Abstract
Among Escherichia coli strains isolated from stool specimens from patients with acute diarrhea, 1.4% were found to harbor cdtB by use of enrichment cytolethal distending toxin (CDT) PCR. These isolates were identified as being enteropathogenic E. coli (EPEC). In a retrospective study using a probe hybridization assay, 6 of 138 EPEC strains were found to harbor the cdtB locus. cdtB-positive isolates mostly belong to the O86a and O127a serogroups, with the former being associated with higher expression of CDT. Pulsed-field gel electrophoresis profiles showed that the EPEC strains harboring cdtB strains are genetically diverse.
Cytolethal distending toxin (CDT) is a novel class of bacterial genotoxin that induces characteristic elongation of eukaryotic cells followed by progressive cellular distention and death (12, 14, 23). CDT is considered to be an important factor in intestinal pathogenesis (3), as this toxin is able to induce tissue damage and fluid accumulation in the descending colon of orally infected suckling mice (21). Three genes, cdtA, cdtB, and cdtC, arranged in an apparent operon are required for the production of active CDT (25). The deduced amino acid sequences of these genes from Escherichia coli strains E6468-62 (serogroup O86) and 9142-88 (serogroup O128) are 38, 56, and 37% homologous, respectively (24, 25), and the corresponding toxins are called Cdt-I and Cdt-II (27). The amino acid sequence of Cdt-III from strain S5 (serogroup O15) has >90% homology to Cdt-II and 55 to 69% homology to Cdt-I (22). The presence of cdt in different bacterial species (8, 20, 24, 28) and the results of analysis of its flanking regions suggest that this gene has been acquired from heterologous species by horizontal gene transfer (7, 18, 22) or through a phage (13). Even though the data on the structural and functional aspects of CDT are expanding, knowledge of the epidemiological association of E. coli harboring cdt remains scanty (1, 15, 17, 19).
To investigate the incidence of cdt-harboring E. coli, a total of 284 stool specimens collected from acute-diarrhea patients of all age groups admitted to the Infectious Diseases Hospital and B. C. Roy Memorial Hospital for Children (Calcutta, India) from May to July 2002 were examined. Relevant clinical information such as presence of fever, vomiting, dehydration status, and type and duration of diarrhea was recorded for each patient. For enrichment CDT PCR, overnight stool cultures in Luria-Bertani broth (Difco, Detroit, Mich.) were directly tested for the presence of the cdtB gene in a standard PCR assay. The primer pair used in this study was based on the cdt nucleotide sequence of E. coli (25) and had the sequences 5′-GATTTTGCCGGGTATTTCT-3′ and 5′-CCCTCAACAGAGGAAGAA-3′. These primers are specific for Cdt-I. After a hot start at 94°C for 5 min, the DNA was subjected to 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min 30 s. The expected size of the PCR amplicon was 707 bp. The sensitivity of the CDT PCR assay was 103 CFU. For confirmation, a PCR amplicon from a wild-type strain was directly sequenced and matched with the sequence of E. coli cdtB (GenBank accession no. U03293) previously published by Scott and Kaper (25). A 100% homology was recorded in the sequence matching, so this strain was used as the positive control.
Except for a few preliminary reports, there is no information on the epidemiology of CDT-producing E. coli in India (4, 5). As detected by CDT PCR, the incidence level of E. coli-harboring cdtB in Calcutta was 1.4% (4 of 284 isolates) among hospitalized patients with acute diarrhea, which is comparable to the results of a Nigerian study (19). There have been reports on the incidence of CDT-producing E. coli among diarrhea patients from several developing countries (1, 9, 17, 19). The PCR-positive cultures were diluted in sterile 10 mM phosphate-buffered saline (pH 7.0), and 100 μl of each dilution was spread onto Luria agar (Difco) plates. Plates with 30 to 300 colonies were selected for the hybridization assay with PCR-amplified ctdB as the probe by using the digoxigenin DNA labeling and detection kit (Boehringer, Mannheim, Germany). The majority of the colonies (∼90%) from each PCR-positive culture hybridized with the cdtB probe. The ctdB-harboring strains were confirmed as being E. coli by an automated identification system (ID 32 GN system; Biomerieux, Marcy l'Etoile, France). None of the strains, except F17290 and F06580, fermented sorbitol.
Since the CDT PCR strains were identified as being E. coli, we checked for other virulence genes specific for diarrheagenic E. coli by using PCR (6). Interestingly, all four strains having the cdtB gene were identified as EPEC since they harbored eae and bfpA (Table 1). As the strains from the diarrhea stool specimens were positive by CDT PCR and confirmed as being EPEC strains, we further screened a total of 138 EPEC strains collected over a period of 4 years (from 1998 to 2001) with the cdtB probe by using colony hybridization assay. Five strains (2.7%) hybridized with the cdtB probe, and two of these gave negative results in the CDT PCR (Table 1).
TABLE 1.
Serotypes and virulence gene profiles of E. coli strains harboring cdtB
| Strain | Serogroup | PCR result
|
Reciprocal titer of Cdt in HeLa cell assay | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cdtB | est | stx2 | elt | eae | bfp | EAF | astA | EAgg | stx1 | |||
| GB 1371 | O86a | + | − | − | − | + | + | + | − | − | − | 128 |
| VTE 1456 | O142 | + | − | − | − | + | + | NDb | − | − | − | 128 |
| VTE 1488 | O86a | + | − | − | − | + | + | ND | − | − | − | 128 |
| GB 1807 | O127a | + | − | − | − | + | + | ND | − | − | − | 132 |
| D05491 | O86a | + | − | − | − | + | + | + | − | − | − | 128 |
| F06580 | OUTa | − | − | − | − | + | − | − | − | − | − | |
| F17290 | O157 | − | − | − | − | + | − | − | − | − | − | |
| GB 469 | O127a | + | − | − | − | + | + | + | − | − | − | 32 |
| NT 3363 | O127a | + | − | − | − | + | + | + | − | − | − | 32 |
OUT, not typeable.
ND, not done.
In the serological analysis with somatic antisera (Denka Seiken, Tokyo, Japan), 33.3% of the strains were identified as being O86a and O127a. Production of CDT seemed to be an exclusive characteristic of these serogroups (1, 5, 9, 11). All of the CDT-expressing E. coli strains belonged to classical EPEC serogroups. Studies conducted in India, Bangladesh, and Brazil have revealed the same trend (1, 4, 9). Since we have not encountered any E. coli strain that harbored only the cdtB locus, it appears that there is a preferential association of cdtB with EPEC strains.
No other enteric pathogen was detected in patients infected with cdtB-harboring E. coli. The present study showed that, except for one, all of the patients infected with E. coli-harboring cdtB were children ranging in age from 4 months to 6 years. It appears that infants are more susceptible to the E. coli harboring cdtB (1, 2). As shown in Table 2, the majority of patients (five of nine) had blood in their stool. The relevance of bloody diarrhea in association with cdtB-harboring EPEC remains to be explored. In an earlier report, hemorrhagic response was observed in a rat ligated ileal loop test with CDT-positive strains of Campylobacter spp. (14).
TABLE 2.
Clinical manifestations of patients infected with E. coli strains harboring cdtB
| Strain | Patient age | Clinical manifestation
|
|||
|---|---|---|---|---|---|
| Stool type | Dehydration | Vomiting | Fever | ||
| GB 1371 | 4 mo | Occult blood | Not severe | Yes | No |
| VTE 1456 | 7 mo | Blood mucous | Not severe | No | No |
| VTE 1488 | 2 yr | Blood mucous | Not severe | Yes | Yes |
| GB 1807 | 5 mo | Watery | Not severe | Yes | No |
| D05491 | 25 yr | Occult blood | Not severe | Yes | No |
| NT 3363 | 7 mo | Watery | Not severe | Yes | No |
| GB 469 | 7 mo | Watery | Not severe | Yes | Yes |
| F06580 | 11 mo | Watery | Severe | Yes | Yes |
| F17290 | 6 yr | Occult blood | Severe | Yes | No |
An antibiotic susceptibility test performed with the ATB-G system (Biomerieux) showed that all of the strains were resistant to amoxicillin and piperacillin and were sensitive to the majority of drugs such as tazobactam, imipenem, cefoxitin, ceftazidime-1, ceftazidime, cefepime, cefpirome, tobramycin, amikacin, gentamicin, netilmicin, and ciprofloxacin. Amoxicillin and ciprofloxacin are used for the treatment of diarrhea in India.
CDT activity on HeLa cells was determined by using a reciprocal dilution of culture filtrates from the overnight growth of strains in Trypticase soy broth (Difco). The toxin titer was expressed as the reciprocal of the highest dilution that caused 50% of the HeLa cells in a well to be distended up to 96 h of incubation. The reciprocal titer for strains belonging to the O86a and O142 serogroups was higher than that for O127a strains (Table 1). The strains that expressed CDT were found to be sorbitol nonfermenters. This trait may be considered while screening for cdt-positive E. coli strains. As shown in Table 1, all of the high-titer CDT-producing O86a and O142 serogroup strains were isolated from patients with bloody diarrhea. Strains F06580 and F17290, in which the CDT PCR did not amplify the target gene, were probe positive but did not produce CDT when examined by tissue culture assay. Even though the gene coding for the B subunit is supposed to be more conserved than the other genes, the PCR and CDT expression assay results indirectly showed that there might be some sequence variation and/or alterations in the functional domain of cdt in these two strains.
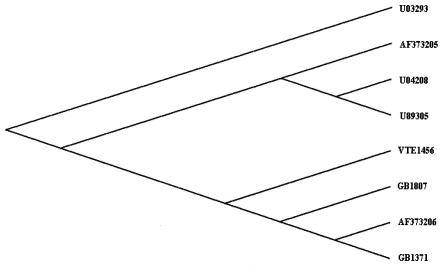
To determine the DNA sequence of the cdtB locus from three representative strains (GB 1371, GB 1807, and VTE 1456), the amplified cdtB gene was purified (Qiagen, Hilden, Germany) and sequenced in both directions on an ABI 310 automated sequencer (Applied Biosystems, Foster City, Calif.). Analysis of the sequences was performed using the ClustalW (version 1.8) multiple-sequence alignment program (27). The cdtB sequence was searched for homologous sequences by using BLAST (10), and the phylogenetic analysis was made using ClustalX software. Analysis of the cdtB sequences of these strains showed that all three sequences were identical and closely related to the sequences of U03293 (25) and AF373206 (S. Bouzari, M. Oloomi, and M. Zarepoor, unpublished data). When the phylogenetic tree was rooted through U03293, all of the Calcutta strains formed one group along with AF373206, which was reported from Iran (Fig. 1).
FIG. 1.
Phylogenetic analysis based on the comparison of cdtB gene sequences of three representative strains (GB 1371, GB 1807, and VTE 1456) with other sequences available in GenBank. The tree was constructed using ClustalX and viewed with TREEVIEW software after rooting through the U03293 cdt sequence.
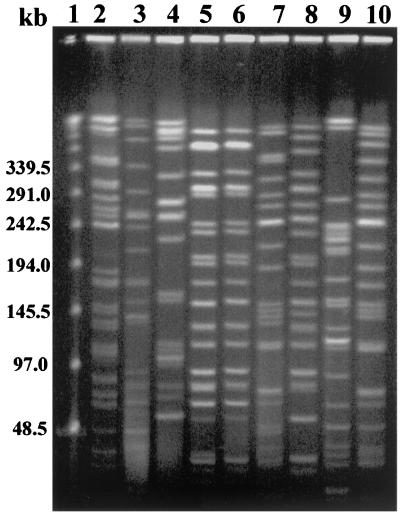
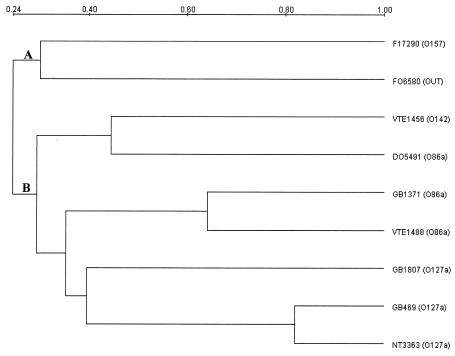
DNA fingerprinting was done using pulsed-field gel electrophoresis (PFGE) with the restriction enzyme XbaI (Takara) according to standard procedures (16) with the CHEF Mapper PFGE system (Bio-Rad, Hercules, Calif.). Except for GB 469 and NT 3363, the remaining strains showed different PFGE profiles (Fig. 2) and could not be considered as clonal according to the PFGE interpretation criteria (26). However, it has been shown by ribotyping that among E. coli strains, CDT production was associated with a clone distributed all over the world and represented by the serotype O86:H34 (9). The clonal relationship between the strains was detected with an unrooted UPGMA (unweighted pair group method with arithmetic averages) method of alignment. Two main clusters were observed: cluster A, with two non-CDT-producing strains, F17290 (serogroup O157) and F06580 (OUT), and cluster B, which combined all of the CDT-expressing strains (Fig. 3).
FIG. 2.
PFGE profiles of E. coli strains harboring ctdB after digestion with XbaI. Lanes (the respective serogroups of the strains are indicated in parenthesis): 1, bacteriophage lambda molecular size marker; 2, D05491 (O86a); 3, F06580 (OUT); 4, F17290 (O157); 5, GB 469 (O127a); 6, NT 3363 (O127a); 7, GB 1371 (O86a); 8, GB 1807 (O127a); 9, VTE 1456 (O142); 10, VTE 1488 (O86a).
FIG. 3.
Dendrogram of the E. coli strains harboring ctdB based on the PFGE profiles by using Diversity Data Base software (Bio-Rad) employing the UPGAMA method. Main lineages A and B are indicated near the nodes.
The present study is the first systematic report applying conventional and molecular approaches to the screening and characterization of E. coli strains harboring the cdtB locus associated with acute diarrhea in India. These strains should be investigated in great detail, especially in relation to bloody diarrhea and with regard to cdt sequence variation among strains that did not express CDT.
Nucleotide sequence accession numbers.
The nucleotide sequences of E. coli strains VTE 1456, GB 1807, and GB 1371 have been deposited in the GenBank database under accession numbers AY351905, AY351906, and AY351907, respectively.
Acknowledgments
This work was supported by the United States-Japan Cooperative Medical Science Program and the Japan International Cooperation Agency (JICA/NICED project 054-1061-E-0).
REFERENCES
- 1.Albert, M. J., S. M. Faruque, A. S. Faruque, K. A. Bettelheim, P. K. Neogi, N. A. Bhuiyan, and J. B. Kaper. 1996. Controlled study of cytolethal distending toxin-producing Escherichia coli infections in Bangladeshi children. J. Clin. Microbiol. 34:717-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. D., A. J. MacNab, W. R. Gransden, S. M. Damm, W. M. Johnson, and H. Lior. 1987. Gastroenteritis and encephalopathy associated with a strain of Escherichia coli O55:K59:H4 that produced a cytolethal distending toxin. Pediatr. Infect. Dis. J. 6:1135-1136. [PubMed] [Google Scholar]
- 3.Aragon, V., K. Chao, and L. A. Dreyfus. 1997. Effect of cytolethal distending toxin on F-actin assembly and cell division in Chinese hamster ovary cells. Infect. Immun. 65:3774-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouzari, S., and A. Varghese. 1990. Cytolethal distending toxin (CLDT) production by enteropathogenic Escherichia coli (EPEC). FEMS Microbiol. Lett. 59:193-198. [DOI] [PubMed] [Google Scholar]
- 5.Bouzari, S., B. R. Vatsala, and A. Varghese. 1992. In vitro adherence property of cytolethal distending toxin (CLDT) producing EPEC strains and effect of the toxin on rabbit intestine. Microb. Pathog. 12:153-157. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty, S., J. S. Deokule, P. Garg, S. K. Bhattacharya, R. K. Nandy, G. B. Nair, S. Yamasaki, Y. Takeda, and T. Ramamurthy. 2001. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J. Clin. Microbiol. 39:3241-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope, L. D., S. Lumbley, J. L. Latimer, J. Klesney-Tait, M. K. Stevens, L. S. Johnson, M. Purven, R. S. Munson, Jr., T. Lagergard, J. D. Radolf, and E. J. Hansen. 1997. A diffusible cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. USA 94:4056-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyigor, A., K. A. Dawson, B. E. Langlois, and C. L. Pickett. 1999. Cytolethal distending toxin genes in Campylobacter jejuni and Campylobacter coli isolates: detection and analysis by PCR. J. Clin. Microbiol. 37:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghilardi, A. C., T. A. Gomes, and L. R. Trabulsi. 2001. Production of cytolethal distending toxin and other virulence characteristics of Escherichia coli strains of serogroup O86. Mem. Inst. Oswaldo Cruz 96:703-708. [DOI] [PubMed] [Google Scholar]
- 10.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 11.Guth, B. E., R. Giraldi, T. A. Gomes, and L. R. Marques. 1994. Survey of cytotoxin production among Escherichia coli strains characterized as enteropathogenic (EPEC) by serotyping and presence of EPEC adherence factor (EAF sequence). Can. J. Microbiol. 40:341-344. [DOI] [PubMed] [Google Scholar]
- 12.Hassane, D. C., R. B. Lee, M. D. Mendenhall, and C. L. Pickett. 2001. Cytolethal distending toxin demonstrates genotoxic activity in a yeast model. Infect. Immun. 69:5752-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and H. Karch. 2003. Cytolethal distending toxin gene cluster in enterohemorrhagic Escherichia coli O157:H7: characterization and evolutionary considerations. Infect. Immun. 71:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, W. M., and H. Lior. 1988. A new heat-labile cytolethal distending toxin (CLDT) produced by Campylobacter spp. Microb. Pathog. 4:115-126. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, W. M., and H. Lior. 1988. A new heat-labile cytolethal distending toxin (CLDT) produced by Escherichia coli isolates from clinical material. Microb. Pathog. 4:103-113. [DOI] [PubMed] [Google Scholar]
- 16.Khan, A., S. C. Das, T. Ramamurthy, A. Sikdar, J. Khanam, S. Yamasaki, Y. Takeda, and G. B. Nair. 2002. Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J. Clin. Microbiol. 40:2009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques, L. R. M., A. T. Tavechio, C. M. Abe, and T. A. T. Gomes. 2003. Search for cytolethal distending toxin production among fecal Escherichia coli isolates from Brazilian children with diarrhea and without diarrhea. J. Clin. Microbiol. 41:2206-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer, M. P., L. C. Bueno, E. J. Hansen, and J. M. DiRienzo. 1999. Identification of cytolethal distending toxin gene locus and features of a virulence-associated region in Actinobacillus actinomycetemcomitans. Infect. Immun. 67:1227-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okeke, I. N., A. Lamikanra, H. Steinrück, and J. B. Kaper. 2000. Characterization of Escherichia coli strains from cases of childhood diarrhea in provincial southwestern Nigeria. J. Clin. Microbiol. 38:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuda, J., H. Kurazono, and Y. Takeda. 1995. Distribution of the cytolethal distending toxin A gene (cdtA) among species of Shigella and Vibrio, and cloning and sequencing of the cdt gene from Shigella dysenteriae. Microb. Pathog. 18:167-172. [DOI] [PubMed] [Google Scholar]
- 21.Okuda, J., M. Fukumoto, Y. Takeda, and M. Nishibuchi. 1997. Examination of diarrheagenicity of cytolethal distending toxin: suckling mouse response to the products of the cdtABC genes of Shigella dysenteriae. Infect. Immun. 65:428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérès, S. Y., O. Marchès, F. Daigle, J. P. Nougayrède, F. Hèrault, C. Tasca, J. De Rycke, and E. Oswald. 1997. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol. Microbiol. 24:1095-1107. [DOI] [PubMed] [Google Scholar]
- 23.Pickett, C. L., and C. A. Whitehouse. 1999. The cytolethal distending toxin family. Trends Microbiol. 7:292-297. [DOI] [PubMed] [Google Scholar]
- 24.Pickett, C. L., D. L. Cottle, E. C. Pesci, and G. Bikah. 1994. Cloning, sequencing, and expression of the Escherichia coli cytolethal distending toxin genes. Infect. Immun. 62:1046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott, D. A., and J. B. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young, V. B., K. A. Knox, and D. B. Schauer. 2000. Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus. Infect. Immun. 68:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]