Abstract
Porphyromonas gingivalis is a major pathogen in destructive periodontal disease in humans. Detection and quantification of this microorganism are relevant for diagnosis and treatment planning. The prevalence and quantity of P. gingivalis in subgingival plaque samples of periodontitis patients were determined by anaerobic culture and real-time PCR amplification of the 16S small-subunit rRNA gene. The PCR was performed with primers and a fluorescently labeled probe specific for the P. gingivalis 16S rRNA gene. By the real-time PCR assay, as few as 1 CFU of P. gingivalis could be detected. Subgingival plaque samples from 259 adult patients with severe periodontitis were analyzed. P. gingivalis was detected in 111 (43%) of the 259 subgingival plaque samples by culture and in 138 (53%) samples by PCR. The sensitivity, specificity, and positive and negative predictive values of the real-time PCR were 100, 94, 94, and 100%, respectively. We conclude that real-time PCR confirms the results of quantitative culture of P. gingivalis and offers significant advantages with respect to the rapidity and sensitivity of detection of P. gingivalis in subgingival plaque samples.
The microflora colonizing the oral cavities of humans consists of numerous bacterial species (15, 25). Most of these species are innocuous, but colonization of the subgingival plaque by certain species can lead to periodontal disease (6, 25, 26, 36). Periodontitis is a chronic, multifactorial inflammatory disease that leads to destruction of the tissues supporting the teeth, and it is a major cause of tooth loss (3). Periodontitis occurs in humans as well as in several animal species (30).
Periodontitis lesions are associated with a complex subgingival microflora which consists mainly of gram-negative bacterial species (37), of which the dark-pigmented organism Porphyromonas gingivalis is considered a major pathogen (2, 6, 20). P. gingivalis is a strict anaerobic, oral microorganism that is involved in periodontitis, endodontic infections, and odontogenic abscesses in humans (34). P. gingivalis is infrequently isolated from individuals with healthy periodontia (4, 5, 33). Anaerobic culture is most commonly used to detect and quantify major components of the subgingival plaque and to determine the in vitro antimicrobial susceptibilities of oral pathogens. Culture, however, has several drawbacks: it is time-consuming and laborious and has a low level of sensitivity. This is due to the extremely slow growth or very specific growth requirements of some oral pathogens. Several alternative methods have been developed for the detection of P. gingivalis, such as immunoassays (9), DNA probe assays (9, 22, 23), and PCR assays (2, 10, 17, 21).
Recently, real-time PCR has been shown to be a sensitive and rapid method for the detection and quantification of individual microbial species (7, 10, 11, 16). Most real-time PCR tests are based on the detection of bacterial small-subunit 16S rRNA sequences (7). This subunit of DNA is present in multiple copies in all bacterial species and contains highly conserved species-specific sequences.
Real-time PCR has also been described for the detection and quantification of P. gingivalis in subgingival plaque samples. However, no attempt was made to compare real-time PCR with the anaerobic culture technique (10) with a significant number of patient samples.
The aim of the present study was to develop a real-time PCR assay for the sensitive, specific, efficient, reproducible, and rapid detection and quantification of P. gingivalis in subgingival plaque samples and to compare the PCR results with anaerobic culture outcomes.
MATERIALS AND METHODS
Study population, sample collection, and bacterial culture.
Subgingival plaque samples from 259 adult patients with periodontitis were collected. Patients were >25 years old and had periodontal pockets >5 mm (mean pocket depth, 6.97 ± 1.18 mm) that showed bleeding upon pocket probing. The patients had not used antibiotics in the past 3 months. Samples were obtained from the deepest periodontal pocket in each quadrant of the dentition by using sterile paper points (12, 13). The samples were pooled in 1.5 ml of reduced transport fluid (28) and were processed for cultivation under anaerobic conditions within 4 h of sampling. Samples were vortexed for 2 min and split. A total of 100 μl of the sample was used for culture by tenfold serial dilution in sterile phosphate-buffered saline solution, and 100 μl was also used for real-time PCR.
A total of 100 μl of the dilutions were plated on blood agar plates (no. 2; Oxoid, Basingstoke, United Kingdom) supplemented with horse blood (5%; vol/vol), hemin (5 mg/liter), and menadione (1 mg/liter) and incubated in 80% N2-10% H2-10% CO2 at 37°C for 7 to 14 days. P. gingivalis was identified on the basis of Gram staining, anaerobic growth, the inability to ferment glucose, the production of indole, and positive hemagglutination with 3% sheep erythrocytes as well as the production of a set of metabolic enzymes (as tested with the Rapid ID kit 32A) (35); and the total number of CFU of P. gingivalis in positive samples was determined
Bacterial strains and growth conditions.
P. gingivalis strain W83 was used as a reference strain. Determination of the number of CFU of the P. gingivalis suspension per milliliter was made by growing the bacteria for 2 to 3 days in brain heart infusion supplemented with 5 mg of hemin per liter and 5 mg of menadione per liter, and serial dilutions were inoculated on blood agar plates as described above.
Table 1 shows the bacterial strains that were used in this study to test the specificity of the P. gingivalis primer-probe set. Bacterial strains were grown as recommended by the American Type Culture Collection (ATCC).
TABLE 1.
Species used to study the specificity of PCR primers and probe for detection of putative P. gingivalis isolates
| Bacteria | Species or type |
|---|---|
| Streptococcus sanguinis | Clinical isolate |
| Bacteroides fragilis | ATCC 25285 |
| Peptostreptococcus micros | Clinical isolate |
| Prevotella melaninogenica | ATCC 25845 |
| Prevotella denticola | Clinical isolate |
| Prevotella intermedia | ATCC 25611 |
| Prevotella nigrescens | NCTC 9338 |
| Porphyromonas endodontalis | Clinical isolate |
| Bacteroides asaccharolyticus | Clinical isolate |
| Bacteroides oralis | Clinical isolate |
Isolation of DNA from plaque samples and bacterial cultures.
The P. gingivalis culture dilution and plaque samples (100 μl) were used for automated DNA extraction and purification with the MagNA Pure DNA Isolation Kit III (Bacteria, Fungi; Roche Molecular Diagnostics). The protocol included 1 h of pretreatment with proteinase K (20 mg/ml) at 56°C. After isolation, the DNA was eluted in 100 μl of elution buffer.
To monitor the efficacy of the DNA isolation method, all samples were spiked with a known amount (1,000 CFU) of an Escherichia coli culture before DNA isolation.
PCR primers and probes.
The 16S rRNA sequences of the genus Porphyromonas were selected from the taxonomy database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/). A sequence alignment by using the multiple-alignment tool in the MegAlign program of the Lasergene system (DNAstar Inc.) was performed to search for homologous sequences within the 16S rRNA. The sequence of P. gingivalis W83 was used to select the primer and TaqMan probe sequences in a region of maximal homology by using Primer Express software (version 2.0; Applied Biosystems, Foster City, Calif.). This software generated series of best combinations for the P. gingivalis primer and probe set. The combinations were checked for primer-dimer or internal hairpin configurations, melting temperature, and percent G+C values.
The sequence of the forward primer, primer P.g.F, was 5′-GCGCTCAACGTTCAGCC-3′ (base pairs 612 to 628); the sequence of the reverse primer, primer P.g.R, was 5′-CACGAATTCCGCCTGC-3′ (base pairs 664 to 679); and the sequence of the Taqman probe, probe P.g.P, was 5′-CACTGAACTCAAGCCCGGCAGTTTCAA-3′ (base pairs 634 to 660). The homologies of the selected primers and the probe with unrelated sequences were checked by a search with the BLAST program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) (1).
The oligonucleotide probe was labeled with the fluorescent dyes 6-carboxyfluorescein at the 5′ end and 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. The E. coli primer-probe combination (8) was labeled with the fluorescent reporter dye VIC at the 5′ end and the quencher dye TAMRA at the 3′ end.
Optimization, sensitivity, and specificity of P. gingivalis-specific primer-probe set.
PCRs were performed by using a matrix of concentrations of the forward primer, the reverse primer, and the probe to determine the optimal concentration yielding the lowest threshold cycle (Ct) values and, hence, the highest amplification efficiencies.
The specificity of the real-time PCR assay was verified with purified genomic DNA from 10 different bacterial strains (Table 1).
The detection limit of the real-time PCR was assessed by determining the Ct values of serial 10-fold dilutions of purified genomic DNA from P. gingivalis strain W83. A standard curve prepared with these dilutions was used in every experiment.
Quantitative PCR assay.
PCR amplification was performed in a total reaction mixture volume of 25 μl. The reaction mixtures contained 12.5 μl of 2× TaqMan universal PCR master mixture (PCR buffer, deoxynucleoside triphosphates, AmpliTaq Gold, an internal reference signal [6-carboxy-X-rhodamine], uracil N-glycosylase, MgCl2; Applied Biosystems), 300 nM each P. gingivalis-specific primer, 100 nM P. gingivalis-specific probe. and 5 μl of purified DNA from plaque samples. Five microliters of the DNA extracted from P. gingivalis W83 was used to prepare the standard curve and as a positive control; the negative control was 5 μl of sterile H2O.
The samples were subjected to an initial amplification cycle of 50°C for 2 min and 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min. The data were analyzed with ABI 7000 Sequence Detection System software.
The degradation of the probe by the DNA polymerase in each elongation step induces an increase in fluorescence that can be monitored during PCR amplification. The fluorescence signal is normalized by dividing the reporter dye emission (6-carboxyfluorescein) by the emission of the passive reference (6-carboxy-X-rhodamine). The higher the starting copy number of the nucleic acid target is, the sooner a significant increase in fluorescence is observed. The Ct parameter is defined as the fractional cycle number at which the fluorescence of the reporter dye generated by cleavage of the probe crosses an arbitrarily defined threshold within the logarithmic phase. Hence, this parameter can be used to compare different amplification reactions.
The results for unknown plaque samples were projected on the standard curve generated with P. gingivalis strain W83.
Statistics.
The specificity was determined as the number of negative results by the real-time PCR assay divided by the number of negative results by the quantitative culture test. The sensitivity was determined as the number of positive results by the real-time PCR divided by the number of positive results by the quantitative culture test.
To compare the number of P. gingivalis cells present in subgingival plaque samples determined by the real-time PCR to the number obtained by culture, the nonparametric procedure sign test (SPSS software package, version 11.0) and a two-by-two matrix were used.
RESULTS
Specificities and sensitivities of PCR primers and TaqMan probe.
The specificity of the P. gingivalis primer-probe set based on the 16S rRNA sequences was determined with various oral and nonoral bacteria (Table 1). The primers specific for P. gingivalis specifically amplified P. gingivalis DNA, whereas PCR products were not obtained with any of the other bacterial species tested (data not shown). We also observed that quantification of DNA from P. gingivalis was not affected when DNA (range, DNA from 104 to 105 bacterial cells) from a variety of other species and/or genera was present in the PCR mixture.
To determine the detection limit of the primer-probe set, serial dilutions of cultures of P. gingivalis were used for determination of the number of CFU.
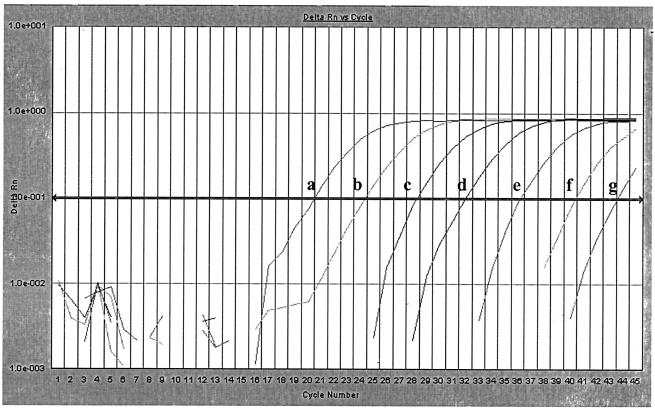
After DNA extraction, six dilutions (range, 0.65 to 650,000 CFU) (Fig. 1) for real-time PCR were prepared and tested. The estimated detection limit for P. gingivalis was 1 CFU.
FIG. 1.
Quantification of P. gingivalis amplification. Serial 10-fold dilutions (a to g, with 650,000 to 0.65 CFU/reaction mixture) of P. gingivalis DNA were amplified with primers P.g.F and P.g.R and detected with TaqMan probe P.g.P. ΔRn, change in fluorescence intensity. The correlation coefficient (R) for the Ct values was 0.999.
To test the reproducibility of the PCR assay, 10 subgingival plaque samples were tested twice. The average Ct values obtained with the dilution containing 65,000 CFU of P. gingivalis was 20.99 (standard deviation [SD] = 0.787).
Extraction of bacterial DNA from plaque samples.
To check the efficacy of DNA isolation from subgingival plaque samples, a known amount of E. coli (K-12) DNA (equivalent to 50 CFU/reaction mixture) was added to each plaque sample before DNA isolation. After isolation, the DNA was analyzed by real-time PCR with an E. coli-specific primer-probe combination (8). The fluorescent signal was compared to the signals on a standard curve generated with E. coli DNA. The efficacy of E. coli DNA isolation (Ct = 35.7 ± 1.2, 2 SDs) was not influenced by the DNA from plaque samples. Possible PCR inhibition was excluded by comparing the Ct values for P. gingivalis-negative samples spiked with 65 CFU of P. gingivalis with the Ct values for 65 CFU of P. gingivalis from a pure culture. The fluorescent signals (Ct = 35.8 ± 1.8, 2 SDs) in the presence or absence of a plaque sample were identical.
Validation of the real-time PCR for analysis of subgingival plaque samples.
The amount of P. gingivalis genome equivalents in each subgingival plaque sample determined by the real-time PCR was calculated and compared to the results obtained by quantitative anaerobic culture. The detection limit of the P. gingivalis real-time PCR for P. gingivalis was 200 cells/ml of subgingival plaque specimen in reduced transport fluid.
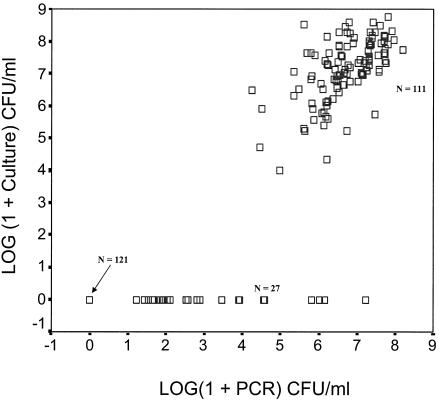
Figure 2 depicts the prevalence of P. gingivalis by real-time PCR and anaerobic culture in the 259 samples. The results obtained by real-time PCR matched the results obtained by anaerobic culture for 97% of the subjects infected with P. gingivalis. The number of positive results determined by both detection methods is summarized in a two-by-two matrix (Table 2). P. gingivalis was cultured from 111 (43%) of the 259 subgingival plaque plaques. All these culture-positive samples also appeared to be positive by the real-time PCR assay (100% sensitivity). In addition, 27 samples were positive for P. gingivalis by the real-time PCR but negative by culture. Twenty of the 27 samples contained <104 CFU/ml. The lowest dilution by culture was 10−3, which results in a detection limit of 10−4 CFU/ml. Seven other PCR-positive samples contained >104 P. gingivalis cells/ml. These samples were thawed and recultured for 14 days. Four of these samples yielded P. gingivalis after this prolonged incubation (32). P. gingivalis could not be retrieved from the other three samples. Of the 128 culture-negative samples, 121 were negative by this PCR assay (94% specificity). In no case (0%) was a PCR-negative, culture-positive result found (Table 2).
FIG. 2.
Scatter plot showing the differences and correlations between the real-time PCR and the anaerobic culture method. Data for P. gingivalis-positive versus P. gingivalis-negative samples by both methods (n = 11 and n = 121, respectively) fall close to the line of equivalence (R2 = 0.977). Samples that were PCR positive and culture negative fall near the x axis (n = 27). Samples which were negative by both methods are shown with an arrow (n = 121). A second linearity coefficient was calculated only for the quantitative results for the 111 samples positive by culture and PCR (R2 = 0.366).
TABLE 2.
Correlation between detection of P. gingivalis by real-time PCR and anaerobic culture in subgingival plaque samples
| Anaerobic culture result | No. (%) of samples with the following real-time PCR resulta:
|
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 111 (46.2) | 0 (0.0) | 111 |
| Negative | 7 (2.9) | 121 (50.4) | 128 |
| Total | 118 | 121 | 239 |
Sensitivity, 100%; specificity, 94%.
Table 3 shows the relationship between the number of P. gingivalis cells determined by anaerobic culture and real-time PCR in the 111 PCR-culture positive samples. There was a difference in cell number of less than 10-fold between PCR and culture for 73% of the samples. For 25.2% of the samples we found 10- to 100-fold differences in cell number. A difference of >100-fold was found for 1.8% of the 111 samples.
TABLE 3.
Real-time PCR versus anaerobic culture for quantification of P. gingivalis in PCR- and culture-positive subgingival plaque samples
| Difference in no. of CFU | No. (%a) of samples for which:
|
||
|---|---|---|---|
| Culture detected more than real-time PCR | Real-time PCR detected more than culture | Total | |
| <10 | 51 (45.9) | 30 (27) | 81 (73) |
| 10-100 | 25 (22.5) | 3 (2.7) | 28 (25.2) |
| >100 | 2 (1.8) | 0 (0) | 2 (1.8) |
| Total | 78 (70.3) | 33 (29.7) | 111 |
Percentage of subgingival plaque samples in which P. gingivalis was detected.
Figure 2 shows the correlation between all positive and negative results by both techniques. There was almost a complete correlation of the positive and negative results between the PCR and culture (R2 = 0.977). Comparison of the quantitative results only for samples positive by culture and PCR revealed a correlation coefficient of 0.366.
DISCUSSION
Microbiological studies have demonstrated that the composition of subgingival plaque is highly complex and variable. So far, about 500 bacterial species have been identified in healthy or diseased periodontal tissues (14, 15, 18, 25). This diversity might, however, even represent an underestimation, since the results of microbial culture are influenced by the sampling methods, the time between sampling and culture, the transport medium used, the choice of culture media and conditions, and identification techniques (27, 29).
In this study, we compared the results of a quantitative anaerobic culture method for the detection and quantification of P. gingivalis in subgingival plaque samples with those of a real-time PCR assay performed with the TaqMan 7000 system.
We found a high linear correlation (R2 = 0.977) between the positive and negative results obtained by real-time PCR and culture for all samples. This result is in accordance with those of other studies (19, 24, 31). Three of 259 samples (1.2%) were positive (>104 CFU) by real-time PCR but negative by culture, even after repeated culturing. Isolation of P. gingivalis was performed on a nonselective medium, which makes the isolation of small numbers of the organism in the presence of a large bacterial cell background difficult. In addition, anaerobiosis is sometimes difficult to maintain during sample collection. Differences between PCR and culture may be also due to insufficient homogenization of the samples, factors affecting the growth of different isolates, or possibly, the presence of antagonistic bacterial species. A major difference between PCR and culture is that PCR also detects nonviable bacterial cells, which may explain in part the higher detection rate by PCR.
Four of the seven PCR-positive, culture-negative samples were confirmed to be P. gingivalis positive by a second prolonged culture. These cultures contained slowly growing P. gingivalis isolates, which may explain the initial negative culture results. Porphyromonas endodontalis was isolated from one of the three culture-negative, PCR-positive samples, but the presence of P. gingivalis was not confirmed.
Twenty samples were PCR positive and culture negative because only 103 to 105 dilutions were used for culture, which affects the results presented in Table 2. The number of samples PCR positive and culture negative for P. gingivalis would change to 27 instead of 7. This would influence the specificity of the real-time PCR, which would decrease from 94 to 82%. However, exclusion of the 20 samples and inclusion of the 2 of 7 samples for which the results were confirmed increase the specificity to 96%. The results of the nonparametric sign test (SPSS software package, version 11.0) confirmed that for 60 plaque samples PCR was more sensitive than culture when P. gingivalis was present in the sample in small amounts.
The number of P. gingivalis CFU present in subgingival samples with at least 104 CFU/ml determined by real-time PCR correlated very well with the numbers of CFU determined by culture. This further confirms that the larger number of positive samples detected by PCR compared to the number detected by culture is due to the detection limit of culture. A linear correlation calculated on the basis of the quantitative results for the positive samples by both techniques (R2 = 0.366) showed that there is also a reasonably good correlation between the techniques at the quantitative level. The differences might be due to the dilution factors used for quantitative culture.
In conclusion, the results of real-time PCR confirm those of quantitative culture of P. gingivalis, and real-time PCR offers significant advantages with respect to the rapidity and sensitivity of detection of P. gingivalis in subgingival plaque samples.
Acknowledgments
We thank Laboral Diagnostics and the Clinic for Periodontology, Amsterdam, for providing patient material and J. Berkhof for help with statistical analysis of the data.
The present study was supported by the Academic Center for Dentistry, Amsterdam.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashimoto, A., C. Chen, I. Bakker, and J. Slots. 1996. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol. Immunol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 3.Brown, L. J., R. C. Oliver, and H. Loe. 1989. Periodontal diseases in the U.S. in 1981: prevalence, severity, extent, and role in tooth mortality. J. Periodontol. 60:363-370. [DOI] [PubMed] [Google Scholar]
- 4.Griffen, A. L., M. R. Becker, S. R. Lyons, M. L. Moeschberger, and E. J. Leys. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 36:3239-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haffajee, A. D., M. A. Cugini, A. Tanner, R. P. Pollack, C. Smith, R. L. Kent, Jr., and S. S. Socransky. 1998. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J. Clin. Periodontol. 25:346-353. [DOI] [PubMed] [Google Scholar]
- 6.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000 5:78-111. [DOI] [PubMed] [Google Scholar]
- 7.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 8.Huijsdens, X. W., R. K. Linskens, M. Mak, S. G. Meuwissen, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2002. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40:4423-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loesche, W. J. 1992. DNA probe and enzyme analysis in periodontal diagnostics. J. Periodontol. 63:1102-1109. [DOI] [PubMed] [Google Scholar]
- 10.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, F. E., M. A. Nadkarni, N. A. Jacques, and N. Hunter. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40:1698-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mombelli, A. 1991. What can the dentist expect in diagnostic procedures in the future? Philip. J. 8:109-114. (In German.) [PubMed]
- 13.Mombelli, A., H. McNabb, and N. P. Lang. 1991. Black-pigmenting gram-negative bacteria in periodontal disease. I. Topographic distribution in the human dentition. J. Periodontal Res. 26:301-307. [DOI] [PubMed] [Google Scholar]
- 14.Moore, W. E., L. V. Holdeman, E. P. Cato, R. M. Smibert, J. A. Burmeister, and R. R. Ranney. 1983. Bacteriology of moderate (chronic) periodontitis in mature adult humans. Infect. Immun. 42:510-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 16.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 17.Nozaki, T., Y. Kusumoto, M. Kitamura, H. Hirano, A. Kohyama, M. Hayakawa, H. Takiguchi, Y. Abiko, S. Murakami, and H. Okada. 2001. A sensitive method for detecting Porphyromonas gingivalis by polymerase chain reaction and its possible clinical application. J. Periodontol. 72:1228-1235. [DOI] [PubMed] [Google Scholar]
- 18.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggio, M. P., T. W. Macfarlane, D. Mackenzie, A. Lennon, A. J. Smith, and D. Kinane. 1996. Comparison of polymerase chain reaction and culture methods for detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in subgingival plaque samples. J. Periodontal Res. 31:496-501. [DOI] [PubMed] [Google Scholar]
- 20.Rodenburg, J. P., A. J. van Winkelhoff, E. G. Winkel, R. J. Goene, F. Abbas, and J. de Graff. 1990. Occurrence of Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in severe periodontitis in relation to age and treatment history. J. Clin. Periodontol. 17:392-399. [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2001. Rapid detection and quantification of five periodontopathic bacteria by real-time PCR. Microbiol. Immunol. 45:39-44. [DOI] [PubMed] [Google Scholar]
- 22.Savitt, E. D., M. W. Keville, and W. J. Peros. 1990. DNA probes in the diagnosis of periodontal microorganisms. Arch. Oral Biol. 35(Suppl.):153S-159S. [DOI] [PubMed] [Google Scholar]
- 23.Savitt, E. D., M. N. Strzempko, K. K. Vaccaro, W. J. Peros, and C. K. French. 1988. Comparison of cultural methods and DNA probe analyses for the detection of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis, and Bacteroides intermedius in subgingival plaque samples. J. Periodontol. 59:431-438. [DOI] [PubMed] [Google Scholar]
- 24.Slots, J., and C. Chen. 1993. Detection of Porphyromonas gingivalis associated with human periodontitis by DNA methods. Clin. Infect. Dis. 16(Suppl. 4):S317-S318. [DOI] [PubMed] [Google Scholar]
- 25.Socransky, S. S., and A. D. Haffajee. 1994. Evidence of bacterial etiology: a historical perspective. Periodontol. 2000 5:7-25. [DOI] [PubMed] [Google Scholar]
- 26.Socransky, S. S., and A. D. Haffajee. 1994. Implications of periodontal microbiology for the treatment of periodontal infections. Compend. Suppl. 18:S684-S685, S688-S693. [PubMed] [Google Scholar]
- 27.Strand, P., R. M. Palmer, and R. F. Wilson. 1987. Sampling of subgingival plaque: a comparison of two methods using darkfield microscopy. Oral Microbiol. Immunol. 2:142-144. [DOI] [PubMed] [Google Scholar]
- 28.Syed, S. A., and W. J. Loesche. 1972. Survival of human dental plaque flora in various transport media. Appl. Microbiol. 24:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner, A. C., and J. M. Goodson. 1986. Sampling of microorganisms associated with periodontal disease. Oral Microbiol. Immunol. 1:15-22. [DOI] [PubMed] [Google Scholar]
- 30.van Foreest, A. 2002. Dentition of the older animal. Tijdschr. Diergeneeskd. 127:554-559. (In Dutch.) [PubMed] [Google Scholar]
- 31.van Steenbergen, T. J., M. F. Timmerman, F. H. Mikx, G. deq Uincey, G. A. van der Weijden, U. van der Velden, and J. de Graaff. 1996. Discrepancy between culture and DNA probe analysis for the detection of periodontal bacteria. J. Clin. Periodontol. 23:955-959. [DOI] [PubMed] [Google Scholar]
- 32.van Winkelhoff, A. J., M. Clement, and J. de Graaff. 1988. Rapid characterization of oral and nonoral pigmented Bacteroides species with the ATB Anaerobes ID system. J. Clin. Microbiol. 26:1063-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Winkelhoff, A. J., B. G. Loos, W. A. Van Der Reijden, and U. Van Der Velden. 2002. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J. Clin. Periodontol. 29:1023-1028. [DOI] [PubMed] [Google Scholar]
- 34.van Winkelhoff, A. J., T. J. van Steenbergen, and J. de Graaff. 1988. The role of black-pigmented Bacteroides in human oral infections. J. Clin. Periodontol. 15:145-155. [DOI] [PubMed] [Google Scholar]
- 35.van Winkelhoff, A. J., T. J. van Steenbergen, N. Kippuw, and J. De Graaff. 1985. Further characterization of Bacteroides endodontalis, an asaccharolytic black-pigmented Bacteroides species from the oral S cavity. J. Clin. Microbiol. 22:75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolff, L. F., D. M. Aeppli, B. Pihlstrom, L. Anderson, J. Stoltenberg, J. Osborn, N. Hardie, C. Shelburne, and G. Fischer. 1993. Natural distribution of 5 bacteria associated with periodontal disease. J. Clin. Periodontol. 20:699-706. [DOI] [PubMed] [Google Scholar]
- 37.Zambon, J. J., and V. I. Haraszthy. 1995. The laboratory diagnosis of periodontal infections. Periodontol. 2000 7:69-82. [DOI] [PubMed] [Google Scholar]