples by direct culture on modified Irgasan-novobiocin (IN) agar plates, which were made by adding novobiocin (2.5 mg), esculin (1 g), and ferric citrate (0.5 g) (all from Wako Pure Chemical Industries, Ltd., Tokyo, Japan) into 1 liter of Yersinia-selective agar base (Difco). The number of Yersinia CFU, as determined by the culture analysis, ranged from 103 to 106 CFU/g. Eight samples that contained more than 103 Yersinia CFU/g were selected out of 30 Yersinia-positive samples from enrichment cultures with phosphate-buffered saline stored at 4°C for 3 weeks (Table 4). These samples were analyzed by LC-PCR with a primer pair of yadA667-F and yadA851-R2 for detection of pathogenic Yersinia strains. Figure 3 shows fluorescent amplification curves and melting curve analyses from 23 swine cecal samples.
TABLE 4.
Swine cecal content analysis made with LC-PCR and bacteriological cultivation
| Pig no. | Species and serotype or biotype | Results with detection method:
|
|||
|---|---|---|---|---|---|
| Direct analysis
|
After enrichment
|
||||
| Isolation (CFU/g) | By LC-PCRa | Isolation | By LC-PCRa | ||
| 1 | Y. enterocolitica O3/B3 | - | - | +b | 2 × 105 |
| 2 | − | − | + | 5 × 104 | |
| 3 | 5 × 103 | − | + | 8 × 106 | |
| 4 | 9 × 103 | − | + | 3 × 106 | |
| 5 | 1 × 104 | − | + | 7 × 105 | |
| 6 | 1 × 105 | − | + | 2 × 107 | |
| 7 | 2 × 105 | − | + | 1 × 107 | |
| 8 | O3/B4 | − | − | + | 4 × 107 |
| 9 | − | − | + | 7 × 105 | |
| 10 | 2 × 103 | − | + | 1 × 107 | |
| 11 | 3 × 104 | − | + | 6 × 104 | |
| 12 | 3 × 105 | 7 × 104 | + | 3 × 105 | |
| 13 | 4 × 106 | 5 × 105 | + | 7 × 107 | |
| 14 | O5/B2 | − | − | + | 5 × 107 |
| 15 | − | − | + | 8 × 106 | |
| 16 | 3 × 103 | − | + | 3 × 107 | |
| 17 | 9 × 103 | 2 × 105 | + | 1 × 108 | |
| 18 | 6 × 105 | 4 × 104 | + | 9 × 107 | |
| 19 | Y. pseudotuberculosis O3 | − | − | + | 3 × 105 |
| 20 | − | − | + | 2 × 105 | |
| 21 | 2 × 105 | − | + | 2 × 107 | |
| 22 | Negative control | − | − | − | − |
| 23 | − | − | − | − | |
The CFU/gram of feces was counted by LC-PCR quantitative analysis.
+, positive result.
FIG. 3.
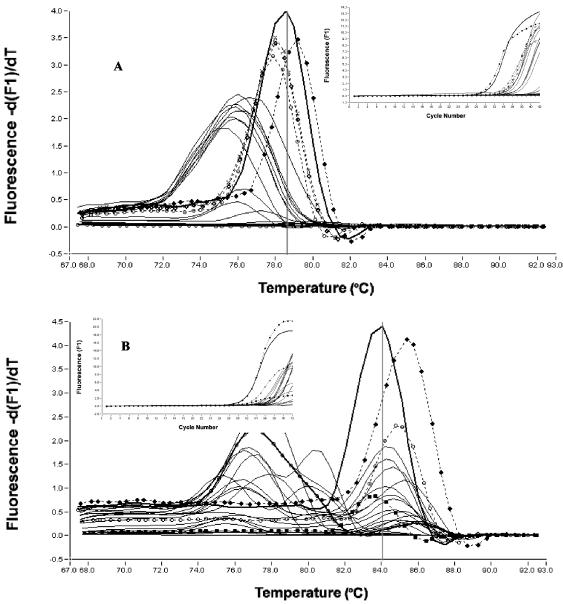
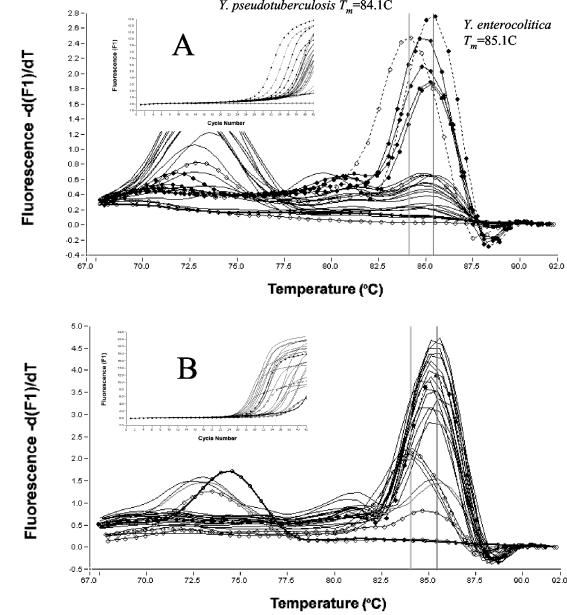
Fluorescent amplification curves and melting curve analysis of real-time SYBR Green LC-PCR products using the yadA primer set for detection of Yersinia spp. from 23 swine cecal samples by direct LC-PCR (A) and by the LC-PCR of cold enrichments (B). Symbols: open circle with thick line, water (negative control); open circle with thin line, cecal content (negative control); closed circle with dashed line, Y. enterocolitica O3/B4; open circle with dashed line, Y. pseudotuberculosis O3; closed circle with thin line, cecal content of pigs 12, 13, 17, and 18; thin line, other pigs.
RESULTS
Development of the LC-PCR procedures for duplex assay.
Real-time LC-PCR procedures which used 20 primer pairs for the detection of 17 bacterial species and five subgroups of E. coli at an annealing temperature of 55°C and a concentration of 3 mM MgCl2 were developed for the duplex assay. The primer sequence, target, LC-PCR product size, Tm values (mean plus standard deviation from a range of 10 to 18 assays), specificities, and references are summarized and listed in Tables 1 and 2. The Shigella spp. primer pair (virA-F and virA-R) cross-reacts with EIEC, the EPEC primer pair (EAE-a and EAE-b) cross-reacts with EHEC, and the EAEC primer (EAST-1S and EAST-1AS) pair cross-reacts with some strains of ETEC. A new primer pair (yadA667-F and yadA851-R2) for Y. enterocolitica was developed for the present study. The specificity of the PCR assay was tested on 19 strains belonging to virulent Y. enterocolitica and Y. pseudotuberculosis and to 11 nonpathogenic Yersinia spp., such as Y. enterocolitica biotype 1A, Y. frederiksenii, Y. intermedia, Y. kristensenii, Y. bercovieri, Y. mollaretii, Y. rhodei, and Y. aldovae, as well as other species. A 203-bp PCR product was obtained in virulent strains of Y. enterocolitica strains O:3, O:5,27, O:8, and O:9 but not in strains belonging to nonpathogenic Yersinia spp. and other species. This primer pair cross-reacted with Y. pseudotuberculosis. The product of this reaction had a larger base pair (>203 bp) than that of Y. enterocolitica but had a Tm (84.1°C) lower than that of Y. enterocolitica. But the procedure did not result in virulent plasmid-cured strains of Y. enterocolitica and Y. pseudotuberculosis and nonpathogenic strains of Yersinia spp. (Fig. 1). These results correlated well with the presence of virulent plasmid in the organism. The V. cholerae primer pair (rtxC-F and rtxC-R) detects V. cholerae strains O1 and O139 as well as non-O1 strains, except for the classical V. cholerae O1 strain (3) and V. vulnificus. The Aeromonas hydrophila primer pair (AHCF1 and AHCR1) detects virulence factors of A. hydrophila as well as Aeromonas trota (16). The Tm values of Aeromonas spp. were slightly different among the species (Table 2), which indicates the specificity of the selected primer sequences (previously noted) with the traditional PCR assay.
Sensitivity of LC-PCR with stool samples.
The concentration of DNA extracted from stool specimens (200 μl of a 10-fold suspension) using the QIAamp DNA Stool Mini kit was finally diluted to 4 × 104-fold in a capillary tube (2 μl of template DNA in a 20-μl PCR mixture). The analytical sensitivity of the assay was approximately >103 CFU per g of stool samples with weak amplification observed for 103 to 104 CFU/g of stool samples. The serial dilutions of each genomic DNA extracted from stool samples were each artificially inoculated with 107 to 109 organism specimens per g, and we used the QIAamp DNA Stool Mini kit (Table 2). As few as 10 bacteria in the reaction mixture yielded a positive result. In the LC-PCR analysis of the stool samples after enrichment, each food- or waterborne bacterium was detected from enrichment of stool samples inoculated with 101 to 103 cells/g. Exceptions were Shigella spp., EIEC, S. aureus, and B. cereus, which were not enriched enough in BPW with stool (Table 2).
Development of duplex LC-PCR procedure for food poisoning outbreaks.
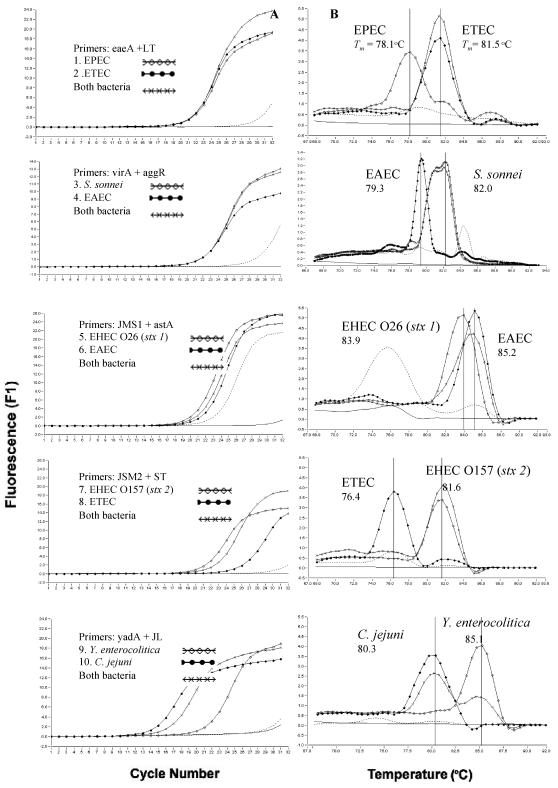
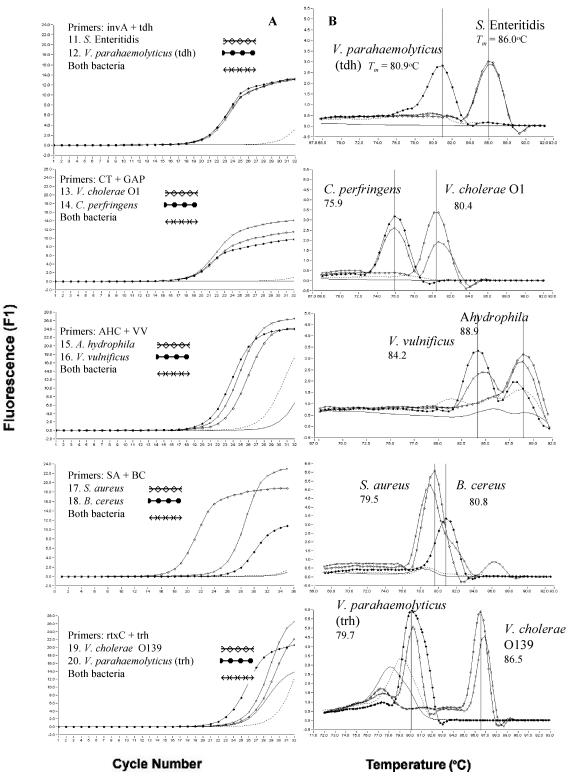
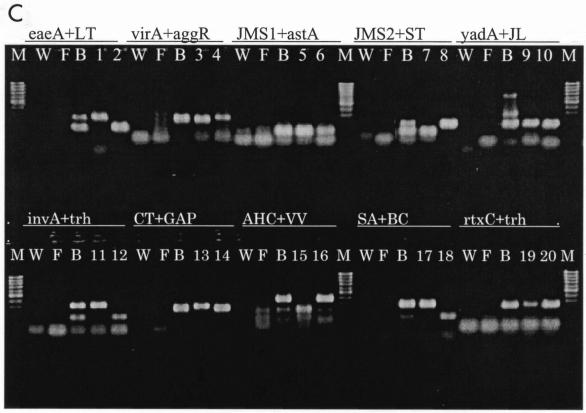
Figure 4 shows the fluorescent amplification curves and Tm curves of the duplex LC-PCR products of the template DNA samples. These were extracted from a stool sample that had been artificially inoculated with target bacteria. We used SYBR Green I fluorescence detection on the F1 channel of the LC instrument and a submarine agarose gel (LC-PCR product). In the SYBR Green PCR assay with two primer pairs, each PCR product was generated with a different Tm curve. These could be resolved in the LC by using Tm curve analysis when a target bacterium was present in the reaction tube. When two target bacteria were present in the same tube, each PCR product, with primer sets of yadA plus JL, CT plus GAP, AHC plus VV, and rtxC plus trh, could be resolved in the LC by using Tm curve analysis. PCR products achieved with other primer pairs of close Tm could not be resolved with Tm curve analysis, although these were separated on a submarine agarose gel.
FIG. 4.
Fluorescent amplification curves (A) and melting curve analysis (B) of duplex LC-PCR products obtained with SYBR Green I fluorescence detection on the F1 channel of the LC and submarine agarose gel of the PCR products (C). - - - - - - - - , negative control; ____, DNA extracted from feces; ••••••, water in panels A and B; lane M, 100-bp marker; W, water as negative control; F, DNA extracted from feces as negative control; B, both bacteria; lanes 1 to 20, foodborne bacteria in panel C.
The duplex LC-PCR procedure for food poisoning outbreak cases was developed by extracting template DNA from stool specimens of five patients with acute severe disease by using QIAamp DNA Stool Mini kits in 1 h or less. These template DNAs were then used for the first analysis of duplex real-time LC-PCR. LC-PCR was performed using 32 glass capillary tubes with four groups of two primer sets on the LC instrument at each run. The analysis of each group of primer pairs was made in eight glass capillary tubes, each of which included one negative DNA control consisting of PCR-grade water, two positive controls, and template DNA from five stool samples. The second analysis of LC-PCR was done with the other primer sets.
Application of the duplex LC-PCR procedure in food poisoning outbreak.
In the screening process of seven stool specimens in a waterborne poisoning outbreak, the eaeA gene of EPEC or EHEC was detected from one stool specimen in the duplex LC-PCR assay, but other virulent genes, including the stx1 and stx2 genes of EHEC, were not detected by using duplex LC-PCR and conventional PCR assays (Table 3). This rapid detection of EPEC and presumptive diagnosis of EPEC infections were carried out within 2 h. The subsequent LC-PCR and conventional PCR carried out with seven primer pairs of E. coli could detect the EPEC eaeA gene in four stool specimens, which contained 105 to 107 CFU/g (LC-PCR quantitative analysis), and from enrichments of five stool specimens and contaminated water. The E. coli astA gene was detected by using the same method from three stool specimens, which contained 105 to 108 CFU/g (LC-PCR quantitative analysis), and from enrichments of four stool specimens and contaminated water. The subsequent bacteriological examinations could isolate EPEC strains from five stool specimens and a water sample and could isolate astA gene-positive E. coli strains from three stool specimens and a contaminated water sample.
Application of LC-PCR to swine cecal samples for detection of pathogenic Yersinia organisms.
Thirteen swine cecal samples containing 103 to 106 Yersinia CFU/g, eight samples containing more than 103 Yersinia CFU/g, and two Yersinia-negative controls were all analyzed using both direct and enriched LC-PCR (Table 4). Four cecal samples contained yadA gene-positive Y. enterocolitica, as determined by direct LC-PCR. The corresponding Y. enterocolitica counts determined by culture analysis ranged from 9 × 103 to 4 × 106 CFU/g, and those determined by LC-PCR quantitative analysis ranged from 4 × 104 to 5 × 105 CFU/g. The other 19 cecal samples did not contain yadA gene-positive Y. enterocolitica or Y. pseudotuberculosis, as determined by direct LC-PCR. The corresponding pathogenic Yersinia counts, as determined by culture analysis, were less than 3 × 104 CFU/g for 16 samples and between 105 and 2 × 105 CFU/g for the other three samples. All samples of the cold enrichments contained yadA gene-positive Y. enterocolitica or Y. pseudotuberculosis, except for the negative controls.
DISCUSSION
Stool specimens from patients infected with enteric pathogens with acute severe disease may contain large numbers of causative bacterial species. In most cases of food- or waterborne poisoning, we found that causative bacteria can be rapidly detected and that a presumptive diagnosis of the causative agent of food- or waterborne poisoning outbreak could be made within 2 h. We used a combination of the duplex real-time SYBR Green fluorescent LC-PCR assay with DNA extraction with the QIAamp DNA Stool Mini kit used for detection. The real-time SYBR Green fluorescent LC-PCR assay is a rapid, specific, and sensitive detection technique. The DNA extraction of five stool specimens with the QIAamp DNA Stool Mini kit was carried out within 1 h. Then, the duplex LC-PCR assay was also carried out within 1 h. The product curves could be monitored in real time without product separation by gel electrophoresis. And we could then specifically identify the products based on a Tm curve analysis.
We developed the ultimately new LC-PCR screening system for food- or waterborne pathogens. One can simultaneously analyze eight pathogenic or specific genes of food- or waterborne pathogens in five stool specimens by using duplex LC-PCR and 32 capillary tubes. Single or multiple real-time PCR assays were reported for detection of one species among food- or waterborne pathogens, such as EHEC O157 (1, 11, 14, 29), Salmonella (5, 10, 17), Y. enterocolitica (30, 35, 36), Campylobacter jejuni (19, 25, 39), V. cholerae (21), V. parahaemolyticus (13), P. shigelloides (20), and S. aureus (28, 32). Nineteen pairs of primers for food- or waterborne pathogens were selected from earlier publications (Table 2), and one pair of primers for Y. enterocolitica and Y. pseudotuberculosis was constructed. This was done to make all 20 LC-PCR methods suitable for the same LC-PCR conditions (an annealing temperature of 55°C and a MgCl2 concentration of 3 mM).
The specificity of these primer pairs was carefully confirmed by studying numerous strains of each bacterial species that had been analyzed with conventional PCR or real-time PCR methods as described in previous reports (Table 2). In the SYBR Green LC-PCR assay, the products formed were identified based on Tm curve analysis. The PCR products from each primer pair were generated based on individual Tm values (Table 2). The Tm values of PCR products of stool samples, including each food- or waterborne bacterium, could be identified with that of control bacteria in the LC in the same run based on a Tm curve analysis. The specificity of the PCR assay of the new primer pair for detection of the yadA gene of Y. enterocolitica was tested on 19 different Yersinia spp. All pathogenic Y. enterocolitica and Y. pseudotuberculosis strains yielded positive PCR products from the yadA gene. However, a 203-bp PCR product was found in the pathogenic Y. enterocolitica strains but with a larger base pair than that of Y. enterocolitica in the pathogenic Y. pseudotuberculosis, which was the same as yadA primers, as described previously (18, 23). The Tm values of this yadA primer pair were also different between the pathogenic Y. enterocolitica (85.1°C) and Y. pseudotuberculosis (84.1°C) strains. Therefore, this primer pair was confirmed to be useful for detection and differentiation of two pathogenic Yersinia species.
In this LC-PCR method, almost all bacterial pathogens are detectable in stool specimens at a concentration of 103 to 105 bacteria per g (Table 2). This is because the concentration of DNA extracted from stool specimens using the QIAamp DNA Stool Mini kit was finally diluted to 4 × 104-fold in the reaction mixture. The PCR sensitivity for bacteria inoculated in stool samples may be as low as the presence of 10 cells in the reaction mixture. Particularly, if the detected species is present in more than 105 CFU/g of the stool specimen, the DNA extracted from the stool specimen should not interfere with the LC-PCR detection of a specific species. Then, the LC-PCR also will not give a nonspecific reaction or a strong Tm curve which could readily differentiate stool specimen reactions. However, if the detected species is present in less than 104 CFU/g of stool specimens, the weak reaction cannot be differentiated from a nonspecific reaction of stool specimens. In the results of our examination of human stools in a waterborne poisoning outbreak with E. coli, greater than 105 bacteria per g of samples were detected and calculated using direct LC-PCR. These samples gave strong Tm curves, which were produced with the primer of the EPEC eaeA gene. They were readily differentiated from nonspecific reactions of stool specimens, but differentiation of the weak Tm curves produced with the primer of the E. coli astA gene from nonspecific reactions of stool specimens was difficult. In a survey of Yersinia in pigs, four samples containing 9 × 103 to 4 × 106 yadA gene-positive Y. enterocolitica CFU/g of cecal contents were detected and calculated by the direct LC-PCR. They gave strong Tm curves, but nine samples containing 2 × 103 to 2 × 105 CFU/g gave weak Tm curves which could not be differentiated from nonspecific reactions. These findings suggest that LC-PCR is useful for certain detections when more than 105 bacteria per g of stool samples is present. Since stools of food poisoning patients usually harbor 106 or more bacteria per g (7, 31), real-time SYBR Green LC-PCR is an appropriate diagnostic tool.
We tentatively conclude that the duplex SYBR Green LC-PCR assay combined with DNA extraction using the QIAamp DNA Stool Mini kit (described in this study) offers rapid cycling with real-time (2 h or less) and non-sequence-specific detection of amplicons. The SYBR Green assays not only provide confidence in identification of target genes but also reduce the risk of experiencing laboratory product contamination. This is because the amplification reaction, detection of PCR products, and their Tm analysis can be performed with a single capillary tube. This protocol for processing stool specimens for less than 104 food- or waterborne pathogenic bacteria per g requires an overnight enrichment step to achieve adequate sensitivity. However, the rapid amplification and reliable detection of specific genes of more than 105 food- or waterborne pathogenic bacteria per g should facilitate the diagnosis and management of food- or waterborne poisoning outbreaks.
REFERENCES
- 1.Bellin, T., M. Pulz, A. Matussek, H.-G. Hempen, and F. Gunzer. 2001. Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J. Clin. Microbiol. 39:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, K. H., T. K. Ng, K. Y. Yuen, and W. C. Yam. 2001. Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, E., M. Glennon, S. Hanley, A.-M. Murray, M. Cormican, T. Smith, and M. Maher. 2001. Evaluation of a PCR/DNA probe colorimetric membrane assay for identification of Campylobacter spp. in human stool specimens. J. Clin. Microbiol. 39:4163-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daum, L. T., W. J. Barnes, J. C. McAvin, M. S. Neidert, L. A. Cooper, W. B. Huff, L. Gaul, W. S. Riggins, S. Morris, A. Salmen, and K. Lohman. 2002. Real-time PCR detection of Salmonella in suspect foods from gastroenteritis outbreak in Kerr County, Texas. J. Clin. Microbiol. 40:3050-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franck, S. M., B. T. Bosworth, and H. W. Moon. 1998. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J. Clin. Microbiol. 36:1795-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukushima, H., M. Tsubokura, K. Otsuki, Y. Kawaoka, R. Nishio, S. Moriki, Y. Nishino, H. Mototsune, and K. Karino. 1985. Epidemiological study of Yersinia enterocolitica and Yersinia pseudotuberculosis infections in Shimane Prefecture, Japan. Zentbl. Bakteriol. 1 Abt Orig. B 180:515-527. [PubMed] [Google Scholar]
- 8.Fukushima, H., and M. Gomyoda. 1999. An effective, rapid and simple method for isolation of Shiga toxin-producing Escherichia coli O26, O111 and O157 from faeces and food samples. Zentbl. Bakteriol. 289:415-428. [DOI] [PubMed] [Google Scholar]
- 9.Holland, J. L., L. Louie, A. E. Simor, and M. Louie. 2000. PCR detection of Escherichia coli O157:H7 directly from stools: evaluation of commercial methods of purifying fecal DNA. J. Clin. Microbiol. 38:4108-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoorfar, J., P. Ahrens, and P. Radstrom. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibekwe, A. M., P. M. Watt, C. M. Grieve, V. K. Sharma, and S. R. Lyons. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, F., T. Hagino, K.-I. Ito, and H. Watanabe. 1992. Differentiation and detection of pathogenic determinants among diarrheagenic Escherichia coli by polymerase chain reaction using mixed primers. Nihonrinshou 50(Suppl.):343-347. (In Japanese.) [PubMed] [Google Scholar]
- 13.Iwade, Y., A. Yamauchi, A. Sugiyama, O. Nakamura, and N. H. Kumazawa. 2001. Quantification of thermostable direct hemolysin-producing Vibrio parahaemolyticus from foods assumed to cause food poisoning using the LightCycler instrument, p. 171-176. In U. Reischl, C. Wittwer, and E. R. Cockerill (ed.), Rapid cycle real-time PCR, methods and applications, microbiology and food analysis. Springer-Verlag, Berlin, Germany.
- 14.Jothikumard, N., and M. W. Griffiths. 2002. Rapid detection of Escherichia coli O157:H7 with multiplex real-time PCR assays. Appl. Environ. Microbiol. 68:3169-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, N., S.-M. Kim, H. Kato, K. Tanaka, K. Watanabe, K. Ueno, and Y. Chong. 1993. Identification of enterotoxin-producing Clostridium perfringens by the polymerase chain reaction. Jpn. J. Infect. Dis. 67:724-729. [DOI] [PubMed] [Google Scholar]
- 16.Kingombe, C. I. B., G. Huys, M. Tonolla, M. J. Albert, J. Swings, R. Peduzzi, and T. Jemmi. 1999. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knutsson, R., C. Lofstrom, H. Grage, J. Hoofar, and P. Radstrom. 2002. Modeling of 5′ nuclease real-time responses for optimization of a high-throughput enrichment PCR procedure for Salmonella enterica. J. Clin. Microbiol. 40:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lantz, P.-G., R. Knutsson, Y. Blixt, W. A. Al-Soud, E. Borch, and P. Radstrom. 1998. Detection of pathogenic Yersinia enterocolitica in enrichment media and pork by a multiplex PCR: a study of sample preparation and PCR-inhibitory components. Int. J. Food Microbiol. 45:93-105. [DOI] [PubMed] [Google Scholar]
- 19.Logan, J. M. J., K. J. Edwards, N. A. Saunders, and J. Stanley. 2001. Rapid identification of Campylobacter spp. by melting peak analysis of biprobes in real-time PCR. J. Clin. Microbiol. 39:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loh, J. P., and E. P. H. Yap. 2001. Rapid and specific detection of Plesiomonas shigelloides directly from stool by LightCycler PCR, p. 161-169. In U. Reischl, C. Wittwer, and E. R. Cockerill (ed.), Rapid cycle real-time PCR, methods and applications, microbiology and food analysis. Springer-Verlag, Berlin, Germany.
- 21.Lyon, W. J. 2001. TaqMan PCR for detection of Vibrio cholerae O1, O139, non-O1, and non-O139 in pure cultures, raw oysters, and synthetic seawater. Appl. Environ. Microbiol. 67:4658-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair, G. B., T. Shimada, H. Kurazono, J. Okuda, A. Pal, T. Karasawa, T. Mihara, Y. Uesaka, H. Shirai, S. Garg, P. K. Saha, A. K. Mukhopadhyay, T. Ohashi, J. Tada, T. Nakayama, S. Fukushima, T. Takeda, and Y. Yoshifumi. 1994. Characterization of phenotypic, serological, and toxigenic traits of Vibrio cholerae O139 Bengal. J. Clin. Microbiol. 32:2775-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neubauer, H., L. D. Sprague, A. Hensel, S. Aleksic, and H. Meyer. 2000. Specific detection of plasmid bearing Yersinia isolates by PCR. Clin. Lab. 46:583-587. [PubMed] [Google Scholar]
- 24.Nishibuchi, M., Y. Takeda, J. Tada, T. Oohasshi, N. Nishimura, H. Ozaki, and S. Fukushima. 1992. Methods to detect the thermostable direct hemolysin gene and a related hemolysin gene of Vibrio parahaemolyticus. Nippon Rinsho 642:348-352. (In Japanese.) [PubMed]
- 25.Nogva, H. K., A. Bergh, A. Holck, and K. Rudi. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahn, K., S. A. De Grandis, R. C. Clarks, S. A. McEwen, J. E. Galan, C. Ginocchio, R. Curtis III, and C. I. Gyles. 1992. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6:271-279. [DOI] [PubMed] [Google Scholar]
- 27.Ratchtrachenchai, O. A., S. Subpasu, and K. Ito. 1997. Investigation on enteroaggregative Escherichia coli infection by multiplex PCR. Bull. Dept. Med. Sci. 39:211-220. [Google Scholar]
- 28.Reischl, U., H.-J. Linde, M. Metz, B. Leppmeier, and N. Lehn. 2000. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J. Clin. Microbiol. 38:2429-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reischl, U., M. T. Youssef, J. Kilwinski, N. Lehn, W. L. Zhang, H. Karch, and N. A. Strockbine. 2002. Real-time fluorescence PCR assays for detection and characterization of Shiga toxin, intimin, and enterohemolysin genes from Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 40:2555-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen, K. 2000. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J. Clin. Microbiol. 38:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shandera, W. X., C. O. Tacket, and P. A. Blake. 1983. Food poisoning due to Clostridium perfringens in the United States. J. Infect. Dis. 147:167-170. [DOI] [PubMed] [Google Scholar]
- 32.Shrestha, N. K., M. J. Tuohy, G. S. Hall, C. M. Isada, and G. W. Procop. 2002. Rapid identification of Staphylococcus aureus and the mecA gene from bacT/ALERT blood culture bottles by using the LightCycler system. J. Clin. Microbiol. 40:2659-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 34.Villalobo, E., and A. Torres. 1998. PCR for detection of Shigella spp. in mayonnaise. Appl. Environ. Microbiol. 64:1242-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vishnubhatla, A., D. Y. C. Fung, R. D. Oberst, M. P. Hays, T. G. Nagaraja, and S. J. A. Flood. 2000. Rapid 5′ nuclease (TaqMan) assay for detection of virulent strains of Yersinia enterocolitica. Appl. Environ. Microbiol. 66:4134-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vishnubhatla, A., R. D. Oberst, D. Y. C. Fung, W. Wonglumsom, M. P. Hays, and T. G. Nagaraja. 2001. Evaluation of a 5′-nuclease (TaqMan) assay for the detection of virulent strains of Yersinia enterocolitica in raw meat and tofu samples. J. Food Prot. 64:355-360. [DOI] [PubMed] [Google Scholar]
- 37.Wang, G., C. G. Clark, and F. G. Rodgers. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, R.-F., W.-W. Cao, and C. E. Cerniglia. 1997. A universal protocol for PCR detection of 13 species of foodborne pathogens in foods. J. Appl. Microbiol. 83:727-736. [DOI] [PubMed] [Google Scholar]
- 39.Wilson, D. L., S. R. Abner, T. C. Newman, L. S. Mansfield, and J. E. Linz. 2000. Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay. J. Clin. Microbiol. 38:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yatsuyanagi, J., S. Saito, H. Sato, Y. Miyajima, K.-I. Amano, and K. Enomoto. 2002. Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J. Clin. Microbiol. 40:294-297. [DOI] [PMC free article] [PubMed] [Google Scholar]