Abstract
We report on the development of a scheme for the typing of Pseudomonas aeruginosa, multiple-locus variable number of tandem repeat (VNTR) analysis (MLVA). We first evaluated the polymorphisms of 201 tandem repeat loci selected from more than 3,000 such sequences present in strain PAO1 with a test collection of 12 genotypically distinct clinical strains. Seven VNTR loci which can be easily scored with the technology used here were identified and used to genotype a collection of 89 clinical isolates that had previously been classified into 46 ribotypes, including 2 widespread ribotypes. Seventy-one different MLVA genotypes could be distinguished. With only two exceptions, strains with identical ribotypes were grouped together upon cluster analysis of the MLVA data. The 27 isolates with the most frequent ribotype were divided into 14 MLVA types, and the 18 isolates with the second most frequent ribotype were divided into 15 MLVA types. Analysis of a subset of 17 strains belonging to the major ribotype by pulsed-field gel electrophoresis with the enzyme SpeI distinguished seven types, identical to the number of MLVA types in this subset. Our data show that MLVA typing of P. aeruginosa based on the first set of loci has a high discriminatory power. Because MLVA is highly reproducible and easily portable among laboratories, it represents a very promising tool for the molecular surveillance of P. aeruginosa. A free, online strain identification service based on the genotyping data produced herein has been developed.
Pseudomonas aeruginosa is a ubiquitous gram-negative bacterium and a common opportunistic pathogen in hospitals. It is the most important cause of lung colonization in patients with cystic fibrosis and has high rates of multidrug resistance (for a review, see reference 14). Accurate typing and characterization of isolates are essential to understanding the epidemiology of this pathogen. Serotyping is traditionally used for strain typing, but it cannot be easily applied to the mucoid strains found in a significant proportion of cystic fibrosis patients. The development of DNA-based typing methods has circumvented this difficulty, as well as the problem of the limited discriminatory power of serotyping. To be useful, a molecular surveillance tool should be highly discriminatory and reproducible. It should also be easy to standardize. Furthermore, the resulting data should be able to be easily stored, retrieved, and compared by use of databases that can be shared between laboratories. Finally, the overall cost (including that of labor) of the methodology should be as low as possible (23). The methods available at this time for the genotyping of P. aeruginosa include pulsed-field gel electrophoresis (PFGE), arbitrarily primed PCR, and ribotyping (1, 10, 19). The first two methods suffer from a lack of interlaboratory reproducibility (5, 9); and furthermore, the present “gold standard” with the greatest discriminatory power, PFGE, is generally too costly and labor-intensive for routine clinical strain typing. On the other hand, automated ribotyping with the RiboPrinter (Qualicon, Wilmington, Del.) showed very high interlaboratory reproducibility (3) but suffered from a lack of discriminatory power when clinical P. aeruginosa strains were investigated (4).
Genetic markers called minisatellites or variable number of tandem repeats (VNTRs) were initially developed as the basis for fingerprinting the DNA of humans (11). More recently, minisatellites have been shown to exist in bacterial genomes as well, and the availability of whole-genome sequence data has opened the way to the systematic evaluation of tandem repeat polymorphisms (13, 24). When applicable, this method has been shown to fulfill most of the criteria required for an “ideal” typing system, including ease of use, speed, high discriminatory power, and reproducibility. The number of repeat units at each locus is usually estimated by measuring the sizes of the PCR products amplified with locus-specific primers flanking the repeat region. One kind of VNTR typing assay, often called multiple-locus VNTR analysis (MLVA), is based on a set of polymorphic tandem repeat loci. Approximately 20 loci are used for MLVA with Bacillus anthracis (13), Yersinia pestis (13), or the Mycobacterium tuberculosis complex (12).
In order to develop an MLVA scheme, one needs to identify polymorphic minisatellite loci, which must then be individually checked for variations of the repeat number among strains. The genome of P. aeruginosa strain PAO1 (20) is relatively rich in tandem repeats (13). We report on the identification of seven polymorphic loci and validation of their applicability in the MLVA scheme. Evaluation of these seven polymorphic loci allows a high degree of discrimination among strains with a high degree of reproducibility and easy scoring.
MATERIALS AND METHODS
Tandem repeat locus identification.
The sequence data (20) for P. aeruginosa PAO1 were obtained from the World Wide Web (http://www.genome.pseudomonas.com) and were processed by using the Tandem Repeats Finder (TRF) software (http://c3.biomath.mssm.edu/trf.html). The output was then imported into a database accessible via the Internet (http://minisatellites.u-psud.fr), as described previously (13, 24). Tandem repeat loci are designated by using the nomenclature described previously (12); for instance, ms173-5186_243bp_14U is the tandem repeat locus named ms173, which appears at position 5186 kb in the PAO1 genome with a 243-bp repeat unit and 14 units in PAO1 (Table 1).
TABLE 1.
Characteristics of tandem repeat loci
| Locus namea | Associated open reading frame | Motif length (bp) | No. of units in PAO1 | % G+C content | % Conservation | Primer sequenceb | Expected PCR product length in PAO1 (bp) | Estimated size range (bp) | No. of alleles | Polymorphism index |
|---|---|---|---|---|---|---|---|---|---|---|
| ms010-0098_6bpc | PA0081 | 6 | 11 | 65 | 100 | L: GCAGGAACGCTTGCAGCAGGT | 167 | 143-233 | 16 | 0.91 |
| R: CTTCGCCGACCCAGGGATCA | ||||||||||
| ms061-1844_6bpa | pscP | 6 | 12 | 65 | 98 | L: CTTGCCGTGCTACCGATCC | 127 | 85-139 | 10 | 0.87 |
| R: CCCCCATGCCAGTTGC | ||||||||||
| ms077-2263_39bpc | pcoA | 39 | 5 | 57 | 93 | L: GCGTCATGGTCTGCATGTC | 442 | 349-520 | 7 | 0.61 |
| R: TATACCCTCTTCGCCCAGTC | ||||||||||
| ms127-3496_15bp | PA3115 | 15 | 8 | 72 | 52 | L: CTCGGAGTCTCTGCCAACTC | 210 | 210-225 | 2 | 0.45 |
| R: GGCAGGACAGGATCTCGAC | ||||||||||
| ms142-3873_115bp | PA3463 | 115 | 7 | 66 | 94 | L: AGCAGTGCCAGTTGATGTTG | 890 | 200-775 | 7 | 0.68 |
| R: GTGGGGCGAAGGAGTGAG | ||||||||||
| ms172-5083_54bp | PA4541 | 54 | 12 | 64 | 72 | L: GGATTCTCTCGCACGAGGT | 789 | 573-843 | 8 | 0.75 |
| R: TACGTGACCTGACGTTGGTG | ||||||||||
| ms173-5186_243bp | PA4625 | 243 | 14 | 61 | 81 | L: CTGCAGTTCGCGCAAGTC | 3,503 | 1,073-4,718 | 10 | 0.82 |
| R: ATTTCAGCCAGCGTTACCAA | ||||||||||
| ms194-5915_12bpc | algP | 12 | 45 | 70 | 64 | L: CCTTAGGAGGCGCTGGTC | 690 | 600-700 | 8 | 0.78 |
| R: AGCTGCTGGCAAGGCTCT |
Loci are listed according to their positions in the PAO1 genome. The proposed reference name includes the size of the repeat unit.
L, left; R, right.
The observed length variations do not fit with the repeat unit proposed in the minisatellite database but, rather, suggest a smaller (ms010, ms061, ms194) or a larger (ms077) unit for tandem repeat variation. This is due to the emergence of a new tandem repeat unit within a larger tandem repeat. The new unit sequence is derived from the preexisting repeat but has a different length. This was checked in particular for ms077 by sequencing the different alleles (data not shown; the data are available on request).
Isolates and DNA preparation.
A total of 102 isolates were included in the present investigation as representatives of (i) all 53 ribogroups previously identified (one isolate per ribogroup) (4) and (ii) the geographic distribution of the two most frequent ribogroups found (87-S-3 [30 additional isolates] and 88-S-2 [19 additional isolates]) (4). The 203 isolates previously studied were clinical isolates collected from 20 European hospitals between 1997 and 1999 and had been analyzed by automated ribotyping (4). Fifty-three ribogroups (a ribogroup being defined as a set of strains showing the same ribotype; i.e., not a single band difference was detected among their profiles obtained by automated ribotyping with PvuII) were initially identified among 203 isolates. Four ribogroups comprised approximately half of the isolates, with the two most frequent ones being ribotype 87-S-3, with 43 isolates, and ribotype 88-S-2, with 28 isolates.
The polymorphisms of candidate VNTR loci were initially evaluated by using a subset of 12 isolates from different ribogroups (see Fig. 2).
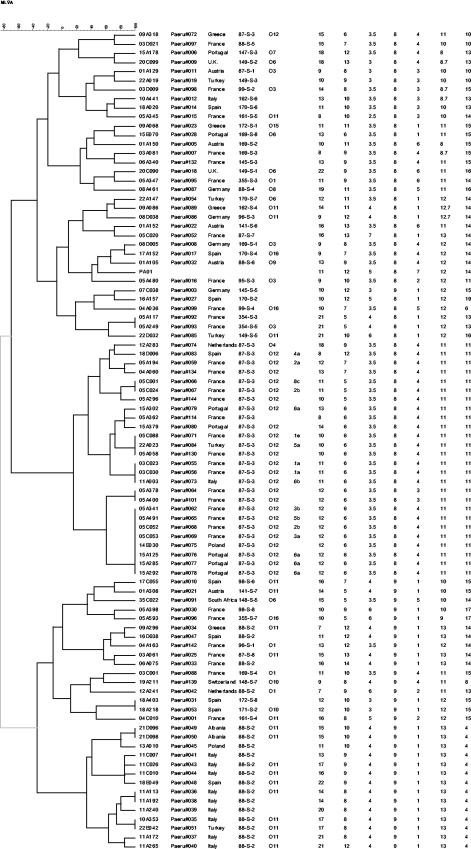
FIG. 2.
Dendrogram deduced from the clustering analysis of the 90 strains (including strain PAO1). The first column on the left identifies the isolates (the screening set comprised strains 03D021, 15A178, 03D009, 09A068, 08A461, 08D005, 01A105, 04A036, 05A400, 35C022, 03C001, 19A211). The second column indicates the DNA batch, and the third column indicates the country of origin of each isolate. The fourth column indicates the ribotype, as reported previously (4). The fifth column contains the serogroup for isolates which could be typed by serotyping. The sixth column indicates the PFGE type and subtype of isolates tested by PFGE, while the last seven columns indicate the MLVA types (repeat copy number) at loci ms010, ms061, ms077, ms127, ms142, ms172, and ms173, respectively. The number of units is deduced from the sizes of the PCR products and was formally checked by sequencing only for the ms077 locus (data not shown).
Isolates were grown at 37°C in Luria-Bertani broth. One milliliter of an overnight culture was centrifuged at 5,000 × g for 10 min. A QIAamp DNA mini kit (Qiagen, Hilden, Germany) was used for DNA extraction, as recommended by the manufacturer, with an extended lysis step (5 h at 55°C).
Minisatellite PCR amplification and genotyping.
The PCR mixtures (15 μl) contained 1 ng of DNA, 1× Taq Reaction Buffer (Qbiogen, Illkirch, France), 1 U of Taq DNA polymerase (Qbiogen), 200 μM each deoxynucleoside triphosphate, and 0.3 μM each flanking primer (for primer sequences, see Table 1). PCRs were performed in an MJ Research PTC200 thermocycler. Initial denaturation at 96°C for 5 min was followed by 30 cycles of denaturation at 96°C for 20 s, annealing at 60°C for 30 s, and elongation at 65°C for 90 s. The final extension step was 5 min at 65°C. Different annealing temperatures and/or elongation times were used for ms173-5186_243bp and ms194-5915_12bp (for ms173, the annealing temperature was 64°C and the extension time was 5 min at 70°C; for ms194, the annealing temperature was 65°C and the extension time was 1 min at 70°C). Five microliters of each of the PCR products was run on standard 1% (ms173), 2% (ms142, ms172, ms194) or 3% (ms010, ms061, ms077, ms127) agarose gels (Qbiogen or ICN, Aurora, Ohio) in 0.5× TBE (Tris-borate-EDTA) buffer at a voltage of 10 V/cm. Gel runs (bromophenol blue position) of 20 cm (ms127, ms142), 30 cm (ms010, ms061, ms077, ms172), or 40 cm (ms173) were used according to the PCR product size and motif length. Gels were stained with ethidium bromide, visualized under UV light, and photographed (Vilber Lourmat, Marne la Vallée, France). The size markers used were a 100- or 20-bp ladder (Bio-Rad, Hercules, Calif.) or a 1-kb ladder plus (Gibco-BRL, Cergy Pontoise, France). Gel images were analyzed with the Bionumerics software package (version 3.0; Applied Maths, Sint-Martens-Latem, Belgium).
PFGE typing and analysis.
DNA for PFGE was prepared as described previously (21) and digested with SpeI (New England Biolabs, Beverly, Mass.). Electrophoresis in a CHEF DRIII apparatus (Bio-Rad, Milano, Italy) was at 6 V/cm and 14°C for 21 h, with switching times linearly ramped from 5 to 23 s. The gels were stained with ethidium bromide and visualized under UV illumination with the E.A.S.Y. Win32 system (Herolab, Wiesloch, Germany).
Data analysis.
Band size estimates were exported from the Bionumerics software and converted to numbers of units. The resulting data were imported back into Bionumerics software for use for clustering analysis with the categorical coefficient and Ward clustering parameter. Use of the categorical coefficient implies that the character states are considered unordered. The same weight is given to a large or a small number of differences in the number of repeats at any locus. The website used for identification (http://bacterial-genotyping.igmors.u-psud.fr/) was developed by using the BNserver application (version 3.0; Applied Maths).
RESULTS
Screening for informative and reliable VNTR loci in the P. aeruginosa genome.
The TRF software (2) identified more than 3,000 tandem repeats within the P. aeruginosa PAO1 genome. Because no general rule has been described to predict tandem repeat polymorphisms directly from the sequence of a single allele (7, 13), present approaches rely upon systematic testing, especially for species in which only one genome has been sequenced (12). Since distinct alleles of VNTR loci with longer repeat units are generally easier to score on agarose gels, we decided to evaluate loci with repeat units more than 9 bp long. A total of 201 such tandem repeats with at least seven units each in the PAO1 sequence were identified with TRF software and were evaluated. Of these, 23 gave weak amplification signals with the set of primers used and were not considered further. One hundred seventy loci within the screening strain collection were monomorphic and were considered of very limited value for P. aeruginosa strain typing. Finally, only eight loci were polymorphic, defined here as showing at least two alleles among the screening strain collection (Table 1). Figure 1 shows examples of the PCR products obtained for the different loci. The number of alleles per locus ranged from 2 to 16. The two most polymorphic loci, at positions 0098 kb (ms010) and 1844 kb (ms061), varied by multiples of 6-bp units instead of the 12-bp repeat unit initially suggested by the database. The locus at position 5915 (ms194) varied by multiples of 12-bp units. Although the alleles at this locus were highly polymorphic, they could not be easily and reproducibly scored with the technology used here (ordinary agarose gel electrophoresis), given the range of allele sizes (600 to 700 bp), and this locus was therefore not characterized further.
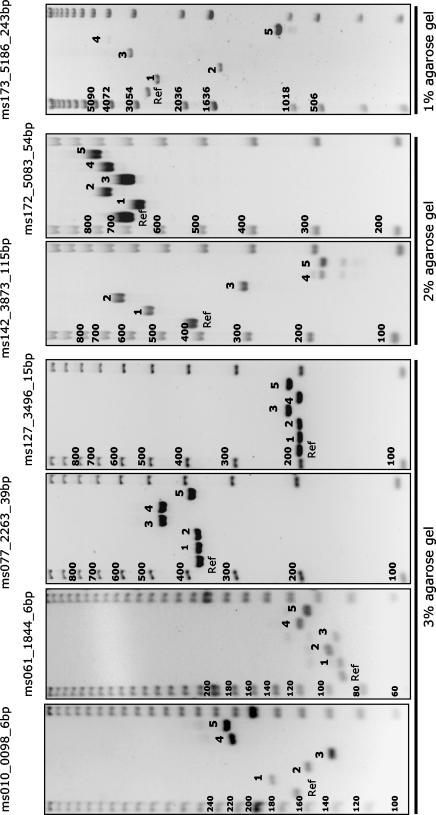
FIG. 1.
MLVA setup on agarose gels. The usual setup for the running of MLVA on agarose gels is shown. Six DNA samples (including one reference DNA control lane on the left [with isolate 05A400; see Fig. 2]) are flanked by size markers (a 20-bp, 100-bp, or 1-kb ladder, according to the locus being typed). The experiment whose results are shown was part of the reproducibility test. Five blind-coded samples are numbered from 1 to 5 (strains 03D021, 04A036, 11C010, 22D032, and 18E049, respectively). The sizes (in base pairs) (indicated on the left of each gel) can be deduced by visual inspection of the patterns observed by taking into account the MLVA type for the reference strain (Ref) used here (isolate 05A400, genotype 12-6-3.5-8-3-11-11).
MLVA of clinical P. aeruginosa isolates.
MLVA analysis with the seven loci selected was then performed with the extended representative collection of 102 strains (Fig. 1 illustrates the setup of the MLVA assay for the seven loci). One or two loci of 13 strains (12%) could not be amplified (total number of missing PCR products, 17; data not shown). The PCR products could therefore be scored for all seven loci in 89 strains. The quality of the data produced was evaluated by retyping 10 coded samples. The correct MLVA genotype (Fig. 2) could be assigned to each coded sample. Figure 2 shows the results of clustering analysis for the 89 strains. PAO1 was included, based on the genome sequence data available and the estimated repeat numbers indicated in Table 1. MLVA discriminated 72 genotypes. This was higher than the number of ribotypes (46 in this set of strains) and serotypes (12 among the 68 typeable strains). Isolates with different ribotypes were all distinguished by MLVA, with only two exceptions: isolate 18A218 (ribotype 171-S-2) and isolate 18A403 (ribotype 172-S-8) from Spain (the ribotype patterns were very similar) (4). All isolates with identical ribotypes were clustered together, with only two exceptions: isolate 09A318 (ribotype 87-S-3) and isolate 12A241 (88-S-2). However, although strain 09A318 clustered in a distinct branch, at five of seven loci its MLVA genotype showed alleles that are generally encountered in the other strains of ribotype 87-S-3. On the contrary, strain 12A241 also had a discrepant serotype (serotype O1) compared to those of all other strains of ribotype 88-S-2 (serotype O11), so one can suspect a strain mix-up in this case. Ribotypes 87-S-3 (27 isolates) and 88-S-2 (18 isolates) were subdivided into 14 and 15 MLVA types, respectively. The subtyping results were in good agreement with the geographic origins of the strains, as isolates with the same MLVA genotype most often originated from the same center (indicated by the two first numbers of the isolate code in Fig. 2).
In order to compare the discriminatory power of MLVA with that of the present gold standard, 17 ribotype 87-S-3 isolates were also typed by PFGE. PFGE resulted in seven distinct types, which were further distinguished into 12 subtypes (indicated in Fig. 2 by Arabic numbers and lowercase letters), according to published criteria (22); among the isolates in this set, seven types were also distinguished by MLVA.
The results of a comparison of the MLVA genotypes with the serotyping data were in agreement with the single origin of serotype O12 postulated earlier (18) as well as with the greater genotypic diversity of serotype O11 isolates (21).
The MLVA genotypes of the strains studied here were loaded onto our web server. They can be accessed and compared with the MLVA genotypes of new, unknown strains. Identification queries can be run from the strain identification page (http://bacterial-genotyping.igmors.u-psud.fr/), as described elsewhere (12).
DISCUSSION
Although the P. aeruginosa genome is relatively rich in tandem repeats, only 8 of the 201 tandem repeats tested by the protocol described here proved to be polymorphic. This is in contrast to the polymorphism found in other bacterial species, including some with low levels of genetic polymorphism at housekeeping loci, such as B. anthracis, Y. pestis, and M. tuberculosis (12, 13). It is very unlikely that the low levels of diversity of VNTR loci found in the present sample are due to the screening strain collection chosen, since these 12 strains could all be distinguished by ribotyping and had originated from seven different countries. Our results show that minisatellite loci are more stable in P. aeruginosa than in other species with lower overall population genetic diversity. This suggests distinct evolutionary mechanisms for tandem repeats, such as various levels of slipped strand mispairing and repair during replication (15). The present investigation evaluated all tandem repeats with a repeat unit of 9 bp or more and at least 7 units. Additional polymorphic markers could be identified by using different queries. For instance, querying of the P. aeruginosa tandem repeat database (http://minisatellites.u-psud.fr) for tandem repeats with an internal conservation of at least 90% identifies 50 such loci, none of which was investigated here. It is likely that more polymorphic loci may be obtained if loci with unit lengths smaller than 9 bp (the threshold used in our screening procedure) are chosen, since in the present study the smallest units were the most polymorphic (Table 1).
One of the eight polymorphic markers identified, ms194 (position 5915), was not used in the final analysis because of the difficulty of scoring the alleles by simple agarose gel electrophoresis. Preliminary sequencing results showed that the level of sequence diversity at this locus is very high and that alleles containing an identical number of repeat units can have distinct sequences (data not shown). This tandem repeat is located within algP, a gene implicated in the regulation of mucoidy in P. aeruginosa, and its associated polymorphism was first described by Deretic and Konyecsni (8). The use of another approach, such as polyacrylamide gel or capillary electrophoresis, which offer higher degrees of resolution, would probably solve this issue, albeit at a higher overall cost.
One (and sometimes two) of the seven loci of 12% of the strains typed failed to be amplified, despite multiple amplification attempts. This may reflect the fact either that the corresponding locus is missing or that sequence divergence results in mispriming. Further investigations will be required (including tests with new primer pairs) before this lack of amplification can be used as additional data. In the course of this preliminary investigation, the corresponding strains were not included in the final analysis (Fig. 2).
The collection of isolates tested here was originally assembled with two distinct but complementary aims: first, to be representative of European clinical P. aeruginosa isolates, and second, to include geographically diverse strains from among the two most frequent ribotypes in order to check if MLVA could subdivide these two groups of isolates. The validity of MLVA for clustering analysis and evaluation of the phylogenetic relationships among strains is not yet formally established. In a recent work, Le Flèche et al. (12) empirically selected clustering parameters to analyze strains from the M. tuberculosis complex. This was made possible by the extensive knowledge of the evolutionary relationships and the underlying epidemiology independently generated with other markers to distinguish M. tuberculosis complex strains. The same parameters have been applied in the present study. The clustering proposed here (Fig. 2) shows similarities with the clustering reported previously, which was based on ribotyping: isolates not distinguished by ribotyping are also generally clustered by MLVA (Fig. 2). This suggests that MLVA does retain some amount of phylogenetic information which can be used to trace the evolutionary histories and relationships of genotypes, as has also been found for other organisms (12). Exceptions to this general rule are easily explained by the fact that cluster analysis is based on only a few informative characters, and therefore, a difference at a single locus can alter the positioning of strains in the dendrogram. Conversely, alleles of different origins may be of identical size (homoplasy), which will also alter the clustering analysis. Allele sequencing, at least with a representative strain collection, may eventually help correct some of these inconsistencies. In any case, for more distantly related genotypes, the relationships depicted by clustering analysis may become increasingly less meaningful, especially in light of the relatively frequent occurrence of horizontal transfer among P. aeruginosa strains (6, 16, 17), which can obscure the evidence of a common ancestral lineage among strains.
Acknowledgments
Work on human bacterial pathogen identification is supported by grants from the Délégation Générale de l'Armement to L.O. and G.V. PFGE typing was performed in the framework of the Genetic Epidemiology Network for Europe (GENE) project (contract QLK2-2000-01404) of the Fifth Framework Program of the European Union.
P.T.T. is grateful to Georgia Diamantopoulou for excellent technical assistance with PFGE. S.B. thanks Jan Verhoef (Utrecht University) for continuous support.
REFERENCES
- 1.Bennekov, T., H. Colding, B. Ojeniyi, M. W. Bentzon, and N. Hoiby. 1996. Comparison of ribotyping and genome fingerprinting of Pseudomonas aeruginosa isolates from cystic fibrosis patients. J. Clin. Microbiol. 34:202-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brisse, S., V. Fussing, B. Ridwan, J. Verhoef, and R. J. Willems. 2002. Automated ribotyping of vancomycin-resistant Enterococcus faecium isolates. J. Clin. Microbiol. 40:1977-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brisse, S., D. Milatovic, A. C. Fluit, K. Kusters, A. Toelstra, J. Verhoef, and F. J. Schmitz. 2000. Molecular surveillance of European quinolone-resistant clinical isolates of Pseudomonas aeruginosa and Acinetobacter spp. using automated ribotyping. J. Clin. Microbiol. 38:3636-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dabrowski, W., U. Czekajlo-Kolodziej, D. Medrala, and S. Giedrys-Kalemba. 2003. Optimisation of AP-PCR fingerprinting discriminatory power for clinical isolates of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 218:51-57. [DOI] [PubMed] [Google Scholar]
- 6.Denamur, E., B. Picard, G. Decoux, J. B. Denis, and J. Elion. 1993. The absence of correlation between allozyme and rrn RFLP analysis indicates a high gene flow rate within human clinical Pseudomonas aeruginosa isolates. FEMS Microbiol. Lett. 110:275-280. [DOI] [PubMed] [Google Scholar]
- 7.Denoeud, F., G. Vergnaud, and G. Benson. 2003. Predicting human minisatellite polymorphism. Genome Res. 13:856-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deretic, V., and W. M. Konyecsni. 1990. A procaryotic regulatory factor with a histone H1-like carboxy-terminal domain: clonal variation of repeats within algP, a gene involved in regulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 172:5544-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foissaud, V., J. M. Puyhardy, J. C. Chapalain, H. Salord, J. J. Depina, M. Morillon, P. Nicolas, and J. D. Perrier-Gros-Claude. 1999. Inter-laboratory reproducibility of pulsed-field electrophoresis for the study of 12 types of Pseudomonas aeruginosa. Pathol Biol (Paris) 47:1053-1059. (In French.) [PubMed] [Google Scholar]
- 10.Grundmann, H., C. Schneider, D. Hartung, F. D. Daschner, and T. L. Pitt. 1995. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J. Clin. Microbiol. 33:528-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffreys, A. J., V. Wilson, and S. L. Thein. 1985. Individual-specific ′fingerprints' of human DNA. Nature 316:76-79. [DOI] [PubMed] [Google Scholar]
- 12.Le Flèche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 15.Lopes, J., H. Debrauwere, J. Buard, and A. Nicolas. 2002. Instability of the human minisatellite CEB1 in rad27Delta and dna2-1 replication-deficient yeast cells. EMBO J. 21:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picard, B., E. Denamur, A. Barakat, J. Elion, and P. Goullet. 1994. Genetic heterogeneity of Pseudomonas aeruginosa clinical isolates revealed by esterase electrophoretic polymorphism and restriction fragment length polymorphism of the ribosomal RNA gene region. J. Med. Microbiol. 40:313-322. [DOI] [PubMed] [Google Scholar]
- 17.Pirnay, J. P., D. De Vos, C. Cochez, F. Bilocq, A. Vanderkelen, M. Zizi, B. Ghysels, and P. Cornelis. 2002. Pseudomonas aeruginosa displays an epidemic population structure. Environ. Microbiol. 4:898-911. [DOI] [PubMed] [Google Scholar]
- 18.Pitt, T. L., D. M. Livermore, D. Pitcher, A. C. Vatopoulos, and N. J. Legakis. 1989. Multiresistant serotype O 12 Pseudomonas aeruginosa: evidence for a common strain in Europe. Epidemiol. Infect. 103:565-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renders, N., Y. Romling, H. Verbrugh, and A. van Belkum. 1996. Comparative typing of Pseudomonas aeruginosa by random amplification of polymorphic DNA or pulsed-field gel electrophoresis of DNA macrorestriction fragments. J. Clin. Microbiol. 34:3190-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 21.Tassios, P. T., V. Gennimata, A. N. Maniatis, C. Fock, N. J. Legakis, and the GreekPseudomonas aeruginosa Study Group. 1998. Emergence of multidrug resistance in ubiquitous and dominant Pseudomonas aeruginosa serogroup O:11. J. Clin. Microbiol. 36:897-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vergnaud, G., and F. Denoeud. 2000. Minisatellites: mutability and genome architecture. Genome Res. 10:899-907. [DOI] [PubMed] [Google Scholar]