Abstract
We constructed a novel tool for genotypic analysis of human immunodeficiency virus type 1 (HIV-1) drug resistance by using an enzyme-linked minisequence assay (ELMA). ELMA is a combination of hybridization and a 1-base extension reaction, and we designed the assay to detect five mutations conferring nucleoside analogue resistance (M41L, D67N, K70R, T215Y, and M184V) and six mutations conferring protease inhibitor resistance (D30N, M46I, G48V, V82A, I84V, and L90M). At all detection points, ELMA demonstrated high sensitivity and specificity, sufficient for clinical use. Compared to that obtained by direct sequencing, the genotypic information obtained by ELMA is limited to the targeted loci for which it was designed. However, ELMA proves advantageous in several respects. The assay does not require expensive equipment, such as an autosequencer, and can be performed in regular clinical diagnostic laboratories. Therefore ELMA can be a candidate for a drug resistance monitoring assay to be introduced in developing countries. In addition, ELMA demonstrated higher sensitivity in the detection of minor resistant populations. We successfully detected a minor virus population (10%) by the assay. The high sensitivity and specificity of the assay recommend it as a first screening assay for drug resistance surveillance.
One of the major causes of human immunodeficiency virus type 1 (HIV-1) treatment failure is the emergence of drug-resistant viruses (10, 12, 18). Each anti-HIV-1 drug induces a specific amino acid mutation pattern responsible for drug resistance expression in its target enzyme, protease or reverse transcriptase (RT) (4). Therefore, the level of drug resistance can be evaluated by nucleotide sequencing of the part of the genome encoding the target enzyme. Several clinical cohort studies have shown that monitoring of the drug resistance genotype during treatment is beneficial to treatment outcome and prognosis—indeed, such testing appears to be necessary in order to proceed with high-quality treatment (1, 6). Nucleotide sequencing technology based on the Sanger method has advanced greatly in the past decade. Fluorerescein-labeled deoxynucleoside triphosphates (dNTP) and the capillary-type autosequencer have made it possible to analyze samples more easily and faster than previously possible. However, the expensive equipment required for such analysis limits its availability, especially in developing countries, where the need for drug resistance genotyping is increasing together with the introduction of generic anti-HIV-1 drugs. Thus, the development of inexpensive and rapid genotypic assays other than those using direct sequencing is eagerly anticipated.
To reduce the cost and increase the availability of drug resistance genotyping, several simplified mutation detection assays, such as line probe assays (19), oligonucleotide ligation assays (7), and mutagenically separated PCR (8, 16), have been developed. Hybridization is the technology commonly used to identify specific nucleotide sequences, by using short complementary oligonucleotide probes, and specificity is controlled by a delicate probe-target annealing interaction. Therefore, unexpected mutations in the target sequence region may cause false results, and standard hybridization may not be a suitable strategy to apply to genes with high polymorphism. To minimize the effect of mutations within probe-targeted sequences and at the same time preserve the simplicity and availability of hybridization, we constructed an HIV-1 genotypic assay, named the enzyme-linked minisequence assay (ELMA), based on a modified hybridization procedure. In ELMA, two modifications to the standard hybridization method were introduced. First, a relatively low annealing temperature was selected for the hybridization reaction. The less-restricted hybridization condition minimized the effect of unexpected mutations within the target sequence and decreased the risk of false-negative results. Second, a 1-base extension reaction of the probe with tagged deoxynucleotide was added after the hybridization step. By this enhanced process, it became possible to control the reaction performance by the 3′ end of the probe. False-positive results due to probe-target misannealing during the hybridization step were eliminated by this additional 3′-end control. By these modifications, we successfully constructed a genotyping assay designed to detect representative drug resistance mutations of zidovudine (AZT) and lamivudine (3TC) and primary mutations of the protease inhibitors.
MATERIALS AND METHODS
Basics of ELMA.
The basic technology of ELMA is a combination of DNA hybridization and point mutation detection by 1-base elongation with a biotinylated deoxynucleotide, i.e., a minisequence (11). The assay consists of four major steps: (i) extraction and amplification, (ii) hybridization, (iii) extension, and (iv) visualization. In the first step, extraction and amplification, target DNA fragments with one or more detection points are amplified from patient plasma viral RNA. In the second step, hybridization, the denatured amplified target DNA fragments are captured by the corresponding oligonucleotide probe applied to an enzyme-linked immunosorbent assay plate. The third step is an extension step. A biotinylated dNTP is incorporated on the 3′ end of each oligonucleotide probe. The final step is visualization, in which the incorporated biotinylated dNTP is visualized by using horseradish peroxidase (HRP)-conjugated avidin and HRP as substrates.
Extraction of viral RNA and amplification of target fragments.
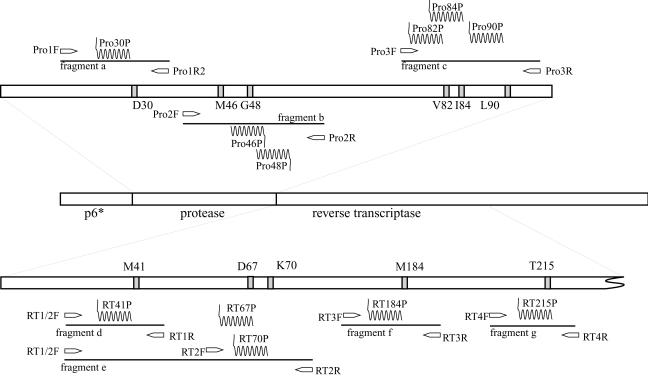
HIV-1 RNA was extracted from 200 μl of patient plasma by using a commercially available viral RNA extraction kit (Roche Diagnostics, Basel, Switzerland). Reverse transcription and the outer PCR were performed by using a one-step RT-PCR system (Takara, Osaka, Japan) with a 30-min reverse transcription step at 60°C, followed by 30 cycles of three-step PCR as follows: 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. With this outer PCR, 480-bp protease fragments and 888-bp RT fragments were amplified independently. The primers used in this outer PCR are shown in Table 1. In the inner PCR step, short target DNA fragments, three in the protease region (Fig. 1, fragments a to c) and four in the RT region (Fig. 1, fragments d to g), were amplified with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, Calif.) and the primers listed in Table 2. The outer PCR products were denatured by 5 min of incubation at 95°C, followed by 40 cycles of three-step PCR as follows: 95°C for 30 s, 60°C for 30 s, and 72°C for30 s. Immediately after PCR termination, 50 μl of denaturation buffer (0.4 M NaOH) was added to the PCR tubes to keep the amplicons as single-stranded DNA and to inactivate residual Taq enzyme. These amplified DNA fragments included 11 drug resistance mutations, as follows: 6 mutations conferring resistance to major protease inhibitors (D30N, M46I, G48V, V82A, I84V, and L90M) (3, 9, 17), 4 mutations conferring resistance to nucleoside analogues (M41L, D67N, K70R, and T215Y) (13), and the 3TC resistance mutation M184V (21). The details of the fragments are summarized in Table 2.
TABLE 1.
Primers used for amplification of the first-strand protease and RT DNA fragments
| Enzyme targeted | Primer designation (orientation) | Sequence |
|---|---|---|
| Protease | PRO5 (sense) | 5′-AGA CAG GYT AAT TTT TTA GGG A |
| PRO2L (antisense) | 5′-TAT GGA TTT TCA GGC CCA ATT TTT GA | |
| RT | RT1L (sense) | 5′-ATG ATA GGG GGA ATT GGA GGT TT |
| RT4L (antisense) | 5′-TAC TTC TGT TAG TGC TTT GGT TCC |
FIG. 1.
PCR primers, amplified target fragments, and detection probes. A total of seven target fragments are amplified in the assay. There are six detection points in the protease region and five detection points in the RT region. Open arrows, PCR primers; solid lines, target fragments, wavy lines, detection primers; shaded boxes, target sites.
TABLE 2.
Primers used for amplification of target DNA and probes for hybridization
| Enzyme targeted | Position | ELMA type | Fragment | Primer for amplification
|
Probe for hybridization
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Directiona | Sequence | Pattern | Name | Sequence | ||||
| Protease | 30 | A | a | PRO1F | Senses | ATA GAC AAG GAA CTG TAT CC | D30 | PRO30P-W | Amine-AGG AAG CTC TAT TAG ATA CAG GAG CAG ATG |
| PRO1R2 | Antisense | AAA TTC ATT TCT TCT AAT ACT GT | N30 | PRO30P-M | Amine-AGG AAG CTC TAT TAG ATA CAG GAG CAG ATA | ||||
| 46 | A | b | PRO2F | Sense | GCC AGG AAG ATG GAA ACC AA | M46 | PRO46P-WC | Amine-TTT GAT AAA ACC TCC AAT TCC CMC TAC CAT | |
| PRO2R | Antisense | TGT AGG TCC TAC TAA TAC TG | M46 | PRO46P-WA | Amine-TTT GAT AAA ACC TCC AAT TCC CMC TAA CAT | ||||
| 146 | PRO46P-MTTG | Amine-TTT GAT AAA ACC TCC AAT TCC CMC TAT CAA | |||||||
| 146 | PRO46P-MATA | Amine-TAC TTT GAT AAA ACC TCC AAT TCC CMC TAT | |||||||
| 48 | A | G48 | PRO48P-W | Amine-TCT TAC TTT GAT AAA ACC TCC AAT TCC CCC | |||||
| V48 | PRO48P-M | Amine-TCT TAC TTT GAT AAA ACC TCC AAT TCC CAC | |||||||
| 82 | A | c | PRO3F | Sense | ATA CCC ATA GAA ATC TGT GG | V82 | PRO8P-2W | Amine-GGT ACA GTA TTA GTA GGA CCT ACA CCT GTC | |
| PRO3R | Antisense | GGA AAA TTT AAA GTG CAA CCA A | A82 | PRO82P-M | Amine-GGT ACA GTA TTA GTA GGA CCT ACA CCT GCC | ||||
| 84 | A | I84 | PRO84P-W | Amine-CAG TAT TAG TAG GAC CTA CAC CTG TCA AYA | |||||
| V84 | PRO84P-M | Amine-CAG TAT TAG TAG GAC CTA CAC CTG TCA AYG | |||||||
| 90 | A | L90 | PRO90P-W | Amine-CAC CTG TCA ACA TAA TTG GAA GAA ATC TGT | |||||
| M90 | PRO90P-M | Amine-CAC CTG TCA ACA TAA TTG GAA GAA ATC TGA | |||||||
| RT | 41 | A | d | RT1/2F | Sense | GTT AAA CAA TGG CCA TTG ACA GA | M41 | RT41P-W | Amine-TAA AAG CAT TAG TAG AAA TTT GTA CAG AAA |
| RT1R | Antisense | GTA TGG ATT TTC AGG CCC AAT T | L41 | RT41P-M | Amine-TAA AAG CAT TAG TAG AAA TTT GTA CAG AAC | ||||
| L41 | RT41P-MT | Amine-TAA AAG CAT TAG TAG AAA TTT GTA CAG AAT | |||||||
| 67 | A | e | RT1/2F | Sense | GTT AAA CAA TGG CCA TTG ACA GA | D67 | RT67P-W | Amine-ATA CTC CAG TGT TTG CCA TAA AGA AAA ARG | |
| RT2R | Antisense | TGA ACT TCC CAG AAG TCT TGA G | N67 | RT67P-M | Amine-ATA CTC CAG TGT TTG CCA TAA AGA AAA ARA | ||||
| 70 | A | K70 | RT70P-W | Amine-GTT CTC TGA AAT CTA CTA ATT TTC TCC ATT | |||||
| R70 | RT70P-M | Amine-GTT CTC TGA AAT CTA CTA ATT TTC TCC ATC | |||||||
| 184 | A | f | RT3F | Sense | AGC ATG ACA AAA ATC TTA GAG CC | M184 | RT1840P-W | Amine-AAA ATC CAG ACA TAG TTA TCT ATC AAT ACA | |
| RT3R | Antisense | TAT TTC TAA GTC AGA TCC TAC ATA | V184 | RT184P-M | Amine-AAA ATC CAG ACA TAG TTA TCT ATC AAT ACG | ||||
| 215 | B | g | RT4F | Sense | GCA GCA TAG AAC AAA AAT AGA GG | T215Y | RT215-P | Amine-CTG AGA CAA CAT CTG TTG AGG TGG GGA TTT | |
| RT4R | Antisense | TAT CAG GAT GGA GTT CAT AAC C | |||||||
Hybridization of amplified targets and determination of alleles by 1-base extension reaction.
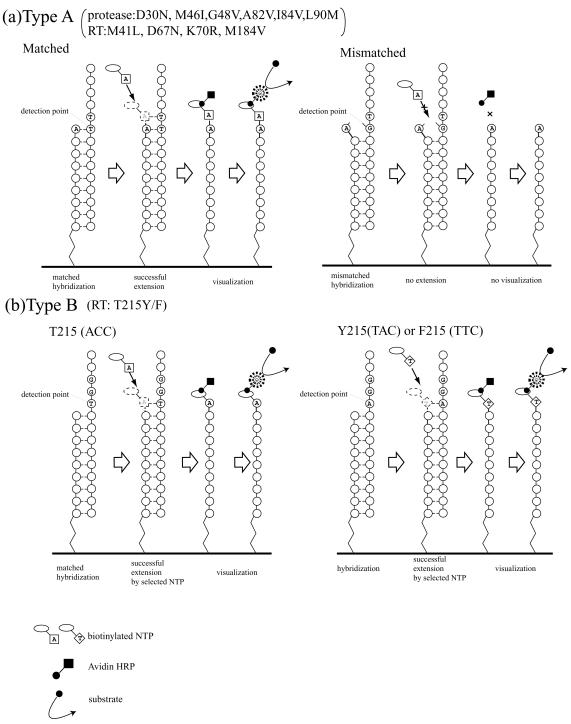
Key to the present assay are the designs of the hybridization probe and the minisequence step following hybridization. Two types of detection strategy were employed (Fig. 2). The first strategy, type A, was to determine the nucleoside pattern of the detection point by annealing of the 3′ end of the hybridization probe. In this strategy, a mutation point locates exactly on or 1 base upstream of the 3′ end of the probe. If the 3′ end of the probe exactly matches the target DNA, then a biotinylated dNTP, complementary to the target locus, will be incorporated in the subsequent extension step. On the other hand, if the 3′ end of the probe does not match the target sequence, the biotinylated dNTP will not be incorporated. Thus, two probes are required in this strategy, one for the wild type and the other for the mutant, and the nucleoside pattern is determined by ascertaining whether the biotinylated dNTP is incorporated or not. This type A strategy is used for determining six protease inhibitor resistance mutations (D30N, M46I, G48V, V82A, I84V, and L90M) and four RT inhibitor resistance mutations (M41L, D67N, K70R, and M184V). In the second strategy, type B, probes were designed to reach exactly 1 base before the detection point, and the nucleoside pattern was defined by analyzing the type of biotinylated dNTP taken up during the extension reaction. Therefore, only one common probe is required for the type B assay. This type B strategy is employed for the T215Y assay, and the wild type and the mutant are distinguished by incorporation of biotinylated dATP or dTTP. Figure 3 shows the alignment of the probes in a 96-plate format.
FIG. 2.
Two ELMA detection strategies. (a) Type A. The 3′ end of the detection primer is designed to reach exactly the detection point. (Left) The corresponding biotinylated dNTP is incorporated at the 3′ end of the probe if the 3′ end of the probe matches with the target DNA. (Right) There will be no incorporation in the case of mismatch. Thus, two probes, a wild-type-specific and a mutant-specific probe, are used for the assay. (b) Type B. The 3′ end of the detection primer is designed to reach 1 base before the detection point. The mutation pattern is determined by the type of biotinylated dNTP (dATP or dTTP) incorporated at the detection point. Therefore, the probe in the type B strategy is not type specific.
FIG. 3.
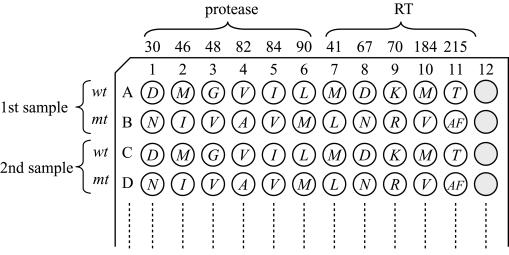
Alignment of hybridization probes in a 96-plate format. There are two rows for each sample. The first row of each sample is coated with wild-type (wt) detection probes, and the second row of each sample is coated with mutant (mt) probes. The italicized letter in each well demonstrates the detectable amino acid pattern.
Oligonucleotide probes were covalently bound to 96-well DNA-binding plates (Corning Costar Corp, Cambridge, Mass.) by a 3-h incubation at 37°C. The wells coated with oligonucleotide probes were filled with 100 μl of hybridization buffer (6× SSPE [pH 7.4] [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 1% [wt/vol] Tween 20, 0.03 M HCl), and subsequently each PCR amplicon was applied to the appropriate well and incubated for 1 h at 55°C. After the hybridization step, AmpliTaq DNA polymerase (Applied Biosystems) and the appropriate biotinylated dNTP were added to each well and incubated for 1 h at 37°C. By this reaction, the corresponding dNTP was incorporated into the 3′ end of the probe. Subsequently, hybridized target DNA was completely detached from the oligonucleotide probes and removed from the wells by three washes with a washing buffer (0.5× phosphate-buffered saline-0.1% Tween 20). Following this step, the covalently linked oligonucleotide probe with or without the corresponding biotinylated dNTP remained in the well. The final step was the determination step. Streptavidin-HRP (Roche Diagnostics, Mannheim, Germany) was added to the well and incubated for 30 min at room temperature. Each well was again washed three times with the washing buffer, followed by the addition of the substrate TMBlue (Cytech) to visualize incorporation of the dNTP at the 3′ ends of the probes.
Evaluation of the sensitivity of the assay by limiting dilution.
The sensitivity of ELMA at each detection point was evaluated by limiting dilution of the template DNA. An HXB2 wild-type clone and 11 recombinant clones, each with a single drug resistance mutation, which were selected as detection points of the assay, were used. The 11 recombinant clones were constructed on an HXB2 backbone as described previously (20). The copy numbers of the fragments were calculated according to the concentration and size of the plasmid DNA. Serial 10-fold dilutions ranging from 106 to 103 were made for each plasmid clone, and ELMA genotyping was performed for all of the dilutions. In these analyses, hybridization cutoff levels were evaluated from the mismatched pairs of target DNA and hybridization probe, i.e., mutant target versus wild-type probe or wild-type target versus mutant probe.
Evaluation of the assay sensitivity for detection of minor mutant populations.
The ability of the assay to detect minor mutant populations was evaluated by analyzing the mixture of wild-type and mutant templates. The ratios of the wild type to the mutant in the mixtures were 1:1, 10:1, and 100:1. The total DNA template amount was fixed at 105 copies. The test was performed for all 11 detection points. The analyses were repeated four times with independently prepared serial dilutions each time.
Evaluation of assay performance against patient samples.
To evaluate the reliability of ELMA, patient samples were analyzed both by ELMA and by standard sequencing, and the results of the two assays were compared. Forty-five samples were chosen randomly from the HIV-1-infected patient samples sent to the National Institute of Infectious Diseases for routine drug resistance genotyping from November 1996 to November 2000.
The details of in-house sequencing have been described elsewhere (15). In brief, HIV-1 RNA was extracted from 200 μl of patient plasma and reverse transcribed to cDNA by using murine leukemia virus RT (Takara). Subsequently, a 480-bp fragment, which covers the whole protease region, and an 888-bp RT fragment including all the known drug resistance mutation points were amplified individually by nested PCR. The nucleotide sequence of each DNA fragment was analyzed by cycle sequencing using Big-Dye terminator chemistry (Applied Biosystems) and an ABI-377 autosequencer (Applied Biosystems). Electropherograms were carefully analyzed using Sequence Navigator (Applied Biosystems).
RESULTS
Evaluation of assay sensitivity and end point level of the assay.
The ELMA data for each detection probe against wild-type and mutant target DNAs are summarized in Tables 3 and 4. In order to determine the limit of the copy number that could be detected by the assay, each target DNA was serially diluted in the range of 106 to 101 copies. Average optical densities (OD) with standard deviations (SD) based on quadruplicated data are shown.
TABLE 3.
Detection end points of wild-type ELMA probes
| Codon | Target | ODa of probe-target pair at the following copy number of the target:
|
Meanc | SDd | Cutoffb (mean + 3 SD) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | 101 | |||||
| 41 | Wild type | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 1.942 ± 0.117 | 0.212 | ||
| Mutant | 0.103 ± 0.010 | 0.103 ± 0.073 | 0.121 ± 0.063 | 0.096 ± 0.005 | 0.096 ± 0.009 | 0.105 ± 0.007 | 0.104 | 0.036 | ||
| 67 | Wild type | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 1.952 ± 0.096 | 0.311 | ||
| Mutant | 0.193 ± 0.027 | 0.210 ± 0.016 | 0.225 ± 0.022 | 0.249 ± 0.009 | 0.233 ± 0.015 | 0.165 ± 0.024 | 0.212 | 0.033 | ||
| 70 | Wild type | 1.553 ± 0.344 | 1.673 ± 0.385 | 1.587 ± 0.349 | 1.828 ± 0.344 | 1.903 ± 0.195 | 1.736 ± 0.260 | 0.125 | ||
| Mutant | 0.093 ± 0.007 | 0.059 ± 0.009 | 0.075 ± 0.007 | 0.056 ± 0.001 | 0.070 ± 0.012 | 0.094 ± 0.010 | 0.074 | 0.017 | ||
| 184 | Wild type | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 1.840 ± 0.321 | 0.560 | ||
| Mutant | 0.088 ± 0.008 | 0.078 ± 0.002 | 0.088 ± 0.007 | 0.091 ± 0.027 | 0.061 ± 0.007 | 0.256 ± 0.369 | 0.110 | 0.150 | ||
| 215 | Wild type | >2.000 | >2.000 | 1.949 ± 0.067 | 0.585 ± 0.097 | 0.085 ± 0.041 | 0.041 ± 0.006 | 0.171 | ||
| Mutant | 0.041 ± 0.007 | 0.078 ± 0.067 | 0.082 ± 0.065 | 0.044 ± 0.004 | 0.029 ± 0.007 | 0.034 ± 0.005 | 0.051 | 0.040 | ||
| 30 | Wild type | >2.000 | >2.000 | >2.000 | >2.000 | 1.810 ± 0.275 | 0.410 ± 0.131 | 0.521 | ||
| Mutant | 0.305 ± 0.031 | 0.269 ± 0.027 | 0.273 ± 0.025 | 0.310 ± 0.033 | 0.150 ± 0.029 | 0.056 ± 0.017 | 0.227 | 0.098 | ||
| 46 | Wild type | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 1.464 ± 0.681 | 0.156 | ||
| Mutant | 0.084 ± 0.011 | 0.094 ± 0.008 | 0.096 ± 0.007 | 0.103 ± 0.018 | 0.076 ± 0.011 | 0.034 ± 0.009 | 0.081 | 0.025 | ||
| 48 | Wild type | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 1.464 ± 0.659 | 1.455 | ||
| Mutant | 0.448 ± 0.014 | 0.411 ± 0.041 | 0.415 ± 0.041 | 0.435 ± 0.057 | 0.735 ± 0.170 | 1.123 ± 0.216 | 0.594 | 0.287 | ||
| 82 | Wild type | >2.000 | >2.000 | 1.682 ± 0.637 | 1.937 ± 0.120 | 0.379 ± 0.376 | 0.602 ± 0.981 | 0.328 | ||
| Mutant | 0.184 ± 0.034 | 0.164 ± 0.013 | 0.166 ± 0.014 | 0.171 ± 0.029 | 0.041 ± 0.005 | 0.036 ± 0.007 | 0.127 | 0.067 | ||
| 84 | Wild type | >2.000 | >2.000 | 1.756 ± 0.489 | >2.000 | 0.611 ± 0.509 | 0.677 ± 0.957 | 0.284 | ||
| Mutant | 0.141 ± 0.019 | 0.134 ± 0.014 | 0.135 ± 0.013 | 0.162 ± 0.004 | 0.229 ± 0.018 | 0.201 ± 0.012 | 0.167 | 0.039 | ||
| 90 | Wild type | >2.000 | >2.000 | >2.000 | 1.936 ± 0.129 | 1.261 ± 0.863 | 0.082 ± 0.055 | 0.213 | ||
| Mutant | 0.134 ± 0.013 | 0.126 ± 0.009 | 0.125 ± 0.011 | 0.086 ± 0.010 | 0.039 ± 0.006 | 0.048 ± 0.005 | 0.093 | 0.040 | ||
Average ± SD based on quadruplicated data.
Determined from OD of mismatched probe-target pairs (wild-type probe and mutant target). The lowest copy number with an OD higher than the cutoff is designated the end point of the detection (indicated by boldfaced OD).
Mean OD of mismatched probe-target pairs.
SD of mismatched probe-target pairs.
TABLE 4.
Detection end points of mutant ELMA probes
| Codon | Target | ODa of probe-target pair at the following copy number of the target:
|
Meanc | SDd | Cutoffb (mean + 3 SD) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | 101 | |||||
| 41 | Wild type | 0.145 ± 0.026 | 0.182 ± 0.025 | 0.167 ± 0.004 | 0.172 ± 0.011 | 0.171 ± 0.119 | 0.191 ± 0.015 | 0.171 | 0.048 | |
| Mutant | >2.000 | >2.000 | >2.000 | 1.531 ± 0.938 | >2.000 | 1.786 ± 0.251 | ||||
| 67 | Wild type | 0.164 ± 0.019 | 0.191 ± 0.016 | 0.194 ± 0.017 | 0.211 ± 0.029 | 0.149 ± 0.008 | 0.176 ± 0.071 | 0.181 | 0.036 | 0.289 |
| Mutant | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 1.995 ± 0.010 | ||||
| 70 | Wild type | 0.169 ± 0.029 | 0.173 ± 0.069 | 0.145 ± 0.022 | 0.208 ± 0.005 | 0.221 ± 0.029 | 0.287 ± 0.071 | 0.200 | 0.061 | 0.383 |
| Mutant | 1.143 ± 0.209 | 0.653 ± 0.248 | 0.659 ± 0.248 | 0.310 ± 0.031 | 0.526 ± 0.015 | 0.632 ± 0.072 | ||||
| 184 | Wild type | 0.096 ± 0.022 | 0.131 ± 0.015 | 0.123 ± 0.004 | 0.119 ± 0.006 | 0.061 ± 0.008 | 0.079 ± 0.013 | 0.101 | 0.028 | 0.185 |
| Mutant | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 1.614 ± 0.772 | ||||
| 215 | Wild type | 0.160 ± 0.024 | 0.204 ± 0.013 | 0.198 ± 0.030 | 0.115 ± 0.005 | 0.028 ± 0.006 | 0.035 ± 0.004 | 0.123 | 0.074 | 0.345 |
| Mutant | 1.157 ± 0.152 | 1.929 ± 0.143 | 1.928 ± 0.144 | 0.716 ± 0.050 | 0.121 ± 0.019 | 0.043 ± 0.013 | ||||
| 30 | Wild type | 0.106 ± 0.023 | 0.107 ± 0.007 | 0.130 ± 0.010 | 0.137 ± 0.010 | 0.054 ± 0.006 | 0.046 ± 0.005 | 0.096 | 0.037 | 0.207 |
| Mutant | 1.963 ± 0.074 | 1.862 ± 0.213 | 1.864 ± 0.208 | 1.800 ± 0.231 | 0.859 ± 0.137 | 0.194 ± 0.085 | ||||
| 46 | Wild type | 0.343 ± 0.035 | 0.295 ± 0.015 | 0.334 ± 0.022 | 0.319 ± 0.022 | 0.380 ± 0.273 | 0.089 ± 0.059 | 0.293 | 0.141 | 0.716 |
| Mutant | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | 0.843 ± 0.457 | ||||
| 48 | Wild type | 0.642 ± 0.068 | 0.526 ± 0.056 | 0.576 ± 0.054 | 0.668 ± 0.130 | 0.539 ± 0.095 | 0.500 ± 0.368 | 0.575 | 0.162 | 1.061 |
| Mutant | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | >2.000 | ||||
| 82 | Wild type | 0.309 ± 0.045 | 0.238 ± 0.050 | 0.222 ± 0.068 | 0.193 ± 0.030 | 0.051 ± 0.012 | 0.172 ± 0.129 | 0.197 | 0.099 | 0.494 |
| Mutant | >2.000 | >2.000 | >2.000 | 1.923 ± 0.154 | 0.064 ± 0.043 | 0.036 ± 0.005 | ||||
| 84 | Wild type | 0.160 ± 0.009 | 0.164 ± 0.039 | 0.199 ± 0.028 | 0.191 ± 0.015 | 0.104 ± 0.010 | 0.168 ± 0.021 | 0.164 | 0.037 | 0.275 |
| Mutant | >2.000 | >2.000 | >2.000 | 1.784 ± 0.302 | 0.150 ± 0.014 | 0.145 ± 0.022 | ||||
| 90 | Wild type | 0.368 ± 0.026 | 0.263 ± 0.068 | 0.310 ± 0.033 | 0.164 ± 0.060 | 0.053 ± 0.023 | 0.040 ± 0.004 | 0.200 | 0.132 | 0.596 |
| Mutant | >2.000 | >2.000 | >2.000 | >2.000 | 0.603 ± 0.664 | 0.050 ± 0.020 | ||||
Average ± SD based on quadruplicated data.
Determined from OD of mismatched probe-target pairs (mutant probe and wild-type target). The lowest copy number with an OD higher than the cutoff is designated the end point of the detection (indicated by boldfaced OD).
Mean OD of mismatched probe-target pairs.
SD of mismatched probe-target pairs.
The data of mismatched target DNA and probe pairs, i.e., wild type probe versus mutant target and mutant probe versus wild type target, were used to define the cutoff OD for each probe. The cutoff was calculated as the average OD + 3 SD. As shown in Tables 3 and 4, each of the probes has a unique cutoff value, which probably reflects the melting temperature of the probes. The highest cutoff value was 1.455 for the protease position 48 wild-type probe, and the lowest cutoff value was 0.125 for the RT position 70 wild-type probe. The detection end point (the lowest copy number for which an OD higher than the cutoff was obtained) for each detection point was determined by using the cutoff values listed in Tables 3 and 4.
Most of the probes were sensitive enough to detect templates of <102 copies. However, one wild-type probe (position 215) and three mutant probes (positions 215, 82, and 84) demonstrated lower sensitivities, with copy numbers at the 103 level. At most detection points, the sensitivities were at the same level for mutant and wild type detection. Three loci, positions 48, 82, and 84, showed different detection limits. The mutant probe was 1 log unit more sensitive than the wild-type probe at position 48, whereas the wild-type probes were 2 log units more sensitive than the mutant probes at positions 82 and 84.
Evaluation of assay sensitivity for detection of mutant populations mixed with wild-type populations.
In Tables 3 and 4 the sensitivities of the probes were evaluated with a clonal DNA target amplified from HXB2 clones. However, virus populations in patients exist as mixed populations in clinical samples. Therefore, the probe sensitivity was evaluated by testing a mixture of wild-type and mutant targets. The same wild-type and mutant target templates used in the end point assay were mixed in three different wild-type/mutant ratios: 1:1, 10:1, and 100:1. All of the mixtures were adjusted to 105 copies of DNA so that the 100:1 mixture would contain more than 103 copies of the mutant template, a number sufficient to be detected at all detection points. Each test was repeated four times. Although the 103 copy level was a sufficient template number for all mutant probes, only three mutants, M41L, V82A, and L90M, were successfully detected in four reproduced tests with a wild-type/mutant ratio of 100:1. For the other eight loci (D67N, K70R, M184V, T215Y, D30N, M46I, G48V, and I84V), the test was not sensitive enough to detect a 1% mutant population in the mixture (detection was consistently unsuccessful at this 100:1 ratio); however, the mutant population was successfully detected at a 10:1 ratio (10%). For single-population detection, the results show that the lowest detectable level was 102 copies. However, with a mixed viral population, 102 copies of mutant clones were not detected when mixed with 106 copies of wild-type clones. In that case, the lowest level of the minor population which could be detected was 103 copies/ml. This discrepancy between the detectable copy number of clonal and mixed target populations may be due to competition between HIV-1 mutant and wild-type target DNAs.
Evaluation of assay performance against patient samples.
The performance of ELMA with clinical samples was evaluated by testing 45 HIV-1 patient samples. The RNA copy number of the 45 patients ranged from 102.6 to 106.3 copies/ml (average, 105.3; median, 104.3). In this study, HIV-1 RNA was extracted from patient plasma, and target DNA was prepared by reverse transcription and nested PCR. The first PCR product was also analyzed by the direct sequencing method, and the result was compared with the ELMA result. The comparison of the direct sequencing results with the ELMA results is summarized in Table 5. The sensitivity and specificity of ELMA were calculated for each detection point, using the sequencing results as the standard. Because ELMA may be used for the first screening of drug resistance, it should capture all possible resistant cases. Therefore, in the calculations of specificity and sensitivity for which formulas are given in the footnotes to Table 5, “mutant” sequencing results and “mixture” ELMA results were considered concordant, as were “mixture” sequencing results and “mutant” ELMA results. In addition, “mixture” sequencing results and “wild-type” ELMA results were considered discordant. Further, cases in which “wild-type” sequencing results and “mixture” ELMA results were obtained were excluded from the calculations, because we could not rule out the possibility that ELMA had detected a minor mutant fraction that the sequencing failed to detect.
TABLE 5.
Summary of comparison between ELMA and sequencing results for clinical samples
| Enzyme targeted | Codon | No. of samples | No. of samples for which the following combination of resultsa was obtained:
|
Sensitivityb | Specificityc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | |||||
| RT | 41 | 43 | 21 | 14 | 1 | 3 | 0 | 2 | 1 | 1 | 0 | 0.947 | 0.955 |
| 67 | 44 | 20 | 10 | 3 | 5 | 0 | 6 | 0 | 0 | 0 | 1.000 | 1.000 | |
| 70 | 44 | 36 | 4 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0.800 | 1.000 | |
| 184 | 42 | 20 | 18 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 0.909 | 1.000 | |
| 215 | 31 | 10 | 18 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0.950 | 1.000 | |
| Protease | 30 | 38 | 22 | 11 | 0 | 0 | 0 | 2 | 0 | 1 | 2 | 0.786 | 1.000 |
| 46 | 45 | 30 | 8 | 3 | 0 | 1 | 1 | 1 | 1 | 0 | 0.923 | 0.968 | |
| 48 | 45 | 44 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.000 | 1.000 | |
| 82 | 43 | 36 | 5 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0.857 | 1.000 | |
| 84 | 45 | 41 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1.000 | 0.976 | |
| 90 | 45 | 23 | 18 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1.000 | 0.958 | |
a, wild type by both ELMA and sequencing; b, mutant by both methods; c, mixture by both methods; d, mixture by ELMA and mutant by sequencing; e, mutant by ELMA and mixture by sequencing; f; mixture by ELMA and wild type by sequencing; g, mutant by ELMA and wild type by sequencing; h, wild type by ELMA and mutant by sequencing; i, wild type by ELMA and mixture by sequencing.
Calculated as (b + c + d + e)/(b + c + d + e + h + i).
Calculated as a/(a + g).
As further shown in Table 5, more than 93% of the samples were analyzed successfully by ELMA at all detection points except position 215. At position 215, ELMA failed to successfully analyze 14 of 45 samples (31%). For some reason the PCR products of these failed samples did not respond to either the wild-type or the mutant probe in the hybridization step. To understand the reason for the unresponsiveness to the codon 215 probes, we compared the target sequences of the position 215 probes of the 31 successfully analyzed samples with those of the 14 failed samples. We found that the frequency of polymorphisms in the target sequences, especially in the 3′ half, was significantly higher in samples for which ELMA analysis failed. High mutation frequencies were observed at the 10th, 11th, 13th, 19th, 20th, and 27th bases of the target sequences in these failed cases.
As shown in Table 5, there were 14 discordant results in total. Among these, we were able to specify the reason for the discordance for three results (two results for M184V and one result for T215Y). For position 184, the assay was constructed to distinguish between methionine (encoded by ATG) (underlining indicates a point targeted by the ELMA probe) and valine (GTG) by targeting the first base of the triplet. However, in the two failed cases, the substitution resulted not in valine (GTG) but in isoleucine (ATA). Thus, in this mutation pattern, the assay could not detect the substitutions. We observed a similar pattern in a position 215 discordant case. The assay was designed to distinguish between threonine (ACC) and tyrosine or phenylalanine (TAC or TTC) by targeting the first base of the triplet. The mutation pattern of the discordant case was isoleucine (ATA); therefore, our assay failed to detect the mutant.
For the other 11 discordant cases, we could not explain the discordance either by substitution pattern or by sequence polymorphisms in the target regions. The most likely explanation is population deviation caused by PCR primer selectivity. In these cases, the DNA population different from that of direct sequencing was preferentially amplified in the nested PCR.
DISCUSSION
Our newly constructed genotyping assay, ELMA, a combination of hybridization and 1-base extension reaction, demonstrated high sensitivity and specificity, sufficient to detect 11 different drug resistance mutations. The most critical point in developing the assay was optimizing the common hybridization condition for 11 different probe-target bindings. Appropriate annealing temperature and hybridization buffer conditions differ according to the length and sequence of the probes, and ideally these should be chosen specifically for each probe. However, as our assay was constructed in a 96-well format, the same buffer and temperature were required for all the probes in order to keep the assay procedure simple. The melting temperatures of the probes ranged from 74 to 88°C according to the targeted sequence, and the final hybridization temperature used for the assay was 74°C, adjusted to the lowest melting temperature of all the probes.
Because the probe-targeted regions of protease inhibitor-resistant mutants had higher GC contents than those of RT inhibitor-resistant mutants, the hybridization condition was less restrictive for the protease inhibitor resistance mutations. This condition is reasonable, because generally protease is highly polymorphic and is expected to have multiple mutations in the probe target regions. In ELMA, the goal of the hybridization step is to capture the target DNA, and the determination of wild type or mutant is made through the binding of the probe 3′ end and the subsequent extension step. Therefore, the 3′-end nucleotide sequence pattern of the probes was critical for assay performance, and the balance between the attractive force of the matched nucleotide pairs and the repulsion force of the mismatched pairs appeared to affect the cutoff OD of the probes. In fact, each probe had a different cutoff value, as shown in Tables 3 and 4. The probes for G48V detection demonstrated significantly high cutoff values: 1.455 for the wild type and 1.061 for the mutant. These high cutoff values can be explained by examination of the sequences of the 3′ ends of the probes. As shown in Table 2, the wild-type and mutant probe sequences were TCCCCC-3′ and TCCCAC-3′, respectively. In the case of a mismatch between a wild-type probe and a mutant target, or between a mutant probe and a wild-type target, the nucleotide pair at the underlined position would be T-C or A-G, respectively. The repulsion forces produced by G-C and A-G mismatches (which may cause the 3′ end of the probe to become detached) are relatively weak compared to the attraction force caused by the surrounding four G-C matched pairs. Therefore, 3′-end cysteine tends to bind to the target even though the next nucleoside does not match with the target, and the high probability of misbinding resulted in a high OD cutoff. Although G48V probes demonstrated high cutoffs, this did not affect assay performance: as shown in Table 5, both the sensitivity and the specificity of ELMA for G48V scored 1.000.
More than the cutoff values, the polymorphisms observed in the target regions are critical for the assay. If there are too many polymorphisms in the target region, probes may not detect the amplified target DNA. In particular, we experienced this problem in designing the probe for the position 215 mutation. Only 31 out of 45 test samples were successfully analyzed by ELMA at position 215. When comparing the sequences of the probe target regions of the 31 successful samples and 14 failed samples, we noted that a significantly higher number of mutations accumulated in the failed samples. To improve the success rate of the assay, it may be necessary to design another probe, taking into consideration the frequency of the accumulated mutations in the probe target region. The limitation of the present probe design can be observed at other detection points as well. There were only four detection points (protease positions 46, 48, 84, and 90) at which all 45 test samples were successfully analyzed. The data suggest a requirement of multiple probes for each detection point to overcome nucleoside polymorphisms in the probe target regions.
Thus, compared to direct sequencing, ELMA is limited in the quality and quantity of the results. Still, the assay is attractive in several respects.
One interesting aspect of ELMA is that the test can detect a minor drug-resistant population equivalent to 10% of the total virus population according to the mixture analyses performed with recombinant clones. This number compares favorably to that for standard direct sequencing, which generally can detect a minor population equivalent to 30 to 50% of the total virus population (22). In the comparison of ELMA and direct sequencing for 45 patient samples, 16 samples tested “wild type” by direct sequencing and “mixture” by ELMA. The data suggest that minor drug-resistant mutant populations might have been detected by ELMA. To confirm the mixture result by ELMA, we performed multiple cloning for the same sequenced samples. Seventeen to 26 clones were sequenced in each sample, and we successfully detected drug-resistant mutant clones in 6 out of 16 samples. The frequencies of the mutant clones ranged from 11.7 to 47.6%. We could not find mutant clones in the remaining 10 samples, but we cannot conclude that these were false-positive results, as a possibility remains that ELMA detected minor populations of <5% in these samples. Another attractive feature of the assay is that the test can be performed in a few hours without the use of expensive equipment.
Taking these qualities into consideration, ELMA can be utilized in a practical manner in the following situations and for the following uses. First, as there is no requirement for expensive equipment such as autosequencers, and considering the high sensitivity of the assay, ELMA is an excellent candidate for drug resistance genotyping to be used in developing countries, where, with the greater availability of generic antiretroviral treatment, the introduction of a drug resistance monitoring system has been an urgent issue. Although specialized training is required to run the assay, a clinical diagnostic laboratory can introduce the assay without an investment in additional equipment. Second, ELMA can be used as a tool for drug-resistant population surveillance. Today, with regard to primary HIV-1 infection, there is an obvious risk of transmission of drug-resistant HIV-1 (2, 23). Because some of the drug-resistant HIV-1 strains demonstrate reduced viral replication activity compared to that of the wild-type virus (14), the resistant viruses can become the minor population upon termination of anti-HIV-1 treatment (5). This is an important issue in understanding the effect of preexisting resistant populations on antiretroviral treatment outcome and in the prognosis of infected patients. Therefore, it is imperative that minor hidden resistant virus populations in treatment-naïve patients be detected, and ELMA as we have described it here has an advantage in the survey.
In conclusion, we successfully constructed a new assay for genotypic analysis of drug resistance, which can be performed in a standard PCR laboratory. However, improvement of the assay through further simplification of the assay procedure, and addition of other important drug resistance mutation points which we have not yet designed, is required for use in clinical studies.
Acknowledgments
We thank the patients and physicians who were involved in our study. We thank the technical staff of Genome Science Inc for assisting in the experiments. We also thank Yumi Kanekama and Mary Phillips for help in preparing the manuscript.
This study was supported by a grant from the Organization of Pharmaceutical Safety and Research (OPSR) of Japan and by the Ministry of Health, Labor and Welfare of the Japanese Government.
REFERENCES
- 1.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, T. C. Merigan, et al. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 2.Boden, D., A. Hurley, L. Zhang, Y. Cao, Y. Guo, E. Jones, J. Tsay, J. Ip, C. Farthing, K. Limoli, N. Parkin, and M. Markowitz. 1999. HIV-1 drug resistance in newly infected individuals. JAMA 282:1135-1141. [DOI] [PubMed] [Google Scholar]
- 3.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Teppler, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 4.D'Aquila, R. T., J. M. Schapiro, F. Brun-Vezinet, B. Clotet, B. Conway, L. Demeter, R. M. Grant, V. Johnson, D. R. Kuritzkes, C. Loveday, R. W. Shafer, and D. D. Richman. 2003. Drug resistance mutations in HIV-1. Top. HIV Med. 11:92-96. [PubMed] [Google Scholar]
- 5.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 6.Durant, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 7.Edelstein, R. E., D. A. Nickerson, V. O. Tobe, L. A. Manns-Arcuino, and L. M. Frenkel. 1998. Oligonucleotide ligation assay for detecting mutations in the human immunodeficiency virus type 1 pol gene that are associated with resistance to zidovudine, didanosine, and lamivudine. J. Clin. Microbiol. 36:569-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frater, A. J., C. C. Chaput, S. Beddows, J. N. Weber, and M. O. McClure. 2001. Simple detection of point mutations associated with HIV-1 drug resistance. J. Virol. Methods 93:145-156. [DOI] [PubMed] [Google Scholar]
- 9.Jacobsen, H., M. Hanggi, M. Ott, I. B. Duncan, S. Owen, M. Andreoni, S. Vella, and J. Mous. 1996. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J. Infect. Dis. 173:1379-1387. [DOI] [PubMed] [Google Scholar]
- 10.Japour, A. J., S. Welles, R. T. D'Aquila, V. A. Johnson, D. D. Richman, R. W. Coombs, P. S. Reichelderfer, J. O. Kahn, C. S. Crumpacker, D. R. Kuritzkes, et al. 1995. Prevalence and clinical significance of zidovudine resistance mutations in human immunodeficiency virus isolated from patients after long-term zidovudine treatment. J. Infect. Dis. 171:1172-1179. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi, S., K. Shimada, H. Suzuki, K. Tanikawa, and M. Sata. 2000. Development of a new method for detecting a mutation in the gene encoding hepatitis B virus reverse transcriptase active site (YMDD motif). Hepatology Res. 17:31-42. [Google Scholar]
- 12.Larder, B. A., G. Darby, and D. D. Richman. 1989. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science 243:1731-1734. [DOI] [PubMed] [Google Scholar]
- 13.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 14.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukaide, M., W. Sugiura, M. Matuda, S. Usuku, Y. Noguchi, K. Suzuki, K. Kawata, A. Ito, H. Sagara, K. Yamada, M. Kondo, and M. Imai. 2000. Evaluation of Viroseq-HIV version 2 for HIV drug resistance. Jpn. J. Infect. Dis. 53:203-205. [PubMed] [Google Scholar]
- 16.Myint, L., K. Ariyoshi, H. Yan, A. J. Frater, W. Auwanit, P. Pathipvanith, K. Yamada, M. Matsuda, T. Chiba, K. Fujita, M. McClure, J. N. Weber, and W. Sugiura. 2002. Mutagenically separated PCR assay for rapid detection of M41L and K70R zidovudine resistance mutations in CRF01_AE (subtype E) human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 46:3861-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrin, L., and A. Telenti. 1998. HIV treatment failure: testing for HIV resistance in clinical practice. Science 280:1871-1873. [DOI] [PubMed] [Google Scholar]
- 19.Stuyver, L., A. Wyseur, A. Rombout, J. Louwagie, T. Scarcez, C. Verhofstede, D. Rimland, R. F. Schinazi, and R. Rossau. 1997. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob. Agents Chemother. 41:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiura, W., Z. Matsuda, Y. Yokomaku, K. Hertogs, B. Larder, T. Oishi, A. Okano, T. Shiino, M. Tatsumi, M. Matsuda, H. Abumi, N. Takata, S. Shirahata, K. Yamada, H. Yoshikura, and Y. Nagai. 2002. Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 46:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Laethem, K., K. Van Vaerenbergh, J. C. Schmit, S. Sprecher, P. Hermans, V. De Vroey, R. Schuurman, T. Harrer, M. Witvrouw, E. Van Wijngaerden, L. Stuyver, M. Van Ranst, J. Desmyter, E. De Clercq, and A. M. Vandamme. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for detection of resistance in mixed HIV-1 genotypic populations. J. Acquir. Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]
- 23.Yerly, S., L. Kaiser, E. Race, J. P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:729-733. [DOI] [PubMed] [Google Scholar]