Abstract
Eight PCR tests were evaluated for their abilities to detect Actinobacillus pleuropneumoniae in swine tonsils. At first they were compared regarding their specificities by using A. pleuropneumoniae and related bacterial species and their analytical sensitivities by using tonsils experimentally infected in vitro. PCRs were carried out both directly with tonsil homogenates (direct PCR) and after culture of the sample (after-culture PCR). Most tests demonstrated good specificities; however, some tests gave false-positive results with some non-A. pleuropneumoniae species. High degrees of variation in the analytical sensitivities among the tests were observed for the direct PCRs (109 to 102 CFU/g of tonsil), whereas those of most of the after-culture PCRs were similar (102 CFU/g of tonsil). In a second phase, the effects of sample storage time and storage conditions were evaluated by using tonsils from experimentally infected animals. Storage at −20°C allowed the detection of the organism for at least 4 months. Finally, the omlA PCR test described by Savoye et al. (C. Savoye et al., Vet. Microbiol. 73:337-347, 2000) and the commercially available Adiavet App PCR test were further validated with field samples. Their effectiveness was compared to those of standard and immunomagnetic separation-based methods of bacterial isolation. In addition, a comparison of tonsil biopsy specimens (from living animals) and whole tonsils (collected at the slaughterhouse) was also conducted. A. pleuropneumoniae was neither isolated nor detected by PCR from a herd serologically negative for A. pleuropneumoniae. PCR was more sensitive than the standard isolation method with whole tonsils from three infected herds. After-culture PCR offered the highest degree of sensitivity (93 and 83% for the omlA and Adiavet App PCRs, respectively). The PCR detection rate was higher with whole tonsils than with tonsil biopsy specimens. Good agreement (κ = 0.65) was found between the presence of A. pleuropneumoniae in tonsils and the individual serological status of the animals.
Actinobacillus pleuropneumoniae, 2 biovars and 15 serotypes of which have been described so far (3, 38), is the causative agent of porcine pleuropneumonia. The disease occurs worldwide and causes important economic losses to the swine industry. The disease course may be acute and fatal, with untreated animals sometimes dying within a few hours. However, in many herds, pigs may be subclinically infected without presenting clinical signs. These apparently healthy animals harbor the organism in their upper respiratory tracts, particularly in their nasal cavities and tonsils (24, 31, 36). It has recently been shown that A. pleuropneumoniae can survive in tonsillar crypts, leading to the carrier state (7). Aerosol transmission under experimental conditions has been documented (12, 34), and it has been proposed that subclinically infected pigs are the main cause of A. pleuropneumoniae dissemination (32).
Early identification of subclinically infected herds is essential for control of the disease. Serology, both standard and immunomagnetic separation-based (IMS) bacterial isolation methods, as well as PCR techniques may be used for this purpose. Serological monitoring has been very helpful in the control of swine pleuropneumonia (5, 11, 14, 22), but it has some limitations. Infected pigs may be serologically negative (6, 31, 36), and sometimes, inconclusive serological results may be observed in the absence of clinical signs or pathological lesions. In these cases, isolation of the organism or its detection by direct methods becomes important for confirmation of the infection. Standard bacteriological isolation from tonsils has a very low sensitivity since swine tonsils are heavily colonized with the commensal flora (including several NAD-dependent bacterial species) that may interfere with the isolation and identification of A. pleuropneumoniae, even with the use of selective media (23, 36). IMS isolation of A. pleuropneumoniae has been shown to have a significantly higher sensitivity compared to that of standard isolation methods (1, 13), but the technique is time-consuming and expensive. Furthermore, by using IMS isolation, a recent study has reported the isolation of A. pleuropneumoniae-like strains, tentatively named “Actinobacillus porcitonsillarum,” that are phenotypically and antigenically very similar to but genetically different from A. pleuropneumoniae (15). These bacteria complicate the interpretation of the results of IMS isolation, as they may be differentiated from A. pleuropneumoniae only by using molecular analysis.
Within the last 12 years several PCR techniques that amplify well-defined sequences of the A. pleuropneumoniae genome have been described as valuable tools for the rapid and affordable detection of the pathogen (8, 10, 17, 18, 21, 28, 34, 35, 37), and a ready-to-use PCR kit has even been commercialized (Adiavet App PCR test; Adiagène, St. Brieuc, France). Some of these tests allow only species detection (8, 17, 34, 35, 37), while others discriminate a particular serotype (28) or groups of serotypes, either directly (18) or after restriction fragment length polymorphism analysis (10, 21). It is claimed that all these tests have good specificities and sensitivities. However, on the basis of published data, a comparison of their performances is almost impossible, since studies vary greatly in the experimental procedures as well as the samples that they use. Some tests have been evaluated only with pure cultures of different bacterial species (18, 21), while others have used mixed bacterial cultures, either from tonsils (17, 19) or from both nasal swabs and tonsils (8). Some tests were conducted directly, without a mixed culture step, with tonsil biopsy specimens and tracheobronchial washes (34), lung tissue (28), or both nasal swabs and lung tissue (10, 35). Moreover, most studies have used experimentally infected animals and have not been validated with field samples. A comparison of the tests by the use of similar conditions thus appears to be essential to evaluate their actual effectiveness and efficacy.
The samples used are critical for the detection of A. pleuropneumoniae. Tonsils have been shown to be better than nasal cavity swabs and tracheobronchial washes for PCR (8, 34). However, some studies were conducted with whole tonsils, while others used tonsil biopsy specimens. It is not known whether a tonsil biopsy specimen is actually representative of the whole tonsil and whether use of the latter does not present a risk of cross-contamination when specimens are collected at the slaughterhouse.
The objectives of the present study were (i) to compare the sensitivities and specificities of eight PCR tests for the detection of A. pleuropneumoniae by using experimentally infected tonsils, (ii) to validate two of these PCR tests regarding their capacities to detect naturally infected A. pleuropneumoniae carrier animals, (iii) to compare the performances of the PCR tests to those of the standard and IMS isolation techniques, and (iv) to compare tonsil biopsy specimens and whole tonsils recovered at slaughter as samples in the PCR tests. In each case, the PCR tests were conducted with and without a previous culture step with the samples. Sample storage conditions were also examined. Finally, the correlation between the presence of the organism in tonsils and individual serological status was also evaluated.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study included strains of the 15 described A. pleuropneumoniae serotypes (reference strains Shope 4074, S4226, ATCC 27090, ATCC 33378, ATCC 33377, Femo, ATCC 33590, 405, CVJ13261, D13039, 56153, 8329, N273, 3906, and HS143); Actinobacillus minor (field strain); Actinobacillus equuli (ATCC 19392); Actinobacillus lignieresii (ATCC 19393); Actinobacillus suis (ATCC 15557 and one field strain); Actinobacillus porcinus (field strain); Haemophilus parasuis (ATCC 19417); Streptococcus suis (S735); and two strains of “A. porcitonsillarum,” an A. pleuropneumoniae-like species described recently (15). All strains with the exception of the A. pleuropneumoniae serotype 13, 14, and 15 reference strains originated from our collection. A. pleuropneumoniae serotype 13 and 14 reference strains were from Ø. Angen, Danish Veterinary Institute, Copenhagen, Denmark; and the serotype 15 reference strain was provided by J. Frey, University of Bern, Bern, Switzerland. All strains were grown as described previously (16, 31, 36).
PCR tests.
The PCR tests evaluated in this study included (i) the test described by Sirois et al. (37), (ii) the omlA test described by Savoye et al. (34), (iii) the dsbE-like test described by Chiers et al. (8), (iv) the aroA test described by Hernanz Moral et al. (21), (v) the apxIVA nested PCR test described by Schaller et al. (35), (vi) the cps-cpx multiplex PCR test described by Lo et al. (28), (vii) the omlA multiplex PCR test described by Gram et al. (18), and (viii) a commercially available multiplex PCR kit (Adiavet App PCR test; Adiagène). The first five PCR tests allow the detection of the species A. pleuropneumoniae only (8, 21, 34, 35, 37) and do not provide information regarding a particular serotype. On the other hand, the multiplex cps-cpx PCR specifically detects most of the A. pleuropneumoniae serotypes by amplification of the cpx gene, while it allows the discrimination of A. pleuropnemoniae serotype 5 by the selective amplification of its cps gene (28). The Adiavet App PCR test enables the detection and typing of the organism into four groups: group I indicates the presence of serotype 1, 9, 11, or 12; group II indicates the presence of serotypes 5a and 5b or 10; group III indicates the presence of serotype 2, 4, 7, 8, or 12; and group IV indicates the presence of serotype 3, 4, 6, or 7 or the NAD-independent serotypes (Adiavet App Technical Manual; Adiagène). Finally, the omlA multiplex PCR test enables the typing of A. pleuropnemoniae serotypes into five groups: group omlAI, serotype 1, 9, 11, or 12; group omlAII, serotype 2 or 8; group omlAIII, serotypes 3, 6, or 7; group omlAIV, serotype 4; and group omlAV, serotype 5a, 5b, or 10 (18).
Comparison of PCR test specificities and analytical sensitivities.
The specificities of the tests were evaluated by performing the PCRs with pure cultures of all the bacterial strains described above. The analytical sensitivities of the tests were compared by experimentally infecting tonsils obtained from animals originating from a herd considered free of A. pleuropneumoniae serotypes 1 to 12 on the basis of regular serological monitoring by the long-chain-lipopolysaccharide enzyme-linked immunosorbent assay (LC-LPS-ELISA) (14) for the previous 3 years. A total of 0.5 g of tonsils was reduced to small pieces with a scalpel and added to 5 ml of serial dilutions (109 to 101 CFU/ml) of a suspension of A. pleuropneumoniae serotypes 1, 5, and 7, which were used separately or in combination. After vortex mixing for 2 min and filtration through filter paper (type 4; Whatman International Limited, Springfield Mill, England), 100 μl of filtrate was plated onto selective PPLO agar and grown overnight (ON) at 37°C (5% CO2). Bacterial growth was harvested by washing the agar plates with 3 ml of sterile Tris-EDTA buffer (pH 8), and the growth was immediately processed as described below. The remaining original filtrate was fractionated into 1-ml aliquots and used as described below.
Template preparation and PCR conditions.
Template DNA from pure cultures was prepared by the guanidinium thiocyanate method (33) and 1 μl/PCR mixture was used for all tests. For PCRs carried out directly with the tonsil filtrate (direct PCRs), template DNA was obtained by a previously described technique by combining diatomaceous earth and guanidinium thiocyanate (4) from the pellet obtained after centrifugation (10,000 × g for 10 min) of 1 ml of the original tonsil filtrate. A total of 10 μl of purified DNA per PCR was used for all tests described here with the exception of those performed with the commercial kit, for which the recommendations of the manufacturer were followed. For PCRs carried out with the mixed cultures (after-culture PCRs), template DNA was extracted from 100 μl of the bacterial wash harvested from the PPLO agars (see above) by using the guanidinium thiocyanate method (33). All after-culture PCRs tests with the exception of those performed with the commercial kit were carried out with 1 μl of purified DNA. For the PCRs performed with the commercial kit, the recommendations of the manufacturer were followed.
All PCR tests were carried out as described previously or as recommended by the manufacturer, with two exceptions. For the cps-cpx test (28), 0.5 μM instead of 480 μM each primers A, B, C, and D was used (T. Inzana, personal communication). The omlA PCR test (34) was performed in a final volume of 50 μl containing the appropriate amount of purified template DNA, 5 μl of 10× buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2), each deoxyribonucleotide triphosphate at a concentration of 100 μM, 0.5 μM each primers LPF and omlAr (34) for the direct PCRs and 0.25 μM each for the after-culture PCRs, and 2.5 U of Taq DNA polymerase. Each sample was subjected to 40 cycles of amplification (denaturation at 94°C for 30 s, annealing at 62°C for 30 s for the direct PCRs and annealing at 64°C for the after-culture PCRs, and extension at 72°C for 90 s). The first cycle was preceded by an initial denaturation step at 94°C for 5 min, and the last cycle was followed by a final extension step at 72°C for 10 min. Primers for all tests were purchased from Invitrogen (Invitrogen, Burlington, Ontario, Canada); deoxyribonucleotide triphosphates, MgCl2, and Taq DNA polymerase were purchased from Amersham (Amersham Biosciences, Piscataway, N.J.). Amplifications were performed either in a DNA Thermal Cycler 480 (Perkin-Elmer Applied Biosystems, Foster City, Calif.) or in a T Gradient 96 thermal cycler (Biometra, Göttingen, Germany). Each test was performed in only one thermal cycler. For each test, the amplified products were separated by electrophoresis in agarose gels (1.2 to 2%, depending of the expected PCR product for each test; Sigma Chemical Co., St. Louis, Mo.) in 1× TAE (Tris-acetate-EDTA) buffer and were visualized by UV transillumination following ethidium bromide staining. A 100-bp DNA ladder (Invitrogen) was used as a molecular size standard in each gel.
IMS isolation.
IMS isolation was performed with 1 ml of original tonsil filtrate as described previously (13) by using immunomagnetic beads coated with rabbit anti-A. pleuropneumoniae serotype 1 immunoglobulin G for artificially infected tonsils and tonsils originating from experimentally infected animals. For field samples, beads were coated with rabbit anti-serotype 5 or 7 immunoglobulin G, depending on the targeted serotype. Colonies suspected of being A. pleuropneumoniae were tested for NAD dependence and the CAMP reaction. NAD-dependent and CAMP reaction-positive colonies were confirmed as being serotype 1, 5, or 7 by dot ELISA with serotype-specific monoclonal antibodies (25-27). Positive strains were confirmed to be A. pleuropneumoniae by the omlA PCR (34) and were serotyped by conventional techniques (30).
Experimental infection and sample storage conditions study.
Four 6-week-old piglets from a herd with minimal disease that was free of infection with A. pleuropneumoniae serotypes 1 to 12 were intranasally infected with 4 ml of a suspension of A. pleuropneumoniae serotype 1 strain Shope 4074 of 2 × 105 CFU/ml. The animals were humanely killed 7 weeks after infection and in the presence of seroconversion. The tonsils were collected and immediately fractionated into 0.5-g pieces that were stored at room temperature, 4°C, or −20°C for different periods of time (see Results). Some pieces were also frozen and thawed once before being analyzed. Detection of A. pleuropneumoniae was carried out by both direct PCR and after-culture PCR by using only the commercial PCR test (Adiavet App).
Validation of omlA (34) and Adiavet App PCR tests with field samples. (i) Animals.
A total of 198 pigs (ages, 6 months) were used and divided into five groups (groups 1 to 5). Animals in groups 1 (n = 54) and 2 (n = 30) were obtained from a herd with minimal disease that was considered free of infection with A. pleuropneumoniae serotypes 1 to 12 on the basis of regular serological monitoring by LC-LPS-ELISA (14). About 24 h before being sent to the slaughterhouse, a blood sample and a tonsil biopsy specimen (2) were taken from each well-identified animal. Early in the morning of the following day, the animals were slaughtered at a commercial slaughterhouse before the slaughtering of any other animal (group 1) or after the slaughtering of more than 500 animals from several origins (group 2) to increase the possibility that the water in the bath used to remove the pigs' hair had been exposed to pigs carrying A. pleuropneumoniae. Whole tonsils were recovered from the carcasses before they were placed in the chilling room and individually identified. The animals in groups 3, 4, and 5 were from farrow-to-finish herds infected or suspected of being infected with one or more serotypes of A. pleuropneumoniae. The pigs in group 3 (n = 36) were from a herd previously identified by pathological and bacteriological examinations to be infected with A. pleuropneumoniae serotype 5b and suffering from a sudden recrudescence of the disease, with several growers presenting clinical signs and/or pathological lesions at the time of sampling. Group 4 was composed of 39 pigs originating from a herd with no clinical signs of pleuropneumonia but in which the disease had been active in the finishing pigs about 6 months before sampling. A. pleuropneumoniae serotype 5b had already been isolated from the lungs of diseased animals, and previous serological examinations (LC-LPS-ELISA) had demonstrated that the herd was positive for serotype 5. The animals in group 5 (n = 39) originated from a herd serologically suspected of being infected with A. pleuropneumoniae serotypes 5 and 7 on the basis of serological examinations but with no clinical signs. Identification of animals in groups 3, 4, and 5 was done; and blood samples, tonsil biopsy specimens, and whole tonsils were obtained as described above for group 1. For all groups, blood samples were used for serology (LC-LPS-ELISA), and tonsil biopsy specimens and whole tonsils were conserved frozen at −20°C until they were analyzed.
(ii) Processing of tonsillar samples.
Each whole tonsil was seared on the surface with a hot spatula. For the standard isolation procedure, three parallel incisions were swabbed and incubated on selective PPLO agar ON at 37°C (5% CO2), as described previously (36). A piece of tonsil weighing 0.5 g was also taken from an open cut and reduced to small pieces with a scalpel, added to 5 ml of phosphate-buffered saline-0.1% bovine serum albumin, and processed (for IMS isolation, direct PCR, and after-culture PCR) as described above for artificially infected tonsils.
All biopsy specimens used weighed between 0.1 and 0.2 g. Due to their small size, the tonsil biopsy specimens were not seared, and neither the standard isolation technique nor IMS isolation was performed with these samples. For after-culture PCR, a selective PPLO agar was inoculated by touching the PPLO agar surface with the biopsy specimen and then streaking the inoculum with a loop. The organisms were allowed to grow on the agar ON at 37°C (5% CO2), and the ON growth was harvested and treated as described above for artificially infected tonsils. After inoculation of the PPLO agar, the biopsy specimens were reduced to small pieces with a scalpel, added to 2 ml of phosphate-buffered saline-0.1% bovine serum albumin, and processed as described above for artificially infected tonsils.
Statistics.
Because a “gold standard” for the detection of A. pleuropneumoniae has not yet been defined, the sensitivity and specificity of each test in the field validation were calculated by using the results of isolation (either the standard or the IMS isolation technique) and both the omlA (34) and the Adiavet App PCR tests (performed either directly or after a culture step for each PCR test used) as independent benchmarks. A cumulative study combining all positive results was also done (9). The agreement between tests was evaluated by calculating Cohen's kappa coefficient (κ). A value of 1 indicates perfect agreement, while a value of 0 indicates that agreement is no better than chance. In general, κ values between 0.3 and 0.5 are considered to represent moderate agreement. The chi-square test was used to demonstrate differences in the results between tonsil biopsy specimens and whole tonsils. The agreement between individual serologically positive status and demonstration of A. pleuropneumoniae in tonsillar samples was evaluated by use of κ.
RESULTS
Preliminary test comparison.
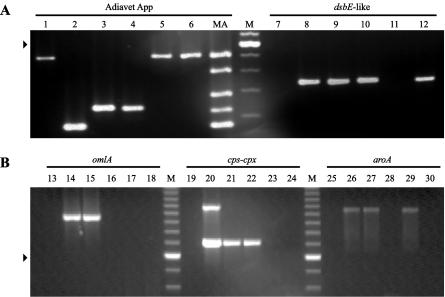
The omlA test (34), the omlA multiplex test (18), and the apxIVA test (35) were strictly specific for A. pleuropneumoniae. By contrast, and confirming the findings presented in previous reports, the test by Sirois et al. (37) and the dsbE-like test (8) also showed positive signals with A. lignieresii. Furthermore, the cps-cpx test (28) and the commercial PCR test (Adiavet App) also showed clear positive signals with this bacterial species (Fig. 1A and B). In our hands, two “A. porcitonsillarum” strains (15) gave a positive signal when they were tested by the dsbE-like test (Fig. 1A). The aroA test reacted with A. equuli, as described previously (21), and also, in our hands, with both reference strain ATCC 15557 (Fig. 1B) and a well-characterized field strain of A. suis. The omlA (18) and the Adiavet App multiplex PCR tests amplified the expected bands described to correspond to those of the different A. pleuropneumoniae serotypes. The latter test, however, presented a band pattern different from the pattern expected for the NAD-independent strains when it was used to test A. pleuropneumoniae serotype 14 reference strain 3906. The cps-cpx multiplex PCR test did not detect A. pleuropneumoniae serotype 4, as reported previously (28); however, it clearly identified the reference strains of serotypes 5a and 5b.
FIG. 1.
Examples of electrophoretic profiles, as observed in 2% (A) or 1.4% (B) agarose gels of the Adiavet App, dsbE-like (8), omlA (34), cps-cpx (28), and aroA (21) PCR tests. Reactions were carried out with no DNA (lanes 1, 7, 13, 19, and 25) or 1 μg of purified genomic DNA from A. pleuropneumoniae serotype 5 (lanes 2, 8, 14, 20, and 26), A. pleuropneumoniae serotype 7 (lanes 3, 9, 15, 21, and 27), A. lignieresii (lanes 4, 10, 16, 22, and 28), A. suis ATCC 15557 (lanes 5, 11, 17, 23, and 29), and “A. porcitonsillarum” (lanes 6, 12, 18, 24, and 30). Lanes M, 100-bp ladder (Invitrogen); arrowheads indicate the position of the 600-bp band of this ladder; lane MA, molecular weight marker provided with the Adiavet App PCR test. Bands of this size marker allow rapid assignment of the amplicons generated to one of the four Adiavet App PCR typing groups. The amplicons observed in lanes 1, 5, and 6 correspond to the internal inhibition control of the Adiavet App PCR test.
The analytical sensitivities of the tests for the detection of A. pleuropneumoniae from experimentally infected tonsils showed that most of them were able to detect the organism after a culture step only if ≥2.2 × 102 CFU of the organism per g of tonsil was present (Table 1). The exception was the aroA test, which could detect the organism only in tonsils infected with a considerably higher concentration of A. pleuropneumoniae. By contrast, great variabilities in analytical sensitivities were observed when detection was performed by direct PCR (Table 1). The apxIVA nested PCR test (35) was the most sensitive test, with analytical sensitivities within the same range as those obtained after a culture step. Other tests were less sensitive when they were performed directly; values ranged from 104 to 107 CFU/g of tonsil. The aroA test was unable to detect A. pleuropneumoniae even when the level of infection was more than 109 CFU/g of tonsil. With the exception of the apxIVA nested PCR test (35), the analytical sensitivity of IMS isolation was higher than that of direct PCR (Table 1).
TABLE 1.
Sensitivities of the different PCR tests and IMS isolation for detection of A. pleuropneumoniae from experimentally infected tonsils
| PCR test (reference) | Sensitivity (CFU/g of tonsil)a
|
|
|---|---|---|
| Direct PCRb | After-culture PCRc | |
| apxIVA (35) | 3.6 × 102 | 2.2 × 102 |
| Adiavet Appd (8) | 1.7 × 104 | 2.2 × 102 |
| omlA (34) | 4 × 104 | 2.2 × 102 |
| dsbE-like (8) | 4.8 × 105 | 5.2 × 102 |
| aroA (21) | <109 | 2.2 × 105 |
| Multiplex omlA (18) | 3.4 × 106 | 2.2 × 102 |
| cps-cpx (28) | 1.8 × 107 | 5.2 × 102 |
| Sirois et al. (37) | 2.1 × 107 | 2.2 × 102 |
A. pleuropneumoniae serotype 1 reference strain Shope 4074 was used to infect the tonsils. The sensitivity of IMS isolation was 4.39 × 103 CFU/g of tonsil.
PCR was carried out with DNA obtained without culture of the sample.
PCR was carried out with DNA obtained after culture of the sample.
Commercial PCR test.
Effects of tonsil storage conditions.
The tonsils of four experimentally infected animals collected immediately after euthanasia (day 0) were positive by both direct and after-culture PCR (by the Adiavet App PCR test only). The results obtained with specimens tested on subsequent days are summarized in Table 2. In general, tonsil storage at −20°C allowed detection of the organism for at least 120 days by both the direct PCR and the after-culture PCR, while in tonsils conserved at 4°C, A. pleuropneumoniae could be detected for at least 2 weeks, also by both direct PCR and after-culture PCR. In samples stored at room temperature, the organism was detected by the direct PCR for at least 8 days, while most animals were negative by the after-culture PCR after 2 days of storage. All animals were positive by both the direct PCR and the after-culture PCR after a freezing (7 days at −20°C) and thawing cycle.
TABLE 2.
Effects of time and storage conditions on Adiavet App PCR detection of A. pleuropneumoniae from whole tonsils of experimentally infected animals
| Storage temp (°C) | Method | No. of animals positive after storage for the following times (days)a:
|
||||
|---|---|---|---|---|---|---|
| 2 | 4 | 8 | 17 | 120 | ||
| RTb | Direct PCRc | 4 | 4 | 3d | ND | NDe |
| After-culture PCRf | 0 | 1 | 0d | ND | ND | |
| 4° | Direct PCR | 4 | 4 | 4 | 4 | ND |
| After-culture PCR | 4 | 4 | 4 | 4 | ND | |
| −20 | Direct PCR | ND | ND | 4 | 4 | 4 |
| After-culture PCR | ND | ND | 4 | 4 | 4 | |
A total of four animals were tested.
RT, room temperature.
PCR was carried out with DNA obtained without culture of the sample.
Only three animals were tested.
ND, not done.
PCR was carried out with DNA obtained after culture of the sample.
Field validation. (i) Serological results.
Sera from animals in groups 1 and 2 did not react in the LC-LPS-ELISA with antigens of A. pleuropneumoniae serotypes 1 to 12. On the other hand, 34 of the 36 animals in group 3 were positive for A. pleuropneumoniae serotype 5, 3 were positive for serotype 12, and 2 were positive for serotype 2. Among the animals in group 2, 16 were seropositive for serotype 5; no reaction was observed with the other serotypes. Among the animals in group 3, 8 were positive by the LC-LPS-ELISA for serotypes 7 and 4 (27, 29), 2 were positive for serotype 12, and 1 was positive for serotype 5.
(ii) Isolation, omlA PCR, and Adiavet App PCR test detection with tonsillar samples.
A. pleuropneumoniae could not be detected in any of the animals in groups 1 and 2, and this was regardless of the method used. A. pleuropneumoniae could be isolated from the tonsils of animals in groups 3, 4, and 5. All the isolates belonged to serotype 5 (groups 3 and 4) or 7 (group 5), as revealed by dot ELISA and serotyping. The results obtained by the Adiavet App PCR test showed that positive samples from animals in groups 3 and 4 belonged to Adiavet App PCR typing group II (which includes serotype 5 of A. pleuropneumoniae). Positive samples from animals in group 5 could be assigned to Adiavet App PCR typing groups III and IV (which include serotype 7). Positive samples from animals in groups 3 and 4 were confirmed as being in fact positive for serotype 5 by using the cps-cpx PCR test, which is specific for serotype 5 (28; results not shown).
Table 3 summarizes the results obtained by isolation (by both the standard technique and IMS isolation), as well as by the omlA PCR (34) and the Adiavet App PCR test, with whole tonsils and tonsil biopsy specimens from animals in groups 3, 4, and 5. When the groups are considered together, the rates of detection by the two PCR tests were in general significantly higher with whole tonsils than with tonsil biopsy specimens. Nevertheless, the rates of detection by the direct Adiavet App PCR test did not show significant differences with these two types of samples. When the groups are considered individually, a significantly higher rate of detection by PCR with whole tonsils than with tonsil biopsy specimens was observed for group 3. For groups 4 and 5, although the differences were not significant for most tests, a smaller number of animals showed a positive result when PCR was performed with tonsil biopsy specimens.
TABLE 3.
Results of standard isolation, IMS isolation, and PCR detection (omlA and Adiavet App PCR tests) of A. pleuropneumoniae with whole tonsils or tonsil biopsy specimens from animals originating from infected herds
| Group and specimen | No. of animals positive
|
|||||
|---|---|---|---|---|---|---|
| Isolation
|
PCR
|
|||||
| Directb
|
After culturec
|
|||||
| Standard | IMSa | omlAd | Adiavet App | omlA | Adiavet App | |
| Group 3 (n = 36) | ||||||
| Whole tonsils | 8 | 23 | 22e | 34 | 35e | 35e |
| Biopsies | NDf | ND | 12e | 28 | 15e | 22e |
| Total positiveg | 8 | 23 | 25 | 35 | 35 | 35 |
| Group 4 (n = 39) | ||||||
| Whole tonsils | 3 | 5 | 8 | 12 | 20e | 12 |
| Biopsies | ND | ND | 3 | 9 | 6e | 10 |
| Total positive | 3 | 5 | 8 | 14 | 21 | 14 |
| Group 5 (n = 39) | ||||||
| Whole tonsils | 0 | 4 | 0 | 3 | 6 | 7 |
| Biopsies | ND | ND | 0 | 0 | 2 | 4 |
| Total positive | 0 | 4 | 0 | 3 | 7 | 9 |
| All positive groups (n = 114) | ||||||
| Whole tonsils | 11 | 32 | 30e | 49 | 61e | 54e |
| Biopsies | ND | ND | 15e | 37 | 23e | 36e |
| Total positive | 11 | 32 | 33 | 52 | 63 | 58 |
IMS for serotype 5 (groups 3 and 4) or serotype 7 (group 5).
PCR was carried out with DNA obtained without culture of the sample.
PCR was carried out with DNA obtained after culture of the sample.
As described by Savoye et al. (34).
Significant differences in detection between whole tonsils and tonsils biopsy specimens (P < 0.05).
ND, not done.
Number of positive animals for each test, regardless of the sample used.
(iii) Sensitivities, specificities, and agreements between tests.
Table 4 shows a comparison of the results of the standard isolation technique, IMS isolation, the omlA PCR (34), and the Adiavet App PCR test when the whole tonsil was used as a sample. By using the cumulative results (all tests) as the benchmark test, the highest sensitivities were obtained by both the after-culture omlA PCR (34) and the after-culture Adiavet App PCR test, followed by the direct Adiavet App PCR test. Very good agreement (κ > 0.70) was found between these three tests. IMS isolation and the direct omlA PCR (34) showed similar moderate sensitivities, very good agreement between themselves, and good agreement with the three PCR tests mentioned in the previous sentence. The standard isolation technique demonstrated the lowest sensitivities and fair or very low levels of agreement with the other tests. However, its specificities were high (99 and 100%), while the others tests showed lower, although very good, specificities.
TABLE 4.
Sensitivities, specificities, and agreement (κ) of standard isolation technique, IMS isolation, and PCR tests (omlA and Adiavet App PCRs) for detection of A. pleuropneumoniae from whole tonsils
| Test | % Positive samples | Results of comparison with the following benchmark tests:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All testsa
|
Isolation
|
PCR
|
||||||||||||||||||||
| Directb
|
After culturec
|
|||||||||||||||||||||
| Standard
|
IMS
|
omlAd
|
Adiavet App
|
omlA
|
Adiavet App
|
|||||||||||||||||
| SEe (%) | SPf (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | ||
| Standard isolation | 10 | 17 | 100 | 0.15 | 31 | 99 | 0.38 | 37 | 100 | 0.46 | 22 | 100 | 0.25 | 18 | 100 | 0.17 | 20 | 100 | 0.21 | |||
| IMS isolation | 28 | 49 | 100 | 0.46 | 91 | 79 | 0.38 | 83 | 91 | 0.73 | 61 | 97 | 0.61 | 49 | 96 | 0.44 | 56 | 97 | 0.53 | |||
| omlA PCR, direct | 26 | 46 | 100 | 0.42 | 100 | 82 | 0.46 | 78 | 94 | 0.73 | 61 | 100 | 0.64 | 49 | 100 | 0.47 | 55 | 100 | 0.57 | |||
| Adiavet App PCR, direct | 43 | 75 | 100 | 0.73 | 100 | 63 | 0.25 | 94 | 77 | 0.61 | 100 | 77 | 0.64 | 77 | 96 | 0.72 | 87 | 97 | 0.84 | |||
| omlA PCR, after culture | 53 | 93 | 100 | 0.93 | 100 | 51 | 0.17 | 94 | 62 | 0.44 | 100 | 63 | 0.47 | 96 | 78 | 0.72 | 98 | 86 | 0.84 | |||
| Adiavet App PCR, after culture | 47 | 83 | 100 | 0.81 | 100 | 58 | 0.21 | 94 | 71 | 0.53 | 100 | 71 | 0.57 | 96 | 89 | 0.84 | 87 | 98 | 0.84 | |||
A sample was considered positive if A. pleuropneumoniae was detected by at least one assay; a sample was considered negative if A. pleuropneumoniae was not detected by any of the assays.
PCR was carried out with DNA obtained without culture of the sample.
PCR was carried out with DNA obtained after culture of the sample.
As described by Savoye et al. (34).
SE, sensitivity. The sensitivity of a test was defined as the number of samples positive by the test that were also positive by the benchmark test divided by the total number of samples positive by the benchmark test.
SP, specificity. The specificity of a test was defined as the number of samples negative by the test that were also negative by the benchmark test divided by the total number of samples negative by the benchmark test.
Table 5 shows a comparison of the performances of both the omlA PCR (34) and the Adiavet App PCR test when tonsil biopsy specimens were used. In general, the sensitivities of these PCR tests were lower than those obtained by PCR with whole tonsils, and the levels of agreement between the tests ranged from moderate to good. Furthermore, the direct omlA PCR (34) showed only a fair level of agreement with the other tests. The specificities were also lower. Both the direct and the after-culture Adiavet App PCR tests showed the best sensitivities, followed by the after-culture omlA PCR (34).
TABLE 5.
Sensitivities, specificities, and agreement (κ) of standard isolation technique, IMS isolation, and PCR tests (omlA and Adiavet App PCRs) for detection of A. pleuropneumoniae from tonsil biopsy specimensa
| Test | % Positive samples | Results of comparison with the following benchmark tests:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All tests
|
PCR
|
|||||||||||||||
| Directb
|
After culturec
|
|||||||||||||||
|
omlAd
|
Adiavet App
|
omlA
|
Adiavet App
|
|||||||||||||
| SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | SE (%) | SP (%) | κ | ||
| omlA PCR, direct | 13 | 31 | 100 | 0.34 | 37 | 97 | 0.41 | 43 | 93 | 0.41 | 29 | 93 | 0.26 | |||
| Adiavet App PCR, direct | 32 | 75 | 100 | 0.78 | 87 | 77 | 0.41 | 77 | 78 | 0.44 | 74 | 87 | 0.61 | |||
| omlA PCR, after culture | 20 | 47 | 100 | 0.49 | 60 | 87 | 0.41 | 46 | 93 | 0.44 | 58 | 99 | 0.64 | |||
| Adiavet App PCR, after culture | 31 | 73 | 100 | 0.76 | 67 | 74 | 0.26 | 72 | 88 | 0.61 | 95 | 83 | 0.64 | |||
(iv) Agreement between serology and direct detection.
When the results for the 114 animals in groups 3, 4, and 5 are considered together, a good agreement (κ = 0.68) was found between the serological status of an individual determined by the LC-LPS-ELISA (14) and the detection of the organism in the tonsils by any of the direct methods (both the standard technique and IMS isolation as well as both the direct and after-culture omlA PCRs [34] and the direct and after-culture Adiavet App PCR tests). A total of 44 animals were positive by both serology and direct detection, while 50 animals were negative by both serology and direct detection. A. pleuropneumoniae was not detected by any of the direct methods in six seronegative animals, while the organism was not detected either by isolation or by PCR in 14 seropositive animals. When the results for the groups are considered individually, a similar correlation was noted for groups 3 and 4. For group 5, too few animals were positive to accurately assess a possible correlation. However, a tendency for a similar association was noted for the last group (data not shown).
DISCUSSION
Numerous PCR tests for the detection of A. pleuropneumoniae in carrier pigs have been described (8, 10, 17, 18, 21, 28, 34, 35, 37) and even commercialized (Adiavet App PCR test). In this study, a comparison of the specificities and the analytical sensitivities of eight of these PCR tests was carried out by using the same experimental procedures and samples. Specificity for the species is essential for validation of a positive result. The apxIVA PCR test and the two omlA PCR tests showed strict specificities for A. pleuropneumoniae. Some other tests also gave positive signals with A. lignieresii (8, 28, 37) (Adiavet App PCR) and A. equuli (21). These unspecific reactions should not necessarily prevent the use of these tests for diagnosis, since it is unlikely that these two bacterial species would be isolated from swine. However, two tests also detected species other than A. pleuropneumoniae likely to be present in swine tonsils. The dsbE-like test gave a positive signal with “A. porcitonsillarum” (15), a species that was not yet described at the time that the dsbE-like test was originally reported. This unspecific reaction may complicate the interpretation of a positive result obtained by this test. It has been found that the aroA test does not detect A. suis (21). Nevertheless, in our hands, a positive signal with two A. suis strains was obtained by this test, which may significantly complicate the interpretation of a positive result. Furthermore, with experimentally infected tonsils, the aroA test showed a very low analytical sensitivity, although a very high sensitivity (12 CFU/PCR assay) had previously been reported for this test when it was performed with pure cultures (21). We hypothesize that the amplification conditions for this test may need to be adapted so that it can be used with DNA samples containing large amounts of foreign DNA and/or potential inhibitors, like those obtained from tonsils. The analytical sensitivities of the other PCR tests were similar, since they showed similar values when amplifications were performed after a culture step. This suggests that the factor limiting the analytical sensitivity for after-culture PCR could be A. pleuropneumoniae itself, as it may be overgrown by the competing flora when it is present in the tonsil at concentrations <102 CFU/g of tonsil.
The results obtained by direct PCR demonstrate that the apxIVA nested PCR test (35) is the best option when a culture step is avoided and maximal analytical sensitivity is desired. However, this test may suffer from cross-contamination in a routine diagnostic laboratory, leading to consideration of a number of samples as false positive. In the absence of clinical signs, the tonsils of carrier pigs are not likely to harbor large numbers of the organism. The Adiavet App PCR test and one omlA PCR test (34) were thus chosen to be validated, as they offered good detection limits for both the direct and the after-culture PCRs in conjunction with a simplicity of execution. The remaining tests presented lower analytical sensitivities when they were performed directly, and their use for diagnosis would increase the risk of missing positive animals. Nonetheless, even if it is less sensitive, the cps-cpx PCR test (26) is the only molecular tool available that could accurately identify a sample positive for A. pleuropneumoniae serotype 5. The serotype 5-infected samples in this study were indeed confirmed to be positive for this serotype by this test (results not shown).
Validation of the omlA PCR test (34) and the Adiavet App PCR test with whole tonsils and tonsil biopsy specimens from field animals showed that these tests did not react with samples from any of the 54 seronegative animals in group 1, further confirming their species specificities with porcine samples. Cross-contamination between carcasses or via the water bath used to remove the pigs' hair at the slaughterhouse was not observed in this study, as negative results were obtained by PCR and by isolation for animals in group 2. Whole tonsils from the animals in this group had been collected at a commercial slaughterhouse after more than 500 animals of different origins had been slaughtered. Although sampling at the slaughterhouse does not seem to present a major risk of obtaining a false-positive result, a larger number of animals should be tested to confirm this hypothesis.
The results obtained with whole tonsils from animals in groups 3, 4, and 5 confirmed the results of previous reports showing that PCR is more sensitive than standard isolation (8, 34, 35). Furthermore, when PCR was performed from whole tonsils after a culture step (and also when it was performed directly by the Adiavet App PCR test), the number of positive animals was higher than that obtained by IMS isolation. On the other hand and as opposed to what was reported in a previous study (34), IMS isolation presented a higher sensitivity than the direct omlA PCR test. It is possible that both techniques have similar sensitivities and that the different results obtained were due to differences in the contaminating flora present in the samples.
The after-culture Adiavet App PCR test has been evaluated in a previous study (8). However, no data concerning its use without culture of the sample have been published. In our hands, this test presented very good specificities and sensitivities with porcine samples when it was performed both directly and after culture. It also provides information regarding the possible serotypes present in tonsils. In fact, this information is important, as differences in virulence among serotypes have been found (20). The subtypes of positive field samples determined by this test correlated with the serotypes of the strains isolated and with the main serotypes identified by serology to be causing the infection. However, for the animals in groups 3 and 5, detection of other serotypes for which a positive result had been obtained by serology was not possible by using the Adiavet App PCR test or bacteriological isolation. It is not clear whether this reflects false-positive serological results, an actual absence of these serotypes in tonsils, or a detection limit problem of isolation and the Adiavet App PCR test. For the latter, if two different serotypes were present in the same tonsil, as might be the case for animals in groups 3 and 5, the risk of missing detection of one of them may exist. With artificially infected tonsils, the Adiavet App PCR test distinguished between groups of serotypes when they were used in combination at equivalent concentrations. By contrast, only the more concentrated serotype was detected when a combination of one serotype at high concentration (107 CFU/g of tonsil) and another serotype at a low concentration (104 CFU/g of tonsil) were tested (unpublished observations).
In general, fewer positive samples were found when tonsil biopsy specimens were used than when whole tonsils were used. This may be explained by the smaller amount of tonsil biopsy specimen tissue used in comparison with the amount of whole tonsils used (34). The different methodologies used to culture both types of samples may account for the lower detection level observed for the after-culture PCRs with tonsil biopsy specimens. As the latter were cultivated by touching the agar surface with the biopsy specimen, it is possible that pathogens located deep in the tonsillar crypts were missed in these cultures (7). With whole tonsils from experimentally infected animals, A. pleuropneumoniae could be detected by both direct and after-culture Adiavet App PCR tests for at least 120 days when the tonsils had been stored at −20°C. However, the same temporal study was not done with tonsil biopsy specimens. Although all biopsy specimens were tested within 2 weeks following sampling, there is a possibility for a loss of viability of the A. pleuropneumoniae organisms present in tonsil biopsy specimens. Even if the detection of the organism from tonsil biopsy specimens was less sensitive than that from whole tonsils, it is still an interesting type of sample, as it allows detection and follow-up of the progression of the infection in the living animal.
The main advantage of the direct PCR is that it greatly reduces the time required to obtain a result. In addition, nonviable microorganisms that may be present in poorly conserved samples may still be detected. This was confirmed in the present study, as samples stored for several days at room temperature tested positive by the direct method and negative by after-culture PCR. On the other hand, with samples conserved at 4°C for at least 2 weeks, A. pleuropneumoniae could be detected by both the direct and the after-culture techniques. Detection of the organism by direct or after-culture PCR from samples that were submitted to one freeze-thaw cycle before being tested did not seem to be affected.
In practice, and even if no published data support this procedure, veterinarians have the tendency to collect samples for isolation or PCR detection of A. pleuropneumoniae from seropositive animals. In fact, serology is an indirect method that allows identification of antibodies against A. pleuropneumoniae in sera from infected animals, while isolation and PCR allow the direct detection of the organism. However, even if neither isolation nor PCR detection in some seropositive animals could be achieved, the results obtained in this study suggest a good agreement between the individual positive serological status and the presence of the pathogen in tonsils.
Bacterial detection by PCR has been proposed as a complementary test to confirm positive serological results for the identification of pigs that are A. pleuropneumoniae carriers (34). While only more extensive epidemiological studies with larger numbers of herds may confirm the suitability of this strategy, the data presented in this study suggest that the application of PCR-based detection tests would increase the accuracy of detecting infected animals by confirming the results of serological assays, especially in conflicting situations. In addition, these assays may have an important role to play in understanding the biological basis of the persistence of the infection in both immunologically positive and immunologically nonresponsive clinically healthy pigs.
Acknowledgments
We are grateful to Réal Boutin for identification of herds and sampling of field animals, Diane Côté for outstanding technical assistance, and Guy Beauchamps for the statistical analysis. We also thank Robert Desrosiers for critical review of the manuscript.
This work was supported by the Conseil de Recherches en Pêche et en Agroalimentaire du Québec and the Canadian Research Network on Bacterial Pathogens of Swine.
REFERENCES
- 1.Angen, O., P. M. Heegaard, D. T. Lavritsen, and V. Sorensen. 2001. Isolation of Actinobacillus pleuropneumoniae serotype 2 by immunomagnetic separation. Vet. Microbiol. 79:19-29. [DOI] [PubMed] [Google Scholar]
- 2.Barrère, E., P. Cabanié, Y. Nicolas, and P. Massabie. 1993. Innocuité d'une technique de biopsie d'amygdale chez le porc, p. 329-334. In Proceedings 25èmes Journées de la Recherche Porcine en France. Paris, France.
- 3.Blackall, P. J., H. L. Klaasen, H. Van Den Bosch, P. Kuhnert, and J. Frey. 2002. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet. Microbiol. 84:47-52. [DOI] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosse, J. T., R. P. Johnson, and S. Rosendal. 1990. Serodiagnosis of pleuropneumonia using enzyme-linked immunosorbent assay with capsular polysaccharide antigens of Actinobacillus pleuropneumoniae serotypes 1, 2, 5 and 7. Can. J. Vet. Res. 54:427-431. [PMC free article] [PubMed] [Google Scholar]
- 6.Chiers, K., E. Donne, I. Van Overbeke, R. Ducatelle, and F. Haesebrouck. 2002. Evaluation of serology, bacteriological isolation and polymerase chain reaction for the detection of pigs carrying Actinobacillus pleuropneumoniae in the upper respiratory tract after experimental infection. Vet. Microbiol. 88:385-392. [DOI] [PubMed] [Google Scholar]
- 7.Chiers, K., F. Haesebrouck, I. van Overbeke, G. Charlier, and R. Ducatelle. 1999. Early in vivo interactions of Actinobacillus pleuropneumoniae with tonsils of pigs. Vet. Microbiol. 68:301-306. [DOI] [PubMed] [Google Scholar]
- 8.Chiers, K., I. Van Overbeke, E. Donne, M. Baele, R. Ducatelle, T. De Baere, and F. Haesebrouck. 2001. Detection of Actinobacillus pleuropneumoniae in cultures from nasal and tonsillar swabs of pigs by a PCR assay based on the nucleotide sequence of a dsbE-like gene. Vet. Microbiol. 83:147-159. [DOI] [PubMed] [Google Scholar]
- 9.De Groote, D., R. Ducatelle, L. J. van Doorn, K. Tilmant, A. Verschuuren, and F. Haesebrouck. 2000. Detection of “candidatus Helicobacter suis” in gastric samples of pigs by PCR: comparison with other invasive diagnostic techniques. J. Clin. Microbiol. 38:1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Puente-Redondo, V. A., N. G. del Blanco, C. B. Gutierrez-Martin, J. N. Mendez, and E. F. Rodriquez Ferri. 2000. Detection and subtyping of Actinobacillus pleuropneumoniae strains by PCR-RFLP analysis of the tbpA and tbpB genes. Res. Microbiol. 151:669-681. [DOI] [PubMed] [Google Scholar]
- 11.Fenwick, B. W., and B. I. Osburn. 1986. Immune responses to the lipopolysaccharides and capsular polysaccharides of Haemophilus pleuropneumoniae in convalescent and immunized pigs. Infect. Immun. 54:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fussing, V., K. Barfod, R. Nielsen, K. Moller, J. P. Nielsen, H. C. Wegener, and M. Bisgaard. 1998. Evaluation and application of ribotyping for epidemiological studies of Actinobacillus pleuropneumoniae in Denmark. Vet. Microbiol. 62:145-162. [DOI] [PubMed] [Google Scholar]
- 13.Gagne, A., S. Lacouture, A. Broes, S. D'Allaire, and M. Gottschalk. 1998. Development of an immunomagnetic method for selective isolation of Actinobacillus pleuropneumoniae serotype 1 from tonsils. J. Clin. Microbiol. 36:251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottschalk, M., E. Altman, N. Charland, F. De Lasalle, and J. D. Dubreuil. 1994. Evaluation of a saline boiled extract, capsular polysaccharides and long-chain lipopolysaccharides of Actinobacillus pleuropneumoniae serotype 1 as antigens for the serodiagnosis of swine pleuropneumonia. Vet. Microbiol. 42:91-104. [DOI] [PubMed] [Google Scholar]
- 15.Gottschalk, M., A. Broes, K. R. Mittal, M. Kobisch, P. Kuhnert, A. Lebrun, and J. Frey. 2003. Non-pathogenic Actinobacillus isolates antigenically and biochemically similar to Actinobacillus pleuropneumoniae: a novel species? Vet. Microbiol. 92:87-101. [DOI] [PubMed] [Google Scholar]
- 16.Gottschalk, M., R. Higgins, and M. Jacques. 1993. Production of capsular material by Streptococcus suis serotype 2 under different growth conditions. Can. J. Vet. Res. 57:49-52. [PMC free article] [PubMed] [Google Scholar]
- 17.Gram, T., and P. Ahrens. 1998. Improved diagnostic PCR assay for Actinobacillus pleuropneumoniae based on the nucleotide sequence of an outer membrane lipoprotein. J. Clin. Microbiol. 36:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gram, T., P. Ahrens, M. Andreasen, and J. P. Nielsen. 2000. An Actinobacillus pleuropneumoniae PCR typing system based on the apx and omlA genes—evaluation of isolates from lungs and tonsils of pigs. Vet. Microbiol. 75:43-57. [DOI] [PubMed] [Google Scholar]
- 19.Gram, T., P. Ahrens, and J. P. Nielsen. 1996. Evaluation of a PCR for detection of Actinobacillus pleuropneumoniae in mixed bacterial cultures from tonsils. Vet. Microbiol. 51:95-104. [DOI] [PubMed] [Google Scholar]
- 20.Haesebrouck, F., K. Chiers, I. Van Overbeke, and R. Ducatelle. 1997. Actinobacillus pleuropneumoniae infections in pigs: the role of virulence factors in pathogenesis and protection. Vet. Microbiol. 58:239-249. [DOI] [PubMed] [Google Scholar]
- 21.Hernanz Moral, C., A. Cascon Soriano, M. Sanchez Salazar, J. Yugueros Marcos, S. Suarez Ramos, and G. Naharro Carrasco. 1999. Molecular cloning and sequencing of the aroA gene from Actinobacillus pleuropneumoniae and its use in a PCR assay for rapid identification. J. Clin. Microbiol. 37:1575-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inzana, T. J., and B. Mathison. 1987. Serotype specificity and immunogenicity of the capsular polymer of Haemophilus pleuropneumoniae serotype 5. Infect. Immun. 55:1580-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsen, M. J., and J. P. Nielsen. 1995. Development and evaluation of a selective and indicative medium for isolation of Actinobacillus pleuropneumoniae from tonsils. Vet. Microbiol. 47:191-197. [DOI] [PubMed] [Google Scholar]
- 24.Kume, K., T. Nakai, and A. Sawata. 1984. Isolation of Haemophilus pleuropneumoniae from the nasal cavities of healthy pigs. Nippon Juigaku Zasshi 46:641-647. [DOI] [PubMed] [Google Scholar]
- 25.Lacouture, S., K. R. Mittal, M. Jacques, and M. Gottschalk. 1997. Serotyping Actinobacillus pleuropneumoniae by the use of monoclonal antibodies. J. Vet. Diagn. Investig. 9:337-341. [DOI] [PubMed] [Google Scholar]
- 26.Lairini, K., E. Stenbaek, S. Lacouture, and M. Gottschalk. 1995. Production and characterization of monoclonal antibodies against Actinobacillus pleuropneumoniae serotype 1. Vet. Microbiol. 46:369-381. [DOI] [PubMed] [Google Scholar]
- 27.Lebrun, A., S. Lacouture, D. Cote, K. R. Mittal, and M. Gottschalk. 1999. Identification of Actinobacillus pleuropneumoniae strains of serotypes 7 and 4 using monoclonal antibodies: demonstration of common LPS O-chain epitopes with Actinobacillus lignieresii. Vet. Microbiol. 65:271-282. [DOI] [PubMed] [Google Scholar]
- 28.Lo, T. M., C. K. Ward, and T. J. Inzana. 1998. Detection and identification of Actinobacillus pleuropneumoniae serotype 5 by multiplex PCR. J. Clin. Microbiol. 36:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mittal, K. R., and S. Bourdon. 1991. Cross-reactivity and antigenic heterogeneity among Actinobacillus pleuropneumoniae strains of serotypes 4 and 7. J. Clin. Microbiol. 29:1344-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittal, K. R., R. Higgins, S. Lariviere, and M. Nadeau. 1992. Serological characterization of Actinobacillus pleuropneumoniae strains isolated from pigs in Quebec. Vet. Microbiol. 32:135-148. [DOI] [PubMed] [Google Scholar]
- 31.Moller, K., L. V. Andersen, G. Christensen, and M. Kilian. 1993. Optimalization of the detection of NAD dependent Pasteurellaceae from the respiratory tract of slaughterhouse pigs. Vet. Microbiol. 36:261-271. [DOI] [PubMed] [Google Scholar]
- 32.Nicolet, J. 1992. Actinobacillus pleuropneumoniae, p. 401-408. In A. D. Leman, B. E. Straw, W. L. Mengeling, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames.
- 33.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 34.Savoye, C., J. L. Jobert, F. Berthelot-Herault, A. M. Keribin, R. Cariolet, H. Morvan, F. Madec, and M. Kobisch. 2000. A PCR assay used to study aerosol transmission of Actinobacillus pleuropneumoniae from samples of live pigs under experimental conditions. Vet. Microbiol. 73:337-347. [DOI] [PubMed] [Google Scholar]
- 35.Schaller, A., S. P. Djordjevic, G. J. Eamens, W. A. Forbes, R. Kuhn, P. Kuhnert, M. Gottschalk, J. Nicolet, and J. Frey. 2001. Identification and detection of Actinobacillus pleuropneumoniae by PCR based on the gene apxIVA. Vet. Microbiol. 79:47-62. [DOI] [PubMed] [Google Scholar]
- 36.Sidibe, M., S. Messier, S. Lariviere, M. Gottschalk, and K. R. Mittal. 1993. Detection of Actinobacillus pleuropneumoniae in the porcine upper respiratory tract as a complement to serological tests. Can. J. Vet. Res. 57:204-208. [PMC free article] [PubMed] [Google Scholar]
- 37.Sirois, M., E. G. Lemire, and R. C. Levesque. 1991. Construction of a DNA probe and detection of Actinobacillus pleuropneumoniae by using polymerase chain reaction. J. Clin. Microbiol. 29:1183-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, D. J. 1999. Actinobacillus pleuropneumoniae, p. 343-354. In A. D. Leman, B. E. Straw, W. L. Mengeling, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine, Iowa State University Press, Ames.