Abstract
A Capnocytophaga sp. was inadvertently isolated from a cat with chronic sinusitis and rhinitis when cytopathic effects were observed in Crandall-Reese feline kidney cells that had been inoculated with oropharyngeal swab samples. Although Capnocytophaga spp. are of considerable zoonotic importance, their clinical relevance for dogs or cats has not been established. However, failure to do so may be attributed to the infrequent use of specialized isolation techniques that are required to grow Capnocytophaga spp. To our knowledge, successful isolation of these organisms from a cat with nasopharyngeal disease has not been reported.
CASE REPORT
A 10-year-old male castrated domestic shorthair cat was referred for further evaluation of chronic bilateral nasal discharge and increased upper respiratory sounds. Five months prior to evaluation at North Carolina State University College of Veterinary Medicine (NCSU-CVM) the cat had developed clear nasal discharge with sporadic episodes of coughing and sneezing. Two months prior to referral the discharge had become purulent. At that time, both nares were patent, and lung sounds ausculted normally. Thoracic radiographs were unremarkable, and Dirofilaria immitis antigen and antibody tests were negative. Following treatment with amoxicillin-clavulanic acid and chlorpheniramine the cough resolved, and the frequency of sneezing decreased. One week later prednisone was added to the treatment regimen, but the owners reported no noticeable improvement. After 17 days amoxicillin-clavulanic acid was discontinued and enrofloxacin was prescribed, immediately after which signs began to recur. At reevaluation 1 week prior to referral, the complete blood count indicated a mature neutrophilia (10,220 cells/μl; reference range, 2,500 to 8,500 cells/μl) and eosinophilia (1,168 cells/μl; reference range, 0 to 1,000 cells/μl); a serum chemistry panel, feline leukemia virus antigen test, feline immunodeficiency virus antibody test, feline coronavirus antibody test, and determination of Toxoplasma gondii immunoglobulin G (IgG) and IgM titers were all unremarkable. Significant environmental history included free access to the outdoors, and there were no newly identifiable allergens in the household environment.
At presentation to NCSU-CVM the cat was alert and responsive, with vital parameters within normal limits. A small amount of dried brown exudate was noted at the lateral aspect of each naris, but there was no obvious nasal discharge. Referred upper airway sounds were auscultable over all lung fields as well as a III/VI systolic heart murmur.
Oral and fundoscopic examinations were unremarkable. Although the segmented neutrophil count was within the reference range, band neutrophils were slightly increased (426 band neutrophils/μl; reference range, 0 to 100 band neutrophils/μl). Serum biochemical abnormalities included a mild hyperglobulinemia (5.4 g/dl; reference range, 2.8 to 5.3 g/dl) and an increase in blood urea nitrogen (40 mg/dl; reference range, 15 to 35 mg/dl). Thoracic radiographs were unremarkable. Fecal floatation did not reveal parasite ova. A Cryptococcus neoformans antigen test was negative. Computerized tomography of the nasal cavity showed bilateral filling of the meatuses with fluid-dense opacity, with extension to the right frontal sinus and sphenopalatine recess; minimal contrast uptake was noted. The nasal septum and all turbinates were intact.
Primary differential diagnoses for chronic rhinitis and sinusitis included chronic viral infection (feline herpesvirus or feline calicivirus) with secondary bacterial infection, fungal infection (cryptococcosis), allergic rhinitis with secondary bacterial infection, and neoplasia.
Prior to endoscopy, sterile swabs were used to obtain samples for virus isolation and bacterial culture from the caudal oropharynx and the nasal passages. Samples were placed in 2 ml of viral transport medium (Dulbecco's minimal essential medium containing 2% fetal bovine serum and 0.2 mg of gentamicin/ml) and submitted for viral isolation. Crandall-Reese feline kidney (CRFK) cells were inoculated with fluid from the swab samples. A cytopathic effect consistent with virus infection was observed in inoculated cells by 5 days postinoculation; however no viruses were identified based on specific immunohistochemical staining for feline herpesvirus 1 and feline calicivirus. Additionally, no virus particles were detected by transmission electron microscopy of cell culture supernatant fluids. Subsequent filtration studies suggested that cytopathic effects were due to a bacterial agent because filtration of cell culture supernatant fluid through 0.22-μm-pore-size filters but not through 0.45-μm-pore-size filters prevented development of cytopathic effects.
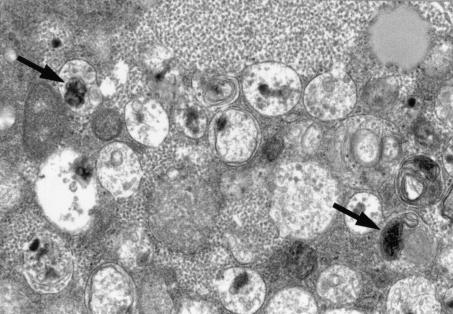
Inoculated CRFK cells were fixed in 2% glutaraldehyde, postfixed in 1% osmium tetroxide, and embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and examined by transmission electron microscopy (Fig. 1). CRFK cells were highly vacuolated, and vacuoles contained particles consistent with bacteria approximately 0.3 by 0.2 μm.
FIG. 1.
Electron micrographs of Crandall-Reese feline kidney (CRFK) cells, 5 days after inoculation with ocular and oropharyngeal fluids from the affected cat. Cells were highly vacuolated, and vacuoles contained approximately 0.3- by 0.2-μm bacteria-like particles (arrows).
Swab samples of the caudal oropharynx and nasal passages were also submitted for aerobic bacterial culture to the NCSU-CVM microbiology laboratory, and results were negative. However, as Gram staining of cell culture supernatant from inoculated CRFK cells revealed gram-negative, filamentous rods, an attempt was made to culture the viral cell culture fluid in addition to the previously mentioned cultures of the swab samples. Material from the fourth and fifth passages on CRFK cells was inoculated onto blood agar plates and placed in a carbon dioxide incubator (37°C, 5% CO2); however, no growth was observed after 5 days of incubation.
Endoscopic examination of the nares revealed a large amount of thick, pale yellow mucous in all meatuses that could not be removed by vacuum suction. No mass lesions were evident, and all visible mucosal surfaces were within normal limits. Cytopathology of the nasal discharge was consistent with epithelial cell hyperplasia, and nasal turbinate histopathology revealed chronic, severe ulcerative rhinitis without visible viral inclusion bodies.
To facilitate identification of the gram-negative filamentous rods observed in the cell culture supernatant from inoculated CRFK cells, a series of PCRs to isolate the 16S ribosomal DNA (rDNA) sequence were performed on a Progene thermocycler (Techne, Inc., Princeton, N.J.) using 8F (5′-AGAGTTTGATCCTGGCTCAG) and 1492R (5′-GGTTACCTTGTTACGACTT) primers (Sigma-Genosys, Inc., The Woodlands, Tex.). PCR products were separated on a 1% agarose gel containing ethidium bromide and were visualized by UV transillumination and compared to DNA size standards (Hyperladder I; Denville Scientific, Inc., Metuchen, N.J.). PCR fragments were purified with a commercial PCR purification kit (QIAquick PCR purification kit; Qiagen, Inc., Valencia, Calif.).
Purified PCR products were ligated into the pGEM-T Easy vector (Promega, Madison, Wis.), followed by transformation of Escherichia coli JM109 high-efficiency competent cells as described by the manufacturer (Promega). Recombinants were selected by blue-versus-white screening of the colonies, and plasmid DNA from multiple clones was isolated with the QIAprep plasmid kit (Qiagen, Inc.). Plasmid DNA was analyzed on a 0.5% agarose gel with a circularized pGEM-T Easy vector (Promega) as a control to confirm that each selected clone was a recombinant. Both (i.e., forward and reverse) strands of the recombinant plasmid DNA were sequenced by cycle sequencing in accordance with the manufacturer's protocol for the Sequitherm EXCEL II DNA sequencing kit (Epicentre, Madison, Wis.) on a PCRexpress thermocycler (Hybaid, Franklin, Mass.). The infrared fluorescently labeled primers used were T7-800 (5′-TAATACGACTCACTATA), M13R-700 (5′-AGCGGATAACAATTTCACACAGGA), 515F-800 (5′-GTGCCAGCMGCCGCGGTAA; M = A or C), and 1391R-700 (5′-GACGGGCGGTGWGTRCA; W = A or T; R = A or G) (Li-Cor, Inc., Lincoln, Neb.). The sequencing reactions were analyzed by polyacrylamide gel electrophoresis (3.75%) on a Li-Cor 4200 automated DNA sequencer. The 16S rDNAs of all five clones demonstrated 97% sequence identity to Capnocytophaga canimorsus and Capnocytophaga cynodegmi when compared to sequence data available in GenBank by using BLAST (1).
Following discharge from NCSU-CVM the cat was treated with doxycycline (5 mg/kg of body weight orally every 12 h) for 30 days. Enrofloxacin and chlorpheniramine were discontinued. Once infection with a member of the genus Capnocytophaga was identified, the cat was treated with amoxicillin-clavulanic acid (Clavamox) for 6 weeks. For 10 weeks after discontinuing the antibiotics the cat was very active and alert but continued to experience occasional sneezing and mild upper respiratory stridor. Five months after evaluation at NCSU-CVM, the cat again developed bilateral nasal discharge, which was managed by the referring veterinarian.
Although C. cynodegmi and C. canimorsus are considered commensal organisms of the oral flora of dogs and cats, having been isolated from 16 and 18% of healthy dogs and cats, respectively (6), it is possible that they may cause opportunistic infections in susceptible individuals. Although Capnocytophaga spp. are of considerable zoonotic importance, their clinical relevance for dogs or cats has not been established (2, 6). Previously, dogs were the only species proven to transmit Capnocytophaga to people, but more recently five cases of cat-associated Capnocytophaga sp. infection have been documented (6). The owners of this cat reported substantial improvement in clinical signs and resolution of nasal discharge following a protracted course of antibiotics, but recurrence of nasal discharge 5 months after isolation of a member of the genus Capnocytophaga supports the continued presence of an as yet unidentified disease process.
Capnocytophaga sp. infections in susceptible humans (notably due to C. canimorsus) are usually caused by a bite wound. The bacteria can cause a variety of clinical manifestations including septicemia, disseminated intravascular coagulopathy, tissue destruction, meningitis, and death (2, 4, 5, 6). Most people who develop fatal complications are immunocompromised or suffer from a chronic disease. C. cynodegmi can cause a localized wound infection. Both C. canimorsus and C. cynodegmi are known to induce cytopathic effects in cell culture (2). Previous studies have not been designed to determine whether cats might suffer adverse clinical effects from infection with Capnocytophaga spp.
Diagnostic evaluation by the referring veterinarian and at NCSU-CVM failed to identify a specific cause of sinusitis or rhinitis; therefore, the detection of cytopathic effect in CRFK cells inoculated for virus isolation purposes provided the only evidence for a nonviral agent as a potential contributor to rhinitis and sinusitis in this cat. As noted by electron microscopy the inoculated CRFK cells contained vacuoles with particles that appeared to be bacterial species; these particles were consistent in size and location in vacuoles with previous reports of Capnocytophaga spp. (2). Both species of Capnocytophaga have been shown to be phagocytized by macrophages, replicate intracellularly within vacuoles, and induce cytopathic effects (2). This description is consistent with the appearance of the CRFK cell culture derived from the nasal swab samples of the cat in this report.
Detection of gram-negative filamentous bacteria in the CRFK cell culture supernatant pointed to the possibility of an infection with a member of the genus Capnocytophaga, which fits this morphological description. Members of the genus Capnocytophaga are gram-negative, facultative anaerobic organisms that grow on brain heart infusion agar containing 5% rabbit serum (2, 3, 4). Capnocytophaga spp. are slow-growing and fastidious bacteria (2, 3, 4, 6), and the methods used in previous studies were not employed during the attempts at isolation from CRFK cells, which most probably explains the absence of growth on blood agar plates.
PCR amplification of the DNA directly from the CRFK cell culture material confirmed the presence of a Capnocytophaga sp. Since all five clones had between 96 and 98% homology with C. cynodegmi and C. canimorsus, it is likely that the organism in the swabs obtained from the caudal oropharynx and nasal passages for virus isolation was a member of the genus Capnocytophaga closely related to C. cynodegmi and C. canimorsus.
The range of sequence homology between the five clones and C. cynodegmi and C. canimorsus can be explained in the following ways. It is unclear at this point how many ribosomal operons (i.e., how many different 16S rDNA copies) are in the genomes of Capnocytophaga spp. E. coli, for example, is known to have seven operons due to microheterogeneity (i.e., sequence differences due to evolution), which causes it to have seven slightly different sequences. Therefore, the sequence ambiguities within the five clones could represent different 16S rDNAs from different operons in the genome of the same strain. Second, the difference in homology could be due to PCR errors or sequencing errors caused by the DNA polymerases. All polymerases can make mistakes, and we are forced to use a Taq polymerase that does not have “proofreading” activity. The A overhangs that the Taq polymerase produces are actually one of those errors, but without them we would be unable to clone our PCR fragments into a T-cloning vector. However, the rate of occurrence of these mistakes is statistically very low and may not be the best way to explain the differences among clones. The third explanation for the differences is that there could be more than one Capnocytophaga strain within the patient (i.e., a coinfection). This depends on how one defines strains or species by using 16S rDNA. This has always been difficult and won't change in the future, but the possibility that, for strains of some species, sequence similarity within a strain's own different operons is lower than that with operons from other strains must be addressed.
In conclusion, Gram staining, electron microscopy, and PCR amplification followed by DNA sequencing facilitated the identification of a strain of Capnocytophaga closely related to C. cynodegmi and C. canimorsus that had been isolated from a cat with chronic nasal discharge by inoculating a sample in CRFK cell culture. Evidence to support other causes of chronic bilateral nasal discharge besides an infection with a member of the genus Capnocytophaga was lacking; therefore, this case may represent an unusual or previously unrecognized presentation of Capnocytophaga sp.-induced disease. Alternatively, Capnocytophaga may not have been the causative agent for the rhinitis in this cat, since a definitive cause for chronic nasal discharge is frequently undetermined in the clinical setting despite extensive diagnostic testing. It is possible that some undetected confounding factor (e.g., occult feline leukemia virus infection, history of prior prednisone usage, or senescence of the immune system in an older cat) may have contributed to immunosuppression, which resulted in secondary colonization of Capnocytophaga. In the future, Capnocytophaga spp. should be considered possible opportunistic pathogens in cats with chronic rhinitis, particularly when there is a poor response to commonly used antibiotics and when viral, fungal, and neoplastic etiologies have been thoroughly investigated.
Nucleotide sequence accession number.
The 1,477-nucleotide consensus sequences of all five clones were deposited under the accession number AF426105 in the GenBank database.
REFERENCES
- 1.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. F. Oulette, B. A. Rapp, and D. L. Wheeler. 2000. GenBank. Nucleic Acids Res. 28:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer, L., R. S. Weyant, E. H. White, and F. D. Quinn. 1995. Intracellular multiplication and toxic destruction of cultured macrophages by Capnocytophaga canimorsus. Infect. Immun. 63:3484-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez-Garces, J.-L., J.-I. Alos, J. Sanchez, and R. Cogollos. 1994. Bacteremia by multidrug-resistant Capnocytophaga sputigena. J. Clin. Microbiol. 32:1067-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greene, C. E. (ed.). 1998. Infectious diseases of the dog and cat, p. 333. W. B. Saunders Company, Philadelphia, Pa.
- 5.Mossad, S. B., A. E. Lichtin, G. S. Hall, and S. M. Gordon. 1997. Diagnosis: Capnocytophaga canimorsus septicemia. Clin. Infect. Dis. 24:123, 267. [PubMed]
- 6.Valtonen, M., A. Lauhio, P. Carlson, J. Multanen, A. Sivonen, M. Vaara, and J. Lähdevirta. 1995. Capnocytophaga canimorsus septicemia: fifth report of a cat-associated infection and five other cases. Eur. J. Clin. Microbiol. Infect. Dis. 14:520-523. [DOI] [PubMed] [Google Scholar]