Abstract
The genetic relatedness of 127 methicillin-resistant Staphylococcus aureus (MRSA) isolates, belonging to five major types as identified by pulsed-field gel electrophoresis (PFGE) and antibiotic resistance profiles, was examined further using phage typing and fluorescent amplified fragment length polymorphism (FAFLP). The MRSA isolates were recovered from patients at the Prince of Wales Hospital (PWH), Hong Kong, over a 13-year period, 1988 to 2000. These strains were also compared with representatives of the well-described MRSA international clones and with epidemic MRSA strains (eMRSA) 1 to 16 from the United Kingdom. Phage typing distinguished two major “clones” at this hospital: all of the phage type 1 (PT1) isolates belonged to PFGE types A, C, D, and E, while most of the PT2 isolates were associated with PFGE type B, which exhibited a unique antibiotic resistance profile. MRSA isolates belonging to PFGE subtype A2 were indistinguishable from the British eMRSA-1, while isolates of PFGE type B were closely related to eMRSA-9 by PFGE. Based on FAFLP, all five predominant PFGE types at the PWH belonged to one group and fell into the same cluster as eMRSA-1, -4, -7, -9, and -11 isolates. Multilocus sequence typing and staphylococcal cassette chromosome mec typing classified representatives of our MRSA isolates as members of the same clone (ST239-MRSA-III). Thus, the predominant MRSA isolates frin the PWH in the last decade are closely related to early United Kingdom eMRSA clones 1, 4, and 11 and are members of a lineage that includes the Brazilian MRSA clone.
Methicillin-resistant Staphylococcus aureus (MRSA) is a major pathogen in many hospitals around the world, and in Hong Kong hospitals MRSA is now endemic (11; D. J. Lyon, Abstr. 1st Int. Congr. Asia Pacific Soc. Infect. Control, abstr. S9.4, 1999). Previous studies have indicated that a diverse number of circulating MRSA strains have been present at the Prince of Wales Hospital (PWH), Shatin, Hong Kong, since it was commissioned in 1984 (5, 10, 24). In time, however, one or more clonal types often predominate within a hospital setting (12). The PWH is a 1,350-bed teaching hospital that serves a population of approximately 1 million (one-sixth of Hong Kong's total population) in the New Territories East region. At the PWH, five major MRSA types have been identified by antibiogram and pulsed-field gel electrophoresis (PFGE) analysis in a 13-year longitudinal study from 1988 to 2000 (M. Ip, D. J. Lyon, F. Chio, L. Tsang, and A. F. Cheng, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-308, p. 121, 2001).
PFGE is often recognized as the “gold standard” for typing MRSA (26). However, because PFGE analyzes genetic markers that undergo rapid evolutionary change, strains exhibiting differences of 1 to 6 fragments in their banding patterns may still be clonally related (3), and this must be considered in evaluating MRSA isolates over a longitudinal period. Clinical epidemiological information is needed to interpret PFGE results, and strains need to be compared to epidemic strains where possible. Strain typing is important for effective infection control, and the choice of method depends on the local strain types and the circumstances of the investigation. In this study, we sought to investigate further the relationship among the five predominant types at the PWH based on PFGE and antibiogram by using a combination of methods including phage typing and fluorescent amplified fragment length polymorphism (FAFLP). These strains were compared with representatives of MRSA clones including the Brazilian (7), Iberian (23), pediatric (HDE288) (22), and New York/Tokyo (BK2464) (6) clones and epidemic MRSA strains 1 to 16 (eMRSA-1 to -16) from the United Kingdom. FAFLP had been shown to be highly discriminatory against strains of eMRSA-15 (15) and was able to classify eMRSA-1 to -16 from the United Kingdom (13) and European isolates into nine clone clusters (14). Hence, FAFLP may be suitable for surveillance of MRSA epidemics at both the local and international levels.
MATERIALS AND METHODS
Bacterial isolates.
One hundred twenty-seven MRSA isolates representing the five predominant types from the PWH as defined by antibiogram and PFGE were examined (Ip et al., 41st ICAAC). These isolates included 44 (35%) from blood cultures and cerebrospinal fluid, 49 (39%) from superficial wounds, 12 (9%) from deep wound swabs, 15 (12%) from respiratory tract specimens, 3 from urine, and 4 from miscellaneous specimens obtained in the 13-year period from 1988 to 2000. The isolates were single-patient, nonduplicate MRSA isolates representative of strains from different clinical units within the hospital during this period. They were stored in the Protect Bead System (Technical Service Consultants Ltd., Heywood, Lancashire, United Kingdom) at −70°C. The identities of the S. aureus isolates were confirmed by colonial morphology, Gram staining, and the tube coagulase test. Methicillin resistance was screened by oxacillin (1-μg) disk susceptibility testing according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (21) and by demonstration of the 304-bp fragment of the mecA gene (bp 32274 to 32577; NCBI nucleotide sequence accession no. AB033763). Representative strains of the United Kingdom eMRSA-1 to -16 clones (a gift from M Enright, University of Bath, Bath, United Kingdom) and the Brazilian (HSJ216) (7), Iberian (HPV107) (23), pediatric (HDE288) (22), and New York/Tokyo clones (BK2464) (6) were included (a gift from H. De Lencastre and A. Tomasz, Laboratory of Microbiology, The Rockefeller University, New York, N.Y.).
Antimicrobial resistance profiles.
Antibiograms were determined by disk diffusion on Mueller-Hinton agar according to NCCLS guidelines (21). The antimicrobial agents tested included tetracycline (30 μg) (T), erythromycin (15 μg) (E), clindamycin (2 μg) (D), gentamicin (10 μg) (G), tobramycin (10 μg) (To), ciprofloxacin (5 μg) (Ci), chloramphenicol (30 μg) (C), cotrimoxazole (25 μg) (S), rifampin (5 μg) (R), netilmicin (30 μg) (N), mupirocin (5 μg) (M), and fusidic acid (10 μg) (F). Results were interpreted according to NCCLS guidelines (21), with the exception of fusidic acid and mupirocin, for which the equivalent breakpoints for resistance were ≥2 and ≥8 mg/liter, respectively (zones of inhibition, ≤29 and ≤21 mm, respectively) (4). S. aureus ATCC 25923 was included as a control. Strains with zones of inhibition falling into the category of intermediate susceptibility to a particular antibiotic were considered resistant.
Phage typing.
All isolates were subjected to phage typing by use of a standardized protocol (17) and according to the recommendations of Aucken et al. (2). A set of 23 international phages was used (a gift from Tyrone Pitt, Central Public Health Laboratory, Colindale, United Kingdom). These included group I (phages 29, 52, 52A, 79, and 80), group II (phages 3A, 3C, 55, 71, and 95), group III (phages 6, 42E, 47, 53, 54, 75, 77, 83A, 84, and 85), and group V (phages 81, 94, and 96) lytic phages. The lytic reactions on the inoculated plates were read visually after overnight incubation at 30°C and were graded as negative, inhibition, weak lytic reaction, or strong lytic reaction. Any strains nontypeable by a 100× routine test dilution (RTD) were tested with a 1,000× RTD.
PFGE.
MRSA was typed by PFGE using the SmaI restriction enzyme, as previously described (18). The fragments were resolved on a 1% gel on a Chef Mapper (Bio-Rad, Richmond, Calif.) PFGE apparatus at 6 V/cm for 22 h, with switching times ramped from 5 to 35 s at 14°C. An including angle of 120°C was used. A lambda DNA PFGE molecular size standard (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) and an ATCC control strain were included in each gel. PFGE patterns were interpreted according to the criteria of Tenover et al. (25).
FAFLP.
Approximately 500 ng of a DNA template was digested with MseI (New England Biolabs, Hitchin, United Kingdom) followed by EcoRI (Gibco BRL, Life Technologies, Paisley, United Kingdom) at 37°C. The DNA fragments were then ligated using double-stranded EcoRI and MseI adaptors (Life Technologies) as previously described (15). PCR was performed in a 25-μl volume containing 2.5 μl of ligated DNA and 22.5 μl of PCR master mix containing 2 μM Cy-5-labeled EcoRI primer (Genset, SA), 5 μM MseI primer containing the extra selective base C (MseI+C) (Life Technologies), 0.2 mM deoxynucleoside triphosphates (Amersham Pharmacia Biotech), 0.75 U of Taq DNA polymerase, and 2.5 μl of 10× PCR buffer (Amersham Pharmacia Biotech). PCR was performed in a DNA thermal cycler (GeneAmp PCR system 9600; Perkin-Elmer, Norwalk, Conn.) under conditions described previously (15). Five microliters of the PCR product was mixed with 3 μl of loading dye (formamide and bromphenol blue dye) and was denatured at 94°C for 3 min in a DNA thermal cycler (GeneAmp PCR system 9600; Perkin-Elmer). The Cy-5-labeled size markers, containing 1 μl of ALFexpress sizer 50-500 bp (5 fmol/μl; Amersham Pharmacia Biotech), 1 μl of an additional 75-bp oligonucleotide marker (25 fmol/μl) (Genset, SA), 3 μl of loading dye, and 3 μl of Tris-EDTA buffer, were denatured in the same way as the PCR products. The amplified DNA fragments were separated through a 6% polyacrylamide-7 M urea denaturing gel (Amresco, Solon, Ohio) in 0.5× Tris-borate-EDTA buffer on an ALFexpress DNA sequencer (Amersham Pharmacia Biotech), and electrophoresis was performed at 1,500 V, 60 mA, and 25 W for 700 min at 55°C. The Cy-5-labeled markers were included in each gel and were run every 4 or 5 lanes as reference points for inter- and intragel alignment.
The raw data from the ALFexpress were converted to TIFF format for analysis by BioNumerics software (version 2.5; Applied Maths, Kortrijk, Belgium). Cluster analysis of the FAFLP patterns was performed with the Dice similarity coefficient by using optimization at 0.1% and position tolerance and change toward the end of the fingerprint at 0.3%. Dendrograms were calculated by the unweighted pair-group method by using average linkages (UPGMA).
RESULTS
Antibiotic resistance profiles and PFGE.
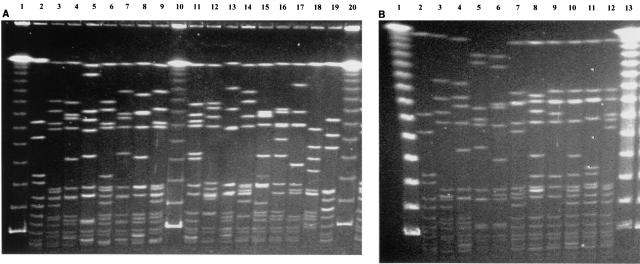
The five predominant MRSA types by PFGE and their respective antibiotic resistance profiles are listed in Table 1. The predominant type, PFGE type A, which includes 21 subtypes, has the antibiotic resistance pattern TEDGTOCi and was present throughout the 13-year period. eMRSA-1 showed a PFGE pattern identical to that of our PFGE subtype A2. Other eMRSA isolates, including eMRSA-4, -7, and -11, were also closely related to our MRSA PFGE type A isolates. The pulsotype of PFGE type B differed from that of PFGE type A by 7 fragments and was present only from 1995 to 2000. These isolates also carried unique antibiotic resistance profiles, and the majority were resistant to netilmicin and mupirocin and susceptible to clindamycin. eMRSA-9 also fell into a subtype of PFGE type B. The fingerprints of PFGE type C isolates differed by 5 bands from those of PFGE group A according to the interpretations of Tenover et al. (25) and are therefore possibly related to PFGE group A isolates. These isolates often have variable antibiotic resistance profiles. PFGE types D and E were minor types during this period of study but were more prevalent at the PWH after the year 2000 (unpublished data). The SmaI-digested PFGE profiles of the five predominant types at the PWH, types A through E, and those of eMRSA-1 to -16 and representatives of international clones are shown in Fig. 1.
TABLE 1.
Antibiotic resistance profiles of the five major Hong Kong MRSA PFGE types at the PWH
| PFGE type | No. of subtypes | Antibiotic resistance patterna | No. of isolates |
|---|---|---|---|
| A | 21 | TEDGToCi | 84 |
| B | 14 | TEGToCi(N)M(FS) | 22 |
| C | 6 | TE(DGToCiNSC) | 16 |
| D | 2 | TE(D)GTo(CiNSC) | 3 |
| E | 1 | TE(D)GToCi(NS) | 2 |
Antibiotics in parentheses are those to which only some of the isolates in a subtype are resistant.
FIG. 1.
PFGE profiles of SmaI macrorestriction fragments of MRSA strains. (A) eMRSA isolates. Lanes 1, 10, and 20, λ DNA PFGE markers; lane 2, ATCC 25923; lanes 3 to 9, eMRSA-1 to -7; lanes 11 to 19, eMRSA-8 to -16. (B) Representatives of international MRSA clones and major PFGE types from the PWH. Lanes 1 and 13, λ DNA PFGE markers; lane 2, ATCC 25923; lane 3, Brazilian clone (HSJ216); lane 4, Iberian clone (HPV107): lane 5, New York/Tokyo clone (BK2464); lane 6, pediatric clone (HDE288); lanes 7 to 11, representatives of major PFGE types of MRSA from the PWH (A1, B1, C1, D, and E, respectively); lane 12, internal control strain (MRSA 3).
Phage typing.
Based on the three-reaction-difference rule (2), two major phage patterns were identified among the 127 MRSA isolates. These were designated phage types PT1 and PT2 and included 96 and 31 isolates, respectively. The major difference between these two phage types was the strength of reaction. Isolates belonging to PT1 mainly had inhibition reactions to lytic group III phages, while isolates belonging to PT2 showed strong lytic reactions to group III phages but also reacted to group I phages. The most common pattern of PT1 was 6/47/54/85 (underlining indicates inhibition reaction). No specific pattern was found to be dominant for PT2 isolates, but they tended to react equally well to all phages in group III (phages 6, 42E, 47, 53, 54, 75, 77, 83A, 84, and 85). More than 50% of PT2 isolates were susceptible to these phages.
MRSA isolates belonging to PT1 included isolates with PFGE types A, C, D, and E, while the majority of PT2 isolates belonged to PFGE type B (Table 2). The phages of the international set itself were not adequate to distinguish all types of eMRSA-1 to -16; in addition, phages 88A, 90, 83C, and 932 were required. The international set was able to phage type representatives of the Iberian clone (85) and the New York/Tokyo clone (47/54/75/83A), but the Brazilian clone gave inhibition reactions only at 1,000× RTD, and the pediatric clone was nontypeable by this set of phages. Based on the reactions of these reference strains, eMRSA-1, -4, -6, -7, -8, -9, and -11 and the representative of the Iberian clone fell into the PT1 category.
TABLE 2.
Correlation of phage types, FAFLP types, and PFGE types
| Phage type | FAFLP type | No. of isolates | PFGE Type (No. of Isolates)
|
||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | |||
| PT1 | I | 96 | 78 | 0 | 13 | 3 | 2 |
| PT2 | I | 31 | 7 | 22 | 2 | 0 | 0 |
FAFLP.
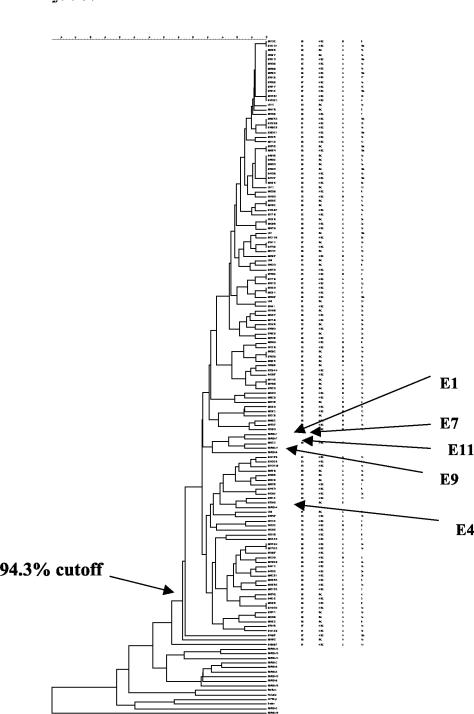
All the MRSA isolates examined were typeable by this method. Each MRSA isolate produced approximately 70 fragments ranging from 50 to 350 bp. About 70% of the fragments were smaller than 200 bp. Isolates of all five PFGE types, A through E, examined by FAFLP were within the similarity level of 94%, which was the similarity cutoff level recommended by Hookey et al. (16); all were considered to be indistinguishable (Fig. 2), and all belonged to type I (Table 2). Representatives of eMRSA-1, -4, -7, -9, and -11 fell within the same cluster as the MRSA isolates at the PWH, while the rest of the eMRSA and international MRSA clones clustered separately. These five eMRSA clones were considered to be closely related to the MRSA isolates at the PWH based on their FAFLP fingerprints.
FIG. 2.
Cluster analysis of FAFLP patterns performed with the Dice similarity coefficient using optimization with a position band tolerance of 0.1%. The dendrogram was calculated by UPGMA. E1, E4, E7, E9, and E11 represent eMRSA of the respective strains from the United Kingdom.
Using the same clustering by UPGMA, the 16 eMRSA clones could be divided into six clusters (Fig. 3). Cluster 1 contained eMRSA-2, eMRSA-5, eMRSA-6, eMRSA-8, and eMRSA-10. Cluster 2 consisted of eMRSA-15 only. Cluster 3 contained eMRSA-12, eMRSA-13, and eMRSA-14. Cluster 4 contained eMRSA-1, eMRSA-4, eMRSA-7, eMRSA-9, and eMRSA-11. eMRSA-3 and eMRSA-16 were clustered into two different groups: cluster 5 and cluster 6. The results were consistent with the clustering of the eMRSA clones by FAFLP performed by Grady et al. (13), except for clusters 1 and 3, which were grouped together in their study. Representatives of the international clones clustered separately from the eMRSA.
FIG. 3.
Cluster analysis of the FAFLP profiles of eMRSA-1 to -16 and representatives of the international clones performed with the Dice similarity coefficient using optimization with a position band tolerance of 0.1%. The dendrogram was calculated by UPGMA. Cluster 1 (CL1) included eMRSA-2, -5, -6, -8, and -10; CL2 consisted of eMRSA-15 only; CL3 included eMRSA-12, -13, and -14; CL4 included eMRSA-1, -4, -7, -9, and -11 and the PWH isolates of PFGE types A through E; CL5 included eMRSA-3; and CL6 included eMRSA 16. Representatives of international clones also clustered separately from eMRSA isolates.
DISCUSSION
All the MRSA isolates from the PWH and the British eMRSA isolates were typeable by the set of international phages at 100 × RTD. Phage typing was able to distinguish two major clones at this hospital, with all the PT1 isolates belonging to PFGE types A, C, D, and E while most of the PT2 isolates were associated with PFGE type B. Indeed, good correlation with the PFGE types would be obtained if 6-fragment differences were used for PFGE interpretations (3). Strains belonging to PFGE types C and D would fall into PFGE A subtypes, while PFGE type E would belong to a PFGE B subtype. This procedure would thus assign PWH MRSA isolates to two predominant PFGE types, A and B, with their subtypes. Although previous studies (10, 24) have found limitations in the use of antibiotic resistance profiles, antibiotic resistance patterns would be useful in distinguishing these two MRSA groups. A unique set of antibiotics including clindamycin, erythromycin, mupirocin, netilmicin, fusidic acid, and rifampin would be able to distinguish between the two groups and could be used in the hospital as part of the infection control strategies to enable early interventions to be made.
Our MRSA isolates of PFGE subtype A2 were indistinguishable from eMRSA-1, while those of PFGE type B related closely to eMRSA-9. Based on the FAFLP fingerprints, all five predominant PFGE types at the PWH belonged to one group and fell in the same cluster as EMRSA-1, -4, -7, -9, and -11 isolates. Other clusters clearly separated the remainder of the eMRSA isolates and representatives of the international clones. Tenover's criteria (25) were useful for interpreting PFGE patterns for small sets of isolates (≤30) and for outbreak studies. In our case, a difference of seven bands in PFGE still fell into the same cluster by FAFLP and supported the notion that within a hospital, a predominant clone exists that varies with time as minor evolutionary changes occur.
Other methods, such as multilocus sequence typing (MLST) (8) and staphylococcal cassette chromosome mec (SCCmec) typing (20), delineated further our MRSA clonal lineage. Preliminary data by MLST classified our MRSA isolates into the same sequence type (ST239) as eMRSA-1, -4, and -11 and the Brazilian clone (M. Enright, unpublished data). Recent evolutionary study of MRSA based on MLST profiles and SCCmec typing postulated that MRSA isolates with ST239 belonged to a large genetic complex, CC8 (9). These isolates were probably derived from ST8 methicillin-susceptible S. aureus isolates through acquisition of type III SCCmec. We have also confirmed that representatives of our PWH MRSA isolates (PFGE types A and B) belonged to type III SCCmec (M. Ip, unpublished data). Thus, the predominant MRSA clone in the PWH in the last decade is closely related to the early United Kingdom eMRSA-1, -4, and -11 clones and is a member of a lineage that includes the Brazilian MRSA clone. eMRSA-1 first appeared in the early 1980s in England (20) and subsequently spread to other countries such as Spain (1) and Australia (19). Our MRSA PFGE type A2 was present in 1988, and earlier strains from the PWH were not available for typing. It appeared that this strain type became established as the predominant clone at the PWH in the last decade.
Antibiotic susceptibility testing, phage typing, and PFGE were useful for studying the MRSA isolates at the PWH. PFGE remained the most discriminatory method, while determination of the antibiotic resistance pattern was the most rapid, easy and inexpensive method for infection control purposes. Phage typing could also distinguish among our local strains but was a labor- and time-consuming task. In this study, ALFexpress (Amersham Pharmacia Biotech) was used for FAFLP, and a 75-bp size marker was introduced to increase the standard points for normalization of the gel. The results were reproducible and consistent with the findings of Grady et al. (13). FAFLP was acceptable for comparison between MRSA isolates obtained from diverse geographic locations but may have limitations in strain typing within one hospital, where strain diversity is limited. The use of advanced electrophoresis equipment, such as the ABI DNA analyzer (Perkin-Elmer Applied Biosystems, Foster City, Calif.), may produce high-resolution genotypes, with detection of fragments that differ by 1 bp, for the study of local hospital outbreaks as well as for comparisons with international clones in MRSA epidemics.
Acknowledgments
We thank T. Pitt, CPHLS, Colindale, United Kingdom, for providing the international set of phages and M. Ganner and M. Desai (CPHLS) for technical advice on phage typing and FAFLP. We also thank H. de Lencastre and A. Tomasz for providing representatives of the international clones.
The work described in this paper was supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region (CUHK direct grant 2001.1.069).
REFERENCES
- 1.Aparicio, P., J. Richardson, S. Martin, A. Vindel, R. R. Marples, and B. D. Cookson. 1992. An epidemic methicillin-resistant strain of Staphylococcus aureus in Spain. Epidemiol. Infect. 108:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aucken, H. M., and K. Westwell. 2002. Reaction difference rule for phage typing of Staphylococcus aureus at 100 times the routine test dilution. J. Clin. Microbiol. 40:292-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc, D. S., M. J. Struelens, A. Deplano, R. De Ryck, P. M. Hauser, C. Petignat, and P. Francioli. 2001. Epidemiological validation of pulsed-field gel electrophoresis patterns for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:3442-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown D. J. F., D. M. Livermore, J. M. Symonds, and R. Wise. 2002. Antimicrobial susceptibility testing: BSAC Working Party Report. J. Antimicrob. Chemother. 48(Suppl. 1):1-102. [Google Scholar]
- 5.Cheng, A. F., and G. L. French. 1988. Methicillin-resistant Staphylococcus aureus bacteraemia in Hong Kong. J. Hosp. Infect. 12:91-101. [DOI] [PubMed] [Google Scholar]
- 6.De Lencastre, H., E. P. Severina, R. B. Roberts, B. N. Kreiswirth, A. Tomasz, et al.. 1996. Testing the efficacy of a molecular surveillance network: methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF) genotypes in six hospitals in the metropolitan New York City area. Microb. Drug Resist. 2:343-351. [DOI] [PubMed] [Google Scholar]
- 7.de Sousa, M. A., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.French, G. L., J. Ling, and M. Farrington. 1986. Typing of Staphylococcus aureus resistant to methicillin. BMJ 293:1438-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French, G. L., J. Ling, T. Ling, and Y. W. Hui. 1988. Susceptibility of Hong Kong isolates of methicillin-resistant Staphylococcus aureus to antimicrobial agents. J. Antimicrob. Chemother. 21:581-588. [DOI] [PubMed] [Google Scholar]
- 12.Givney, R., A. Vickery, A. Holliday, M. Pegler, and R. Benn. 1998. Evolution of an endemic methicillin-resistant Staphylococcus aureus population in an Australian hospital from 1967 to 1996. J. Clin. Microbiol. 36:552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grady, R., G. O'Neill, B. Cookson, and J. Stanley. 2000. Fluorescent amplified-fragment length polymorphism analysis of the MRSA epidemic. FEMS Microbiol. Lett. 187:27-30. [DOI] [PubMed] [Google Scholar]
- 14.Grady, R., D. Blanc, P. Hauser, and J. Stanley. 2001. Genotyping of European isolates of methicillin-resistant Staphylococcus aureus by fluorescent amplified-fragment length polymorphism analysis (FAFLP) and pulsed-field gel electrophoresis (PFGE) typing. J. Med. Microbiol. 50:588-593. [DOI] [PubMed] [Google Scholar]
- 15.Grady, R., M. Desai, G. O'Neill, B. Cookson, and J. Stanley. 1999. Genotyping of epidemic methicillin-resistant Staphylococcus aureus phage type 15 isolates by fluorescent amplified-fragment length polymorphism analysis. J. Clin. Microbiol. 37:3198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hookey, J. V., V. Edwards, S. Patel, J. F. Richardson, and B. D. Cookson. 1999. Use of fluorescent amplified fragment length polymorphism (fAFLP) to characterise methicillin-resistant Staphylococcus aureus. J. Microbiol. Methods 37:7-15. [DOI] [PubMed] [Google Scholar]
- 17.Laboratory of Hospital Infection. 1995. Standard operating procedure for phage typing of Staphylococci, p. 1-4. SOP no. L-4302/01-95. Laboratory of Hospital Infection, Central Public Health Laboratory, Colindale, United Kingdom.
- 18.Laboratory of Hospital Infection. 1999. Standard operating procedure for pulsed field gel electrophoresis, p. 1-10. SOP no. L4510/02-99. Laboratory of Hospital Infection, Central Public Health Laboratory, Colindale, United Kingdom.
- 19.Lim, T. T., Geoffrey W. Coombs, and Warren B. Grubb. 2002. Genetic organization of mecA and mecA-regulatory genes in epidemic methicillin-resistant Staphylococcus aureus from Australia and England. J. Antimicrob. Chemother. 50:819-824. [DOI] [PubMed] [Google Scholar]
- 20.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial susceptibility testing. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Sa-Leao, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanches, I. S., M. Ramirez, H. Troni, M. Abecassis, M. Padua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheel, O., D. J. Lyon, V. T. Rosdahl, F. A. Adeyemi-Doro, T. K. Ling, and A. F. Cheng. 1996. In-vitro susceptibility of isolates of methicillin-resistant Staphylococcus aureus 1988-1993. J. Antimicrob. Chemother. 37:243-251. [DOI] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]