Abstract
Human group A rotavirus (HRV) is the major cause of severe gastroenteritis in infants worldwide. HRV shares the feature of a high degree of genetic diversity with many other RNA viruses, and therefore, genotyping of this organism is more complicated than genotyping of more stable DNA viruses. We describe a novel microarray-based method that allows high-throughput genotyping of RNA viruses with a high degree of polymorphism by multiplex capture and type-specific extension on microarrays. Denatured reverse transcription (RT)-PCR products derived from two outer capsid genes of clinical isolates of HRV were hybridized to immobilized capture oligonucleotides representing the most commonly occurring P and G genotypes on a microarray. Specific primer extension of the type-specific capture oligonucleotides was applied to incorporate the fluorescent nucleotide analogue cyanine 5-labeled dUTP as a detectable label. Laser scanning and fluorescence detection of the microarrays was followed by visual or computer-assisted interpretation of the fluorescence patterns generated on the microarrays. Initially, the method detected HRV in all 40 samples and correctly determined both the G and the P genotypes of 35 of the 40 strains analyzed. After modification by inclusion of additional capture oligonucleotides specific for the initially unassigned genotypes, all genotypes could be correctly defined. The results of genotyping with the microarray fully agreed with the results obtained by nucleotide sequence analysis and sequence-specific multiplex RT-PCR. Owing to its robustness, simplicity, and general utility, the microarray-based method may gain wide applicability for the genotyping of microorganisms, including highly variable RNA and DNA viruses.
Vaccinations and antimicrobial chemotherapy are examples of interventions against microbiological systems that may induce changes in the diversity of species and circulating strains. Given the complexity of many microbiological systems, methods with high throughputs are needed to describe the genetic diversity before and after an intervention. Human rotavirus is an example of a genetically diverse virus for which large vaccine trials are being conducted.
Rotaviruses are classified into G and P types according to the antigenic and genetic diversity of their capsid proteins, VP7 (G types) and VP4 (P types). Combinations of five G types, and four P types constitute the majority of the different human group A rotavirus (HRV) genotypes commonly found in most parts of the world (7, 8). The possible diversity of human rotaviruses is significantly greater, as 10 G types and 9 P types have been described among human strains. Comparative analysis of the VP7 and VP4 genes of the various types shows several short regions with significant type-specific nucleotide sequence divergence. These regions are highly conserved within each type and can be used to differentiate between types by sequencing and multiplex genotype-specific reverse transcription (RT)-PCR (7, 9, 10). Alternatively, monoclonal antibodies have been used to differentiate G and P types (17, 25). The diversity of cocirculating rotavirus strains should ideally be determined before, during, and after intervention with a rotavirus vaccine. A low level of vaccine efficiency may depend on a predominance of strains that remain inefficiently neutralized by the vaccine-evoked immune response. During immunization with viable vaccine strains, reassortment and recombination with coinfecting wild-type strains and the reversion of the vaccine strains to virulence may occur (1, 27). Furthermore, vaccination may induce selective effects on the diversity of strains and cause the emergence of vaccine-escape strains of viruses and bacteria (1, 3, 29). In the case of HRV, the availability of type-specific monoclonal antibodies is limited, and a multiplex approach to RT-PCR for genotyping is complex and time-consuming when large numbers of isolates are processed (13). To avoid these limitations and to improve the throughput, we have developed a new flexible microarray-based method that allows detailed dissection of the viral genotypes in a single reaction after RT-PCR.
MATERIALS AND METHODS
Specimens.
Fecal specimens from 40 children with severe diarrhea determined to be rotavirus antigen positive by enzyme-linked immunosorbent assay were collected from 1998 to 2001 at the community hospital and at health centers in León, Nicaragua, and at the Uppsala University Hospital, Uppsala, Sweden, and were used as reference samples for this study. Stool samples were suspended in phosphate-buffered saline to approximately 10% (vol/vol) and clarified by centrifugation at 800 × g in an Eppendorf microcentrifuge for 5 min. Rotavirus RNA was purified from the fecal suspensions by using the QIAamp RNA Blood Mini kit (Qiagen), as recommended by the manufacturer, with a sample volume of 100 μl eluted in 60 μl. Genotypes were defined by sequence-specific multiplex RT-PCRs, performed as described previously (6, 9). A subset of 26 VP4 genotypes and 15 VP7 genotypes were confirmed by sequencing of one or both strands, followed by alignment with the ClustalW program and analysis with the BLAST program (2, 7, 9, 26). The reference samples had the following genotype distributions: 6 samples, P[8]G1; 24 samples, P[4]G2; 1 sample, P[9]G3; 6 samples, P[6]G4; 1 sample, P[8]G4; and 2 samples, P[8]G9.
Internal control.
The single-stranded RNA phage MS2 (ATCC 15597-B1) was produced in Escherichia coli strain c3000 by a standard procedure. Phage MS2 RNA was purified from the culture medium as described above for HRV RNA. The MS2 phage RNA was subjected to RT-PCR, and the PCR product was used as an internal control (for the primers used in the study, see Table 1).
TABLE 1.
PCR primers and capture oligonucleotides
| Sequence type and name | Sequence (5′ → 3′) | Positiona | Accession no.b |
|---|---|---|---|
| PCR Primers | |||
| VP7 | |||
| Beg9 | GGC TTT AAA AGA GAG AAT TTC CGT CTG G | 1-28 | M21843 |
| VP7 | ACT GAT CCT GTT GGC CAT CCT TT | 395-373 | M21843 |
| VP4 | |||
| HC5 | GTC TAG ATG GTC CAT ATC AAC C | 200-221 | M32559 |
| HC3 | TTC CTT GTA TTC TGA ATT GGT | 701-681 | M32559 |
| MS2 | |||
| MS25 | CAT GGC TAT CGC TGT AGG TAG C | 79-100 | J02467 |
| MS23 | CGC GAA CGG AGG GGA CGA A | 187-169 | J02467 |
| Capture oligonucleotidesc | |||
| G type | |||
| G*A | GTC TGG CTA GCG GTT AGC TC | 23-42 | M21843 |
| G*A NC | GTC TGG CTA GCG GTT AGC TT | 23-42 | M21843 |
| G*B | TTA TAT TAT CCA ACW GAA GC | 295-314 | M21843 |
| G*B NC | TTA TAT TAT CCA ACW GAA GT | 295-314 | M21843 |
| G1-1 | GTA TTA TCA AAC TGA AGC AAG | 297-317 | M21843 |
| G1-2d | TGA TGG TGA ATG GAA AGA CT | 330-349 | M21843 |
| G1-3d | GTA TTA TCC AAC TGA AGC AA | 297-316 | M21843 |
| G2-1 | TAC TAT CCA GCA GAA GCT AA | 298-317 | U73947 |
| G2-2 | ATA CTA TCC AGC AGA AGC TA | 297-316 | U73947 |
| G2-3 | GAA GCT AAA AAT GAG ATT TC | 310-329 | U73947 |
| G3-1 | GCT GCT TCA GTT GGG TAA TA | 298-317 | D86284 |
| G3-2 | TAA TGG ACT TTA TTA TTT AC | 134-153 | D86284 |
| G3-3 | ATT ATG GAA TAA ATC TTC CG | 203-222 | D86284 |
| G4-1 | GAC ACT GAA TGG AAA GAT A | 331-349 | AF161823 |
| G4-2d | ACA CTG AAT GGA AAG ATA CA | 332-351 | AF161823 |
| G4-3d | ACA GCT GAG ATA GTG TAT CT | 338-357 | AF161823 |
| G9-1 | ATA TTA TCC TAC AGA AGC AT | 297-316 | AF060487 |
| G9-2 | CTT TTA CTT ATT GTT ATT GC | 160-179 | AF060487 |
| G9-3 | ATA TGC AAA TTC ATC ACA GC | 246-265 | AF060487 |
| P type | |||
| P* | CTT GTA TTC TGA ATT GGT GG | 698-679 | M32559 |
| P*NC | CTT GTA TTC TGA ATT GGT GT | 698-679 | M32559 |
| P4-1 | AAT TGA TAT ATT ATT TAA ATC C | 611-590 | M32559 |
| P4-2 | TGA TTT TTG GAC AGC AGT TA | 306-325 | M32559 |
| P4-3 | TGA TTA TTG GCT GCT TAT TAG | 243-263 | M32559 |
| P6-1 | TCA GAT AAA TTT GAA ATT G | 593-578 | U16299 |
| P6-2d | TCA GAC AAG TTT GAA GTT G | 593-578 | U16299 |
| P6-3d | TCA GAT AAG TTT GAA GTT G | 593-578 | U16299 |
| P8-1 | CCT TGG AAT AAT GTA AAA TTC TGA | 636-613 | M96825 |
| P8-2 | CTA TCT ACT GGG TTA ACG TGC | 359-339 | M96825 |
| P8-3 | TGA ACC GCA CGT TAA CCC AGT | 333-353 | M96825 |
| P9-1 | TAA ACC ATT ATT TAT ATA TTG TG | 681-659 | M28349 |
| P9-2 | CGT TGC TGT TAT CAT TGT TTA | 604-584 | M28349 |
| MS2 | |||
| MS2 | GGT ACT AAA AGC TCG CAC AG | 147-128 | J02467 |
Nucleotide positions relative to reference sequences.
Accession numbers of reference VP4 and VP7 genes and phage MS2.
Capture oligonucleotides all have a terminal amino group and 15 T residues at their 5′ ends. The first number in the names of the specific oligonucleotides indicates the genotype with which it is designed to hybridize. The oligonucleotides denoted with an asterisk are generic capture oligonucleotides, and an additional NC (negative control) indicates an intentional mismatch (T) at their 3′ ends. P8-1 is complementary to the sequences of both the P[4] and the P[8] genotypes.
Capture oligonucleotides added after the expansion of the array.
Oligonucleotides.
PCR primers and capture oligonucleotides were designed on the basis of published sequences of HRV and the MS2 phage, respectively, by using the MACAW program (22). The sequence alignments of the VP7 and VP4 genes of reference strains used in previous studies were used for a first, preliminary primer selection (4, 6). The primers were then tested for their 3′ specificities by using them as query sequences in a search of the sequences in GenBank with the BLAST program, which confirmed that high-scoring sequences belonged to the intended genotype. In the selection of several primers corresponding to the same genotype, the effort was focused on obtaining high levels of genotype specificity, although no effort was made to define specific subtypes. The sequences of primers Beg9 and VP7 have been published previously (9, 28). The capture oligonucleotides have a terminal amino group and 15 T residues as a spacer at their 5′ ends. The oligonucleotide sequences and explanations are given in Table 1.
RT-PCR amplification.
The VP7, VP4, and MS2 genes were amplified by RT-PCR with the Amp Taq Gold RNA PCR kit (Applied Biosystems). Purified RNA (5 μl) was heated in a final volume of 10 μl together with one of the gene-specific primer pairs for 10 min at 97°C, followed by 5 min at 42°C to allow the primers to anneal. Either 15 pmol of primers Beg9 and VP7, 15 pmol of primer HC3 and 45 pmol of primer HC5, or 25 pmol of primer MS25 and 10 pmol of primer MS23 were used in a final volume of 50 μl of 30 mM Tris-HCl (pH 8.3), 20 mM KCl, and 1.75 mM MgCl2 with a 0.2 mM concentration of each deoxynucleoside triphosphate, 10 U of RNasin, 2.5 U of AmpliTaq Gold DNA polymerase, and 15 U of MultiScribe reverse transcriptase (Applied Biosystems). The RT reaction was performed at 42°C for 12 min, followed by one cycle of 95°C for 10 min. Immediately thereafter the VP7 and MS2 genes were amplified for 35 cycles of 94°C for 60 s, 42°C for 60 s, and 72°C for 30 s. The VP4 gene was amplified for 40 cycles of 94°C for 60 s, 50°C for 15 s, and 72°C for 30 s. Finally, the PCR products derived from VP4 and VP7 in each sample were combined, precipitated with ethanol, and dissolved in 18 μl of water.
Preparation of oligonucleotide arrays.
Microscope slides with 10 individual reaction wells (diameter, 7 mm) formed by a Teflon lining (Erie Scientific Company) or, alternatively, standard microscope slides (Menzel-Gläser, Braunschweig, Germany) with custom-made silicon rubber grids, which were placed on the slide to form individual reaction wells (19), were used. The slides were activated essentially by the procedure described by Guo et al. (11), except that 3-aminopropyltriethoxysilane (Sigma) instead of its methoxy derivative was used for silanization (16). A custom-built arrayer with TeleChem CMP-3 (TeleChem) pins was used to print the arrays with a center-to-center distance between spots of 250 μm (see Fig. 2) (18). Prior to printing of the arrays, the oligonucleotides were dissolved in 400 mM sodium carbonate buffer (pH 9.0) to a final concentration of 25 μM. After printing of the arrays, the slides were exposed to vaporized 25% ammonia for 1 h, followed by thorough rinsing in distilled water. The arrays were dried by centrifugation at 30 × g for 5 min and were then stored at 4°C for up to 12 weeks. Before use, the slides were rinsed in solution 1 (100 mM NaCl, 5 mM Tris-HCl [pH 8], 0.5 mM EDTA, 0.1% Triton X-100) and preheated at 65°C under humid conditions.
FIG. 2.
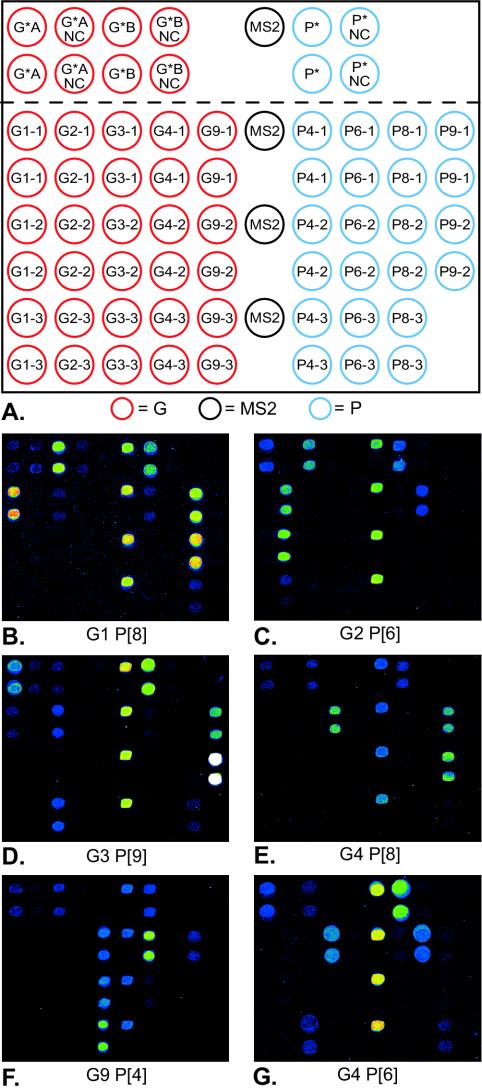
The oligonucleotide pattern printed onto the arrays (A) and examples of genotyping results (B to G). The capture oligonucleotides printed on the array are indicated by their names in the circles of panel A; their sequences are given in Table 1. The four black circles represent spots of the MS2 phage capture oligonucleotide used as a procedural control and a reference for normalization. The two rows above the dotted line shows vertical duplicates of the generic VP7- and VP4-specific oligonucleotides (indicated by asterisks) as well as their corresponding 3′ sequence-mismatched oligonucleotides, used as negative controls (NC). Below the dotted line, the red circles to the left are G-type-specific capture oligonucleotides spotted as vertical duplicates in columns. Each column represents one G type (G1, G2,G3, G4, and G9, from left to right, respectively), as indicated by the first number in the oligonucleotide designation. The blue circles to the right represent P-type-specific capture oligonucleotides spotted in the same manner described for the G types, with the columns representing P4, P6, P8, and P9 from left to right, respectively. Panels B to G shows five different G- and P-type combinations by using primer extension with oligonucleotide arrays obtained by scanning of the slides.
Genotype-specific primer extension.
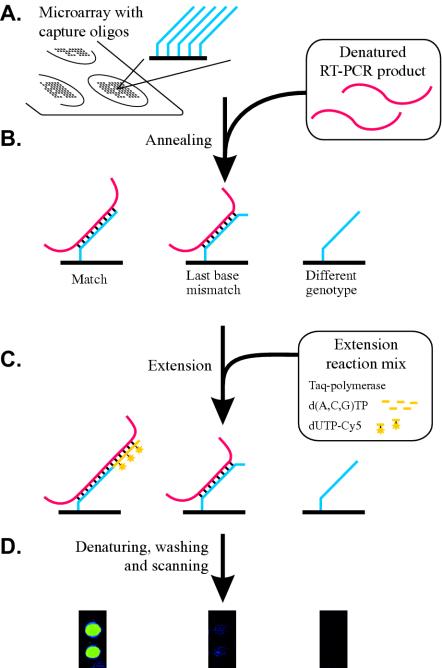
The MS2-derived PCR product (100 ng) was added to the combined VP7 and VP4 PCR products to give a final volume of 30 μl in 200 mM NaCl and 10 mM Tris-HCl [pH 8], 0.1 mM EDTA. The mixture was denatured at 95°C for 1.5 min and directly applied to two wells in the slide. The slide was placed in a humid chamber, and the annealing reaction was allowed to proceed for 15 min at 37°C. The arrays were quickly rinsed with solution 1 and heated to 60°C in a humid chamber. The primer extension reaction mixture (20 μl) contained 1.0 μM (each) dATP, dCTP, and dGTP; 0.6 μM cyanine 5-labeled dUTP (Amersham Pharmacia Biotech); and 1.25 U of Taq DNA polymerase (Amersham Pharmacia Biotech) in a solution containing 10 mM Tris-HCl (pH 9), 50 mM KCl, and 1.5 mM MgCl2 and was added to each microarray well on the slides. The slides were then incubated at 60°C for 10 min. The microarrays were rinsed with water, washed at room temperature for 2 min in 50 mM NaOH with a shaker, and rinsed with water. The glass slides were washed twice in 30 mM NaCl-3 mM sodium citrate (pH 7.0)-0.1% sodium dodecyl sulfate for 5 min at 65°C and, finally, were rinsed with water. See Fig. 1 for an outline of the method.
FIG. 1.
Principle and steps of the microarray procedure for genotyping of HRV. Sets of capture oligonucleotides (oligos) specific for the G and P types of HRV are covalently immobilized on glass microscope slides (A). With these oligonucleotides as probes, the RT-PCR products containing type-specific regions of the VP7 and VP4 genes are captured on the microarrays by hybridization under low-stringency conditions (B). Oligonucleotides with 3′ ends that are matched to the sequences of the captured templates are extended with a mixture of deoxynucleoside triphosphates containing cyanine 5 (Cy5)-labeled dUTP by using a thermostable DNA polymerase (C). The fluorescence incorporated in the sequence-specific primer extension reaction is measured in a microarray scanner (D). The results are interpreted by visual inspection of the arrays or by calculation of the relative fluorescence intensity of the individual spots.
Array scanning and interpretation of results.
The fluorescence signals were measured with a laser fluorescence scanner (ScanArray 5000; GSI Lumonics) with a Red HeNe laser (excitation wavelength, 632.8 nm). The genotypes of the samples were deduced by visual examination of the spot pattern, as shown in Fig. 2. QuantArray analysis software (GSI Lumonics) was used to record and transfer the signal intensities for the individual spots to a spreadsheet computer program for further processing. The background for the corresponding spots for a negative control consisting of water instead of the PCR product was subtracted. To normalize the results between arrays and experiments, the ratios between the mean signals of the duplicate capture oligonucleotides and the quadruplicate MS2 capture oligonucleotides in each array were calculated (see Table 2). The ratios were then sorted, and the G and P genotypes were defined by the highest ratios obtained for one of the P- and G-type-specific capture oligonucleotides on the array.
TABLE 2.
Examples of interpretation of results
| Panela (genotype) | Capture oligonucleotide | Signal | Ratiob | Note |
|---|---|---|---|---|
| B (P[8]G1) | P8-2 | 41,950 | 1.42 | Highest P |
| G1-1 | 37,810 | 1.28 | Highest G | |
| P* | 11,440 | 0.39 | VP4-positive control | |
| P*NC | 320 | 0.01 | VP4-negative control | |
| G*C | 25,040 | 0.85 | VP7-positive control | |
| G*CNC | 780 | 0.03 | VP7-negative control | |
| F (P[4]G9) | P4-1 | 18,760 | 3.65 | Highest P |
| G9-3 | 9,620 | 1.87 | Highest G | |
| P* | 2,670 | 0.52 | VP4-positive control | |
| P*NC | 80 | 0.02 | VP4-negative control | |
| G*B | 1,080 | 0.21 | VP7-positive control | |
| G*BNC | 250 | 0.05 | VP7-negative control |
Refers to the panels in Figure 2.
Ratio between the mean signal intensities, measured as arbitrary fluorescence units and indicated in the column labeled “signal”, for duplicate spots of type-specific capture oligonucleotides and quadruplicate spots of the MS2-specific capture oligonucleotide. The mean MS2 signal intensities were 29,510 for the well shown in Fig. 2B and 5,140 in for the well shown in Fig. 2F. The background from the corresponding spot in a negative control reaction well (water instead of the PCR product) was subtracted from all signal intensities.
DNA sequencing.
Sequencing was performed when the results of the genotype-specific multiplex RT-PCRs showed a small amount of PCR product. Sequencing was also conducted with all samples not initially genotyped by the microarray method. The amplicons obtained in the VP7- and VP4-specific RT-PCRs were purified with the QIAquick PCR purification kit (Qiagen). Direct cycle sequencing analysis of one or both strands was performed on an ABI 310 automated sequencer (Perkin-Elmer Applied Biosystems) by using Big Dye chemistry.
RESULTS
Design of the assay.
We have devised a novel method for dissection of the diverse genotypes of HRV. It is based on multiplex capture and type-specific primer extension on microarrays. Figure 1 illustrates the principle of the method. The method combines G- and P-type-specific capture of amplified viral sequences by using a panel of immobilized oligonucleotides with genotype-specific extension of these capture oligonucleotides with DNA polymerase and fluorescent deoxynucleotides. The specificity of the primer extension reaction depends, first, on the presence of a viral sequence that is complementary to that of the capture oligonucleotide and, second, on the presence of a 3′ genotype-specific nucleotide in this sequence.
Each array included (i) generic capture oligonucleotides recognizing the VP4 and VP7 genes independently of the genotype, (ii) the same generic oligonucleotides but with mismatches at the 3′ ends, (iii) genotype-specific capture oligonucleotides designed to target 20-bp sequences within the amplified VP4 or VP7 gene segments, and (iv) MS2 phage-specific capture oligonucleotides (Table 1). Figure 2A shows the design of the oligonucleotide microarray. The signals generated from the generic capture oligonucleotides serve as positive controls, indicating the presence of the HRV template independently of the genotype. Thus, they detect unusual genotypes not recognized by the genotype-specific oligonucleotides and prevent false-negative results. The mismatched generic capture oligonucleotides serve as negative controls to ensure that the stringency of the reaction conditions allowed discrimination of a 3′ end mismatch. The VP4- and VP7-specific capture oligonucleotides generate a genotype-specific fluorescence pattern on the array after primer extension. The PCR products derived from phage MS2 were added to the HRV-derived PCR product and served as controls for the spotting procedure, determination of the efficiency of the hybridization, and the primer extension reaction for each sample. The MS2 control was also used for normalization of the signals between arrays and experiments. Figure 2 outlines the array design and shows representative examples of the genotyping results. The numerical results calculated for panels B and F are shown in Table 2. The results for panels B and F, with high and low signal intensities, respectively, are shown to illustrate the effect of normalization against the MS2 control. As can be seen, the genotyping result is unequivocal by using the strategy of defining the genotypes as the highest ratio between the signals from specific oligonucleotides and the signal from the MS2 control reaction.
Genotyping of HRV.
Forty fecal specimens collected in Leon, Nicaragua, and Uppsala, Sweden, were determined to be rotavirus antigen positive and were analyzed by the microarray-based method for genotyping of HRV. Primer site mutations may result in failed RT-PCRs and an inability to genotype the virus (12). In order to positively identify the presence of adequate amounts of the targeted VP7 and VP4 regions, generic capture oligonucleotides were included on the array. The sequences of all 40 isolates were successfully amplified, and all samples were shown to contain HRV by the generic capture oligonucleotides on the array.
The method assigned both G and P genotypes to 35 of the 40 strains analyzed. In three samples only either the G type or the P type was defined, and in two samples no specific genotype was defined. Failure to define genotypes was found to correspond to the presence of viral sequences not represented by capture oligonucleotides with sufficient homology in the array. The sequences of isolates for which a genotype was not defined showed three internal mismatches at positions corresponding to a range from 4 to 14 nucleotides from the 3′ end of the capture oligonucleotides. The array was expanded by the addition of two capture oligonucleotides specific for each of types G1, G4, and P[6] (six nucleotides in all) to fit an extended range of isolates, including the previously missed strains of types G4 and P [6] not detected by the original array (see footnote d of Table 1). Due to a shortage of clinical samples, the functionality of the array for these genotypes was shown by using synthetic templates corresponding to previously sequenced HRV strains, with unambiguous results. Our results thus illustrate the advantage of using the generic capture oligonucleotides to detect unusual genotypes not included in the original array. Due to the flexibility of the array format, it is also easy to expand and modify the capture oligonucleotides included. The results of genotyping obtained with the microarray were in full agreement with the results obtained by nucleotide sequence analysis and genotype-specific multiplex RT-PCR (6, 9). All G and P types represented by capture oligonucleotides in the array were represented among the 35 strains that were fully typed (P[8]G1, P[4]G2, P[9]G3, P[6]G4, P[8]G4, P[8]G9).
DISCUSSION
In epidemiological genotyping of microorganisms, multiple polymorphisms need to be targeted to obtain a sufficient resolution for strain or type discrimination, and typically, large numbers of isolates collected from clinical samples should be analyzed. Microarrays are an attractive format for such studies because they allow highly multiplexed assays and because the miniaturized microarray-based assays deploy very small reaction volumes. These features of the microarray format serve to increase the throughput and decrease reagent costs, which are both important considerations in large-scale epidemiological and other clinical studies.
In the present study we devised a DNA polymerase-assisted method in a multiplex microarray format which is particularly well suited for epidemiological studies of rapidly evolving RNA viruses, such as HRV. The strategy of our method uses two levels of specificity. Initially, HRV G- and P-type-specific oligonucleotides are used as capture probes in a low-stringency hybridization reaction. The same oligonucleotides then serve as primers for a sequence-specific extension reaction catalyzed by a DNA polymerase in a second step of the assay, which allows dissection of the G and P subtypes of HRV. In addition to genotyping of previously known G and P subtypes for which specific probe-primer oligonucleotides have been included in the arrays, the design of our array with generic G- and P-type-specific oligonucleotides allows positive identification of novel or rare variants of HRV. Thus, potential false-negative results due to the inability to detect newly emerged genotypes of HRV are avoided. Novel variants detected by the generic oligonucleotides alone can then be sequenced, and the arrays can subsequently be complemented with additional capture oligonucleotides specific for the novel subtypes. A similar approach has previously been used for HRV genotyping by RT-PCR and gel electrophoresis (5, 12). To maintain a high level of specificity, which serves to avoid cross-priming events, the use of degenerated capture oligonucleotides has not been exploited. When an algorithm is used to identify the highest signal ratios for assignment of the HRV genotypes, the ability of our system to detect double infections is limited. However, samples with double infections with roughly equal amounts of each genotype will be identified visually by double genotype patterns with signal levels within the same order of magnitude on the microarray, analogous to the identification of double bands in ethidium bromide-stained agarose gels after RT-PCR.
The use of high-density microarrays with sequence-specific hybridization probes (GeneChips) synthesized in situ was proposed for large-scale analysis of the genetic variation of human immunodeficiency virus (HIV) 6 years ago (15). A limitation of using hybridization with short sequence-specific oligonucleotide probes in multiplexed genotyping assays is, however, that the reaction conditions that allow specific genotyping depend strongly on the sequence context of a variant site. Therefore, the design of a single set of optimal reaction conditions for the simultaneous detection of multiple sequence variants is difficult. Instead, multiplexed genotyping by hybridization with sequence-specific probes may be accomplished by using redundant sets of probes for each sequence variant to be analyzed. For example, in the GeneChip assay for HIV, approximately 30 probes are used for each variable site analyzed in the HIV genome (15). A recent study describes a microarray-based hybridization method for genotyping of HRV, in which five common HRV G types are detected by using 9 or 10 probes specific for each genotype (4). Due to the limitation in specificity and the complexity of the assay design, methods based on the hybridization reaction alone have not become widely used in multiplexed genotyping, while enzyme-assisted methods are gaining acceptance as the methods of choice (for a review, see reference 24). The concept of DNA polymerase-assisted primer extension in the microarray format was first introduced for the detection of point mutations in human DNA (18). In a side-by-side comparison, the primer extension reaction was found to provide more than 10-fold better discrimination between homozygous and heterozygous genotypes than hybridization with short genotype-specific oligonucleotide probes (18). The major advantage of DNA polymerase-assisted microarray methods over hybridization methods is that a single set of optimal reaction conditions can be used to genotype all sequence variants, which simplifies assay design and optimization.
The genotyping system established for HRV in the present study is based on an array-of-arrays conformation on either Teflon-lined microscope slides (20) or standard slides covered with custom-made silicon rubber grids (19). For large-scale studies this array-of-arrays format, which allows analysis of multiple samples on each slide, is more advantageous than the traditional microarray format that was originally designed to profile the transcripts of thousands of genes in one sample per slide (4, 15, 21). By increasing the number of capture probe-primers used on the same type of array of arrays, it would be feasible to increase the resolution of our method to enable subtyping. At the present spotting density, 10 samples can be analyzed with approximately 700 oligonucleotides on each slide, or alternatively, 80 samples can be analyzed with 200 oligonucleotides on each slide. At this resolution the genotyping system could well be applied to the clinically important genotyping of hepatitis C virus and human papillomavirus and, hence, provide an accurate, high-throughput alternative to the methods used at present (14, 23).
In conclusion, because all reagents and equipment required to perform the multiplex capture and type-specific primer extension method described here are generally available and it is easy to the set up the assay and interpret the results, we believe that our method has the potential to become a widely applied tool for laboratories performing large-scale analysis of variable genes for strain definition and genotyping of microorganisms.
Acknowledgments
The work described in this report was supported by grants from the Swedish International Development Cooperation Agency (grant 0.75007 109) and NeTropica (grant 2001-3) (to F.E. and K.B.) and by the Swedish Research Council and the K&A Wallenberg Foundation (WCN) (to A.-C.S.).
REFERENCES
- 1.Abraham, R., P. Minor, G. Dunn, J. F. Modlin, and P. L. Ogra. 1993. Shedding of virulent poliovirus revertants during immunization with oral poliovirus vaccine after prior immunization with inactivated polio vaccine. J. Infect. Dis. 168:1105-1109. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carman, W. F., A. R. Zanetti, P. Karayiannis, J. Waters, G. Manzillo, E. Tanzi, A. J. Zuckerman, and H. C. Thomas. 1990. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325-329. [DOI] [PubMed] [Google Scholar]
- 4.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentsch, J., R. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. Das, and M. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 8.Glass, R. I., P. E. Kilgore, R. C. Holman, S. Jin, J. C. Smith, P. A. Woods, M. J. Clarke, M. S. Ho, and J. R. Gentsch. 1996. The epidemiology of rotavirus diarrhea in the United States: surveillance and estimates of disease burden. J. Infect. Dis. 174(Suppl. 1):S5-S11. [DOI] [PubMed] [Google Scholar]
- 9.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, K. Y., J. F. Sears, K. Taniguchi, K. Midthun, Y. Hoshino, M. Gorziglia, K. Nishikawa, S. Urasawa, A. Z. Kapikian, R. M. Chanock, et al. 1988. Prediction of human rotavirus serotype by nucleotide sequence analysis of the VP7 protein gene. J. Virol. 62:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, Z., R. Guilfoyle, A. Thiel, R. Wang, and L. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22:5456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iturriza-Gomara, M., J. Green, D. W. G. Brown, U. Desselberger, and J. J. Gray. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38:898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iturriza-Gomara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleter, B., L.-J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, M. Burger, and W. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 16.Lindroos, K., U. Liljedahl, M. Raitio, and A. C. Syvanen. 2001. Minisequencing on oligonucleotide microarrays: comparison of immobilisation chemistries. Nucleic Acids Res. 29:E69. [DOI] [PMC free article] [PubMed]
- 17.Matson, D. O., M. K. Estes, J. W. Burns, H. B. Greenberg, K. Taniguchi, and S. Urasawa. 1990. Serotype variation of human group A rotaviruses in two regions of the USA. J. Infect. Dis. 162:605-614. [DOI] [PubMed] [Google Scholar]
- 18.Pastinen, T., A. Kurg, A. Metspalu, L. Peltonen, and A.-C. Syvanen. 1997. Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 7:606-614. [DOI] [PubMed] [Google Scholar]
- 19.Pastinen, T., M. Raitio, K. Lindroos, P. Tainola, L. Peltonen, and A.-C. Syvanen. 2000. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 10:1031-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raitio, M., K. Lindroos, M. Laukkanen, T. Pastinen, P. Sistonen, A. Sajantila, and A.-C. Syvanen. 2001. Y-chromosomal SNPs in Finno-Ugric-speaking populations analyzed by minisequencing on microarrays. Genome Res. 11:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schena, M., D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuler, G. D., S. F. Altschul, and D. J. Lipman. 1991. A workbench for multiple alignment construction and analysis. Proteins 9:180-190. [DOI] [PubMed] [Google Scholar]
- 23.Stuyver, L., A. Wyseur, W. van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation line probe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syvänen, A.-C. 2001. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2:930-942. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi, K., T. Urasawa, Y. Morita, H. B. Greenberg, and S. Urasawa. 1987. Direct serotyping of human rotavirus in stools by an enzyme-linked immunosorbent assay using serotype 1-, 2-, 3-, and 4-specific monoclonal antibodies to VP7. J. Infect. Dis. 155:1159-1166. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. G. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group a rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ushijima, H., H. Koike, A. Mukoyama, A. Hasegawa, S. Nishimura, and J. Gentsch. 1992. Detection and serotyping of rotaviruses in stool specimens by using reverse transcription and polymerase chain reaction amplification. J. Med. Virol. 38:292-297. [DOI] [PubMed] [Google Scholar]
- 29.van Loo, I. H., H. G. van der Heide, N. J. Nagelkerke, J. Verhoef, and F. R. Mooi. 1999. Temporal trends in the population structure of Bordetella pertussis during 1949-1996 in a highly vaccinated population. J. Infect. Dis. 179:915-923. [DOI] [PubMed] [Google Scholar]