Abstract
We sequenced the 16S rRNA and groEL genes of Aegyptianella pullorum, a small bacterium that infects and replicates only in avian red blood cells. A specific PCR test was developed to analyze A. pullorum DNA. Phylogenic analysis revealed A. pullorum is most closely related to Anaplasma spp.
In 1928, Carpano (6) first described an infectious agent that induced intraerythrocytic inclusions in blood smears of domestic fowls in Egypt and named it Aegyptianella pullorum. This bacterium is transmitted by the soft tick Argas (Persicargus) persicus (7, 15, 17). The inclusions are 0.3 to 4 μm in diameter and purple when stained with the Romanowsky method. Similar inclusions have been observed in avian red blood cells in other parts of Africa, Asia, Europe, and South and North America (8, 23, 26). A large variety of bird species appear to be susceptible to infection with this agent (8, 9, 10, 16, 21, 25). Electron microscopy analysis of the South Africa Onderstepoort strain (14) and the Rhodesia strain (3) of A. pullorum in chicken blood revealed that the membrane-bound inclusions contain between 1 and 26 pleomorphic cocci 0.25 to 0.4 μm long. The bacterial cytoplasm includes ribosomes and fine DNA strands and is enveloped within inner and outer trilaminar membranes (3, 14). Castle and Christensen (8) described the ultrastructure of similar organisms in wild turkeys from North America in 1985. In 1992, a strain with the proposed designation “Aegyptianella botuliformis” was described. This strain differs from A. pullorum in its ultrastructure, host bird species specificity, and tick vectors (19). In addition, there are several reports of unconfirmed species of Aegyptianella infecting amphibians and reptiles such as frogs, tortoises, snakes, and lizards (1, 2, 4, 11, 12, 22, 27). Currently, no laboratory isolate and no molecular or antigenic data are available for A. pullorum or other Aegyptianella species. In 1974, the eighth edition of Bergey's Manual of Determinative Bacteriology (24) included A. pullorum in the family Anaplasmataceae, based on its phenotypic similarity to Anaplasma marginale (an intracellular parasite of bovine red blood cells). However, the classification of this genus was recently redesignated as uncertain, due to the lack of molecular information (13). Therefore, in the present study, we analyzed the 16S rRNA and the groEL gene sequences of A. pullorum, in order to better characterize this group of bacteria.
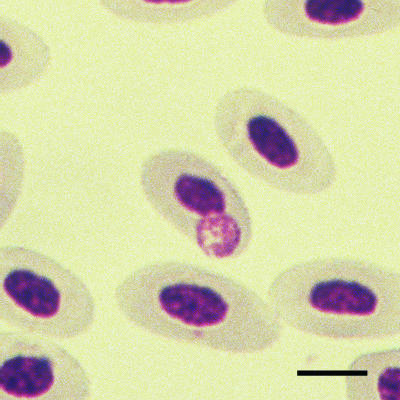
Ten glass slides with Romanowsky-stained blood smears from domestic broad-breasted white turkey poults inoculated with the blood from Rio Grande wild turkeys in southern Texas (A. pullorum Texas strain) were obtained from a study carried out in 1983 and 1984 (8). On these slides, A. pullorum appears as purple compact inclusions 0.3 to 4 μm in diameter (Fig. 1). In larger inclusions, clearly defined small cocci of 0.25 to 0.4 μm that resembled those of Anaplasma spp. could be distinguished (Fig. 1). No other cell types contained these inclusions, and no other bacteria or parasites were visually detected within the blood smear.
FIG. 1.
A. pullorum inclusion in the cytoplasm of a turkey red blood cell stained with the Romanowsky method. Numerous small (<0.5-μm) reddish purple stained cocci are seen within a single round inclusion. Bar, 4 μm.
The slides were extensively washed with TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0), and cells were scraped off with a sterile scalpel blade into a microcentrifuge tube. DNA was extracted with Chelex-100 (Bio-Rad, Hercules, Calif.). Since the DNA had been severely fragmented and very small amounts of target DNA were present, we devised six nested or seminested PCRs with 12 pairs of primers, as shown in Table 1. This approach yielded overlapping ∼100-bp fragments, which we assembled to map the 607 bp of the partial 16S rRNA gene sequence. The nested touchdown PCR (18) protocol included incubation at 94°C for 3 min, followed by 10 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 2 min, with the annealing temperature decreased by 1°C in each cycle. Samples were then subjected to 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, followed by 7 min of extension at 72°C. The 448-bp partial groEL sequence was obtained by assembling four ∼100-bp overlapping fragments amplified by heminested PCR with eight pairs of primers, as shown in Table 1. The PCR products were cloned, and multiple clones were sequenced on both strands (Table 1). Single unique sequences were obtained for the 16S rRNA and groEL genes, indicating that only one bacterial species was present in the specimen. The forward and reverse primers located on different fragments were used in PCR to further verify the structure of these genes.
TABLE 1.
Primers used for each step of nested (seminested) PCR to obtain A. pullorum DNA fragments and sequences and species-specific PCRa
| Gene or procedure (bp sequenced) | First PCR
|
Second PCR
|
||||
|---|---|---|---|---|---|---|
| Primer pairb | Amplicon size (bp) without primers | Nucleotide position corresponding to A. marginale (M60313 or AF165812) | Primer pair | Amplicon size without primers (bp) (no. of clones sequenced) | Nucleotide position corresponding to A. marginale (M60313 or AF165812) | |
| 16S rRNA (607) | EU-F2 + Bird-R1 | 141 | 15-155 | EU-F2 + Bird-R2 | 132 (9) | 15-146 |
| EU-F1 + EU-R1 | 168 | 115-282c | Anap-F11 + Anap-R4 | 108 (7) | 139-245c | |
| Bird-F1 + Anap-R2 | 156 | 203-358 | Bird-F2 + Anap-R2 | 139 (7) | 220-358 | |
| Anap-F3 + EU-R2 | 105 | 357-461 | Anap-F3 + EU-R3 | 87 (7) | 357-443 | |
| Bird-F3 + Anap-R7 | 115 | 414-528 | Bird-F4 + Anap-R7 | 88 (7) | 441-528 | |
| Bird-F5 + EU-R4 | 103 | 517-620 | Bird-F6 + EU-R4 | 91 (6) | 529-620 | |
| groEL (448) | Agro-F3 + Bird-gro-R1 | 120 | 153-272 | Agro-F3 + Bird-gro-R2 | 100 (8) | 153-252 |
| Agro-F1 + Agro-R1 | 100 | 252-351 | Agro-F1 + Agro-R2 | 73 (5) | 252-324 | |
| Bird-gro-F1 + Agro-R4 | 179 | 305-483 | Bird-gro-F2 + Agro-R4 | 160 (8) | 324-483 | |
| Bird-gro-F3 + Agro-R6 | 138 | 463-600 | Bird-gro-F4 + Agro-R6 | 118 (8) | 483-600 | |
| Species-specific PCRa | AP-F1 + AP-R1 | 173d | 50-221c | AP-F2 + AP-R2 | 113d | 72-183c |
Species-specific primers were designed based on the newly obtained 16S rRNA gene sequence of A. pullorum.
Primers with Bird were A. pullorum-specific primers; primers with Anap were designed based on conserved sequences between Anaplasma marginale and Anaplasma phagocytophilum; primers with EU were designed based on conserved sequences of 14 related Ehrlichia species.
There is one base insertion between bases 178 and 179 corresponding to Anaplasma marginale (M600313).
Amplicon size including primers.
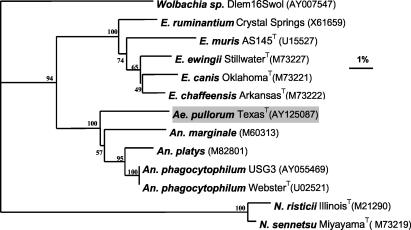
DNA extracted independently from different blood smear slides several months later yielded the identical sequences, indicating that these sequences were derived from the blood smear. Of note, it is unlikely that these sequences were derived from environmental contaminants, such as water or air, since the localization of A. pullorum is exclusively intracellular and these sequences are quite unique. It is also unlikely that these sequences are from our laboratory, since we have never used this bacterium or analyzed DNA of these base sequences. Alignment of 16S rRNA gene sequences (including the gaps) corresponding to nucleotide positions 15 to 620 of Anaplasma marginale and our subsequent phylogenic analysis revealed that the new sequence belonged to a member of the genus Anaplasma clade within the family Anaplasmataceae (Fig. 2). The 16S rRNA gene sequence of A. pullorum had 93.4, 93.2, 93.2, and 92.7% identity with the sequences of Anaplasma platys (strain name unavailable, an intracellular parasite of canine platelets), Anaplasma phagocytophilum USG3 (an intracellular parasite of human, horse, goat, mouse, and sheep granulocytes), Anaplasma phagocytophilum WebsterT, and Anaplasma marginale (strain name unavailable), respectively. These similar levels of identity indicate that A. pullorum is a distinct species that exists at a nearly equal distance from all known Anaplasma spp. Members of the next-closest clade, the genus Ehrlichia, had 16S rRNA gene sequences with 86.3 to 88.4% identity with that of A. pullorum.
FIG. 2.
Phylogram of A. pullorum and other members of the family Anaplasmataceae based on comparison of 16S rRNA gene sequences. GenBank accession numbers are shown in parentheses. Numbers above internal nodes indicate the percentages of 1,000 bootstrap replicates that supported the branch. In cases where bacterial names do not include strain names, the strain names are unavailable. Bar, percentage of divergence. The DNADIST, NEIGHBOR, CONSENSE of PHYLIP (version 3.6; http://evolution.genetics.washington.edu/phylip.html), and TreeView (version 1.5.2) programs were used for the sequence analysis and phylogram construction. Internal nodes were verified with SEQBOOT of PHYLIP with 1,000 replicates. N. sennetsu, Neorickettsia sennetsu; N. risticii, Neorickettsia risticii.
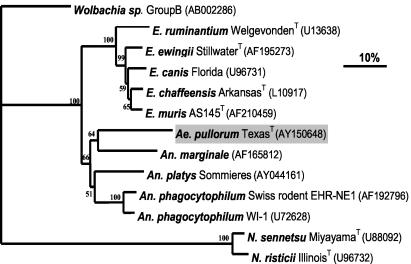
Alignment of groEL gene sequences (including the gaps) that were available from the GenBank database corresponding to nucleotide positions 153 to 600 of Anaplasma marginale (strain name unavailable) and subsequent phylogenetic analysis revealed that the sequence of the groEL gene of A. pullorum was novel. This sequence was from a member of the clade that included Anaplasma species within the family Anaplasmataceae (Fig. 3). The groEL gene sequence of A. pullorum had 73.3, 72.8, and 71.4% identity with the sequences of Anaplasma phagocytophilum Swiss rodent, Anaplasma phagocytophilum WI-1, and Anaplasma marginale (strain name unavailable), respectively. These similar levels of identity further suggest that A. pullorum is at almost equal distances from Anaplasma marginale and Anaplasma phagocytophilum. The levels of identity of the groEL sequence of A. pullorum with the sequences of members of the next-closest clade, the genus Ehrlichia, were 68.3 to 69.2%.
FIG. 3.
Phylogram of A. pullorum and other members of the family Anaplasmataceae based on comparison of groEL gene sequences. GenBank accession numbers are shown in parentheses. Numbers above the internal nodes indicate the percentages of 1,000 bootstrap replicates that supported the branch. For Anaplasma marginale, the strain name is unavailable. Bar, percentage of divergence. Phylograms were constructed as described in the legend to Fig. 2. N. sennetsu, Neorickettsia sennetsu; N. risticii, Neorickettsia risticii.
Based on the newly obtained 16S rRNA gene sequence, we developed an A. pullorum-specific nested-PCR protocol. Primers for specific detection of A. pullorum were designed based on comparison of the A. pullorum 16S rRNA gene sequences with those of the most closely related species: Anaplasma marginale, Anaplasma phagocytophilum, Anaplasma platys, Ehrlichia ruminantium, Ehrlichia chaffeensis, Ehrlichia muris, Ehrlichia ewingii, and Ehrlichia canis (Table 1). Specificities of these primers were verified by a BLAST search. In the PCR, the 50-μl reaction mixture contained a template DNA (in the second round of PCR, the template DNA used was 0.5 μl of PCR product from the first round of PCR), PCR buffer (10 mM Tris-HCl [pH 8.4], 50 mM KCl), 2 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphate, 2.5 U of Taq polymerase, and 20 pmol of each primer. The three-step program PCR cycle conditions were 94°C for 3 min, followed by 35 cycles of 94°C for 50 s, 54°C for 1 min, and 72°C for 1 min, and finally extension at 72°C for 7 min. Only A. pullorum DNA from 20-year-old slides yielded a single band of the predicted size of 113 bp (Fig. 4). In the Anaplasma marginale Florida and Anaplasma phagocytophilum HZ specimens, DNA could be amplified by a single-step PCR based on the p44 (msp2) genes, as described previously (20). Neither the first-round nor the nested-PCR negative controls amplified any signals, showing that there was no DNA contamination from the environment. Eleven of the PCR product clones were sequenced, and all had sequences identical to the sequence obtained above (GenBank accession no. AY125087). These findings also indicate that this method is sufficiently sensitive to detect A. pullorum DNA from a 20-year-old stained blood smear on a slide.
FIG. 4.

Analyses of A. pullorum-specific PCR. Lanes 1 through 3, A. pullorum Texas DNA from the 20-year old slides, Anaplasma marginale Florida strain DNA, and Anaplasma phagocytophilum HZ strain DNA, respectively; lanes 4 and 5, first-step PCR and the nested-PCR template-less negative controls; lane M, 1-kb Plus DNA ladder.
The present molecular phylogenetic study revealed that the A. pullorum Texas strain is most closely related to the Anaplasma species, which is consistent with the previous decision to include this bacterium in the family Anaplasmataceae (24). This classification was based on its ultrastructure and other phenotypic characteristics. These include such observations as (i) A. pullorum does not multiply in cell-free media or in tissue cultures (15), (ii) attempts at continuous propagation of the organism in chicken embryos have not been successful (15), (iii) tetracyclines are effective in treating A. pullorum infection (24), and (iv) A. pullorum is transmitted by ticks (15, 17).
The 16S rRNA and groEL base sequences and A. pullorum-specific PCR developed in this study should advance our understanding of this elusive parasite in birds and ticks and facilitate the diagnosis and characterization of the diseases that are associated with it. Analysis of strains from Egypt, South Africa, and other parts of the world will clarify whether A. pullorum is distinct from proposed species “A. botuliformis” and whether it belongs to the genus Anaplasma or remains in a distinct genus. In general, it is extremely difficult to amplify DNA from old fixed and stained blood or tissue specimens, since such DNA is usually severely fragmented and tightly bound to dye molecules (5). Thus, the strategy and method developed in the present study may be useful for detecting other types of bacteria and their DNA sequences, in cases when fresh specimens are not readily available.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rRNA and groEL gene sequences of A. pullorum Texas are AY125087 and AY150648, respectively.
Acknowledgments
This work was partially funded by a grant R01 AI47885 from the National Institutes of Health.
REFERENCES
- 1.Barta, J. R., Y. Boulard, and S. S. Desser. 1989. Blood parasites of Rana esculenta from Corsica: comparison of its parasites with those of eastern North American ranids in the context of host phylogeny. Trans. Am. Microsc. Soc. 108:6-20. [Google Scholar]
- 2.Batteli, C. 1947. Si di un. piroplasma della Naia nigrocollis (Aegyptianella carpani n. sp.). Riv. Parassitol. 28:205-212. [Google Scholar]
- 3.Bird, R. G., and P. C. C. Garnham. 1969. Aegyptianella pullorum Carpano 1928—fine structure and taxonomy. Parasitology 59:745-752. [PubMed] [Google Scholar]
- 4.Brumpt, E., and G. Lavier. 1935. Sur un piroplasmide nouveau, parasite de tortue Tunetella emydis N. G., N. Sp. Ann. Parasitol. Hum. Comp. 13:544-550. [Google Scholar]
- 5.Burton, M. P., B. G. Schneider, R. Brown, N. Escamilla-Ponce, and M. L. Gulley. 1998. Comparison of histologic stains for use in PCR analysis of microdissected, paraffin-embedded tissues. BioTechniques 24:86-92. [DOI] [PubMed] [Google Scholar]
- 6.Carpano, M. 1928. Piroplasmosis in Egyptian fowles (Egyptianella pullorum). Vet. Serv. Bull. Egypt. Minist. Agric. Sci. Technol. Serv. 86:1-7.
- 7.Carpano, M. 1929. Su di un Piroplasma osservato nei polli Egitto (“Aegyptianella pullorum”). Nota preventiva. Clin. Vet. Milano 52:339-351. [Google Scholar]
- 8.Castle, M. D., and B. M. Christensen. 1985. Isolation and identification of Aegyptianella pullorum (Rickettsiales, Anaplasmataceae) in wild turkeys in north America. Avian Dis. 29:437-445. [PubMed] [Google Scholar]
- 9.Curasson, G. 1938. Notes sur la piroplasmose aviaire en E. O. F. Bull. Serv. Zootechnol. Epiz. A. O. F. 1:33-35. [Google Scholar]
- 10.Curasson, G., and P. Andrjesky. 1929. Sur les ′corps de Balfour' du sang de la poule. Bull. Soc. Pathol. Exot. 22:316-317. [Google Scholar]
- 11.Desser, S. S. 1987. Aegyptianella ranarum sp. n. (Rickettsiales, Anaplasmataceae): ultrastructure and prevalence in frogs from Ontario. J. Wildl. Dis. 23:52-59. [DOI] [PubMed] [Google Scholar]
- 12.Desser, S. S., and J. R. Barta. 1989. The morphological features of Aegyptianella bacterifera: an intraerythrocytic rickettsia of frogs from Corsica. J. Wildl. Dis. 25:313-318. [DOI] [PubMed] [Google Scholar]
- 13.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia, and Ehrlichia with Neorickettsia; description of six new species combinations; and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. E vol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 14.Gothe, R. 1967. Ein Beiträg zür systematischen Stellung von Aegyptianella pullorum Carpano 1928. Z. Parasitenkd. 29:119-129. [PubMed] [Google Scholar]
- 15.Gothe, R. 1971. Wirt-Parasit-Verhaltnis von Aegyptianella pullorum Carpano 1928, im biologischen Übertrager Argas (Persicargas) persicus (Oken, 1818) und im Virbeltierwirt Gallus gallus domesticus L. Fortschr. Veterinaermed. 16:(Suppl):1-144. [Google Scholar]
- 16.Gothe, R., and J. P. Kreier. 1984. Genus II. Aegyptianella Carpano 1929, 12AL, p. 722-723. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 17.Hadani, A., and Y. Dinur. 1968. Studies on the transmission of Aegyptianella pullorum by the tick Argas persicus. J. Protozool. 15(Suppl.):45.
- 18.Hecker, K. H., and K. H. Roux. 1996. High and low annealing temperatures increase both specificity and yield in ‘TouchDown’ and ‘StepDown’ PCR. BioTechniques 20:478-485. [DOI] [PubMed] [Google Scholar]
- 19.Huchzermeyer, F. W., I. G. Horak, J. F. Putterill, and R. A. Earle. 1992. Description of Aegyptianella botuliformis n. sp. (Rickettsiales: Anaplasmataceae) from the helmeted guineafowl, Numida meleagris. Onderstepoort J. Vet. Res. 59:97-101. [PubMed] [Google Scholar]
- 20.Kim, H.-Y., J. Mott, N. Zhi, T. Tajima, and Y. Rikihisa. 2002. Cytokine gene expression by peripheral blood leukocytes in horses during experimental infection with Anaplasma phagocytophila. Clin. Diagn. Lab. Immunol. 9:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laird, M., and F. A. Lari. 1957. The avian blood parasite Babesia moshkovskii (Shchurenkova, 1938), with a record from Corvus splejdens Vieillot in Pakistan. Can. J. Zool. 35:783-795. [Google Scholar]
- 22.Mutinga, M. J., and O. O. Dipeolu. 1989. Saurian malaria in Kenya: description of new species of haemoproteid and haemogregarine parasites Anaplasma-like and Pirhemocyton-like organisms in the blood of lizards in West Pokot district Kenya. Insect. Sci. Appl. 10:401-412. [Google Scholar]
- 23.Peirce, M. A. 1999. A new species of Aegyptianella from south-east Asia. Vet. Rec. 145:288. [DOI] [PubMed] [Google Scholar]
- 24.Ristic, M., and J. P. Kreier. 1974. Anaplasmataceae Philip 1957, p. 906-914. In R. E. Buchanan and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology, 8th ed. Williams & Wilkins, Baltimore, Md.
- 25.Schurenkova, A. 1938. Sogdianella moshkovskii gen. nov. sp. nov.—a parasite belonging to the Piroplasmidea in a raptororial bird—Gypaëtus barbatus L. Med. Parazitol. Parazit. Bolezni. 7:932-937. [Google Scholar]
- 26.Tarello, W. 2001. Aegyptianella-like organisms and microfilariae in a severely diseased bittern (Botautus stellaris stellaris). Rev. Med. Vet. 152:189-193. [Google Scholar]
- 27.Werner, J. K. 1993. Blood parasites of amphibians from Sichuan province, People's Republic of China. J. Parasitol. 79:356-363. [PubMed] [Google Scholar]