Abstract
As part of a national surveillance program on invasive group A streptococci (GAS), isolates that caused specific manifestations of invasive GAS disease in The Netherlands were collected between 1992 and 1996. These site-specific GAS infections involved meningitis, arthritis, necrotizing fasciitis, and puerperal sepsis. An evaluation was performed to determine whether GAS virulence factors correlate with these different disease manifestations. PCRs were developed to detect 9 genes encoding exotoxins and 12 genes encoding fibronectin binding proteins. The genetic backgrounds of all isolates were determined by M genotyping and pulsed-field gel electrophoresis (PFGE) analysis. The predominant M types included M1, M2, M3, M4, M6, M9, M12, and M28. Most M types were associated with all manifestations of GAS disease. However, M2 was found exclusively in patients with puerperal sepsis, M6 predominated in patients with meningitis, and M12 predominated in patients with GAS arthritis. While characteristic gene profiles were detected in most M types, the resolution of detection of different gene profiles within M genotypes was enhanced by PFGE analysis, which clearly demonstrated the existence of some clonal lineages among invasive GAS isolates in The Netherlands. M1 isolates comprised a single clone carrying highly mitogenic toxin genes (speA, smeZ) and were associated with toxic shock-like syndrome. Toxin profiles were highly conserved among the most virulent strains, such as M1 and M3.
Since the mid-1980s, a conspicuous increase in the incidence of invasive infections caused by group A streptococci (GAS) has been observed worldwide (3, 9, 10, 18, 27, 37, 38, 53, 70). Invasive GAS diseases include various clinical syndromes such as bacteremia, arthritis, pneumonia, puerperal sepsis, meningitis, and necrotizing fasciitis. These infections are often of a fulminant nature and can be complicated by streptococcal toxic shock-like syndrome (TSS). This clinical entity is particularly associated with necrotizing fasciitis but can occur as a complication of any GAS infection (14, 45, 53, 67). The cause of the resurgence of severe GAS infections remains enigmatic (14, 49). It has been shown that the immunogenetic background of the host influences the outcome of invasive streptococcal infection (4, 39, 48). In addition, predisposing factors such as trauma, diabetes, alcoholism, or varicella have been documented. Nonetheless, otherwise healthy subjects can be affected as well (15, 38, 41, 62). This observation suggests a relevant role for changes in bacterial virulence, which was the focus of this study. The pathogenicity of GAS disease has been associated with the expression of a plethora of gene products that comprise extracellular and cell-associated proteins.
Extracellular proteins that reportedly play a role in the pathogenesis of GAS are represented by streptococcal exotoxins. They include SPE-A, SPE-B, SPE-C, SPE-F, SPE-G, SPE-H, SPE-J, SSA, and SMEZ, most of which have superantigenic properties that allow them to activate large subsets of T cells, with massive cytokine release as a consequence (1, 23, 33, 43, 47, 50, 56, 57). They are associated with pyrogenicity and enhancement of endotoxic shock. Recently, more streptococcal exotoxins have been described (6), and SMEZ has been identified as the major immunologically active agent (76).
Bacterial attachment to host tissue is the first step leading to colonization and subsequent development of invasive disease. Binding of fibronectin promotes adherence to epithelial cells. Several studies have suggested an association between fibronectin binding and the pathogenic potential of GAS (22, 25, 42, 46, 61, 74, 77). Invasion of streptococci into deeper tissues may be facilitated by specialized adhesion mechanisms. In addition, a particular adhesion factor might confer a certain site or tissue specificity to the GAS (54). Although many adhesins have been identified, an association of these with clinically defined syndromes has not been investigated.
The aim of the present study was to further investigate possible relationships between genes encoding M proteins, exotoxins, and protein binding factors and the clinical manifestations of invasive Streptococcus pyogenes disease. Between 1992 and 1996, all Dutch patients with microbiologically documented invasive GAS disease were registered in a national surveillance study. We selected all GAS isolates from Dutch patients with necrotizing fasciitis, puerperal sepsis, septic arthritis, and meningitis; determined their M genotypes; and evaluated them for the presence of 9 exotoxin and 12 fibronectin binding protein genes. This is the first report on an extensive gene analysis of all clinically well-documented GAS isolates obtained in one country over a limited time span.
MATERIALS AND METHODS
Surveillance.
From 1992 to 1996, Dutch patients with microbiologically defined invasive GAS disease were registered in a nationwide laboratory-based surveillance system (62, 63). Isolates were considered pathogenic and invasive when they were obtained from normally sterile body sites. Upon receipt of an invasive strain, extensive clinical and demographic data for the patients involved were obtained by mailing a standardized questionnaire to the treating physician; the response rate was 57%. For this study, we selected all invasive GAS isolates that had given rise to meningitis, septic arthritis, necrotizing fasciitis, or puerperal sepsis in patients for whom corresponding clinical and demographic data were known. GAS meningitis was defined on the basis of GAS-positive cultures of either cerebrospinal fluid or blood, together with clinically defined meningitis. GAS arthritis was defined on the basis of GAS-positive cultures of either synovial fluid or blood, combined with clinically defined arthritis. Puerperal sepsis was defined as postpartum endometritis in combination with positive blood cultures. The occurrence of clinical complications was registered as TSS and was defined as the presence of hypotension in combination with at least two of the following: acute renal failure, coagulation or liver abnormalities, rash, or necrotizing fasciitis (2).
Typing.
All GAS isolates were subjected to M genotyping. The M genotype was determined by hybridization of the denatured emm amplicon with a panel of 26 emm type-specific probes in a reverse line blotting system, as described previously (36).
Bacterial strains and culture conditions.
Strains were grown on blood agar plates at 5% CO2 and 37°C overnight. Genomic DNA as the target for PCR assays was extracted by heating bacterial suspensions for 10 min at 95°C (64).
Identification of toxin and fibronecting binding protein genes.
Sequences specific for speA (32), speB (34), speC (24), speF (28), speG (56), speH (56), speJ (56), smeZ (21), ssa (59), cpa (55), cpa-1 (T. Miyoshi-Akiyama, N. Wakisaka, J. Zhao, and T. Uchiyama, unpublished data), fba (75), fbp-54 (Miyoshi-Akiyama et al., unpublished), pfbp (61), prtf-1 (66), prtf-2 (29), prtf-15 (35), sciA (58), sciB (78), sfb (72), and sfb-II (40) were detected by PCR on a model 9600 thermocycler (Perkin-Elmer, Gouda, The Netherlands) with the primers listed in Table 1. Amplification of all genes was carried out under the following conditions: an initial 5-min denaturation step at 96°C, followed by 30 cycles of denaturation at 96°C for 55 s, 65 s of annealing at the appropriate temperature for each gene specified in Table 1, and 70 s of extension at 72°C, with a final extension step at 72°C for 5 min. All PCR products were subjected to electrophoresis on 1.5% agarose gels and then stained with ethidium bromide and visualized under UV light. In another study (64), this PCR protocol had a sensitivity of 96%. To minimize the risk of possible sequence variation affecting the primer binding site, for each gene we aligned all published sequences and selected a conserved region for primer design. Furthermore, partial sequencing of the first amplification products yielded no nonspecific sequences for any of the 21 genes under investigation, indicating the complete specificities of the primers used.
TABLE 1.
Oligonucleotide primers and reference strains used for gene detection
| Gene | Reference strain | Annealing temp (°C) | Amplicon size (bp) | Primer sequence (5′-3′) |
|---|---|---|---|---|
| speA | 05A338 | 44 | 248 | TAA GAA CCA AGA GAT GG |
| ATT CTT GAG CAG TTA CC | ||||
| speB | 01A188 | 42 | 955 | AAG AAG CAA AAG ATA GC |
| TGG TAG AAG TTA CGT CC | ||||
| speC | 01A188 | 42 | 584 | GAT TTC TAC TTA TTT CAC C |
| AAA TAT CTG ATC TAG TCC C | ||||
| speF | 01A188 | 42 | 782 | TAC TTG GAT CAA GAC G |
| GTA ATT AAT GGT GTA GCC | ||||
| speG | 01A188 | 42 | 155 | AGA AAC TTA TTT GCC C |
| TAG TAG CAA GGA AAA GG | ||||
| speH | 01A091 | 42 | 416 | AGA TTG GAT ATC ACA GG |
| CTA TTC TCT CGT TAT TGG | ||||
| speJ | 01A188 | 44 | 535 | ATC TTT CAT GGG TAC G |
| TTT CAT GTT TAT TGC C | ||||
| smeZ | 01A335 | 44 | 391 | TAA CTC CTG AAA AGA GGC T |
| CAT TGG TTC TTC TTG ATA AG | ||||
| ssa | 06A047 | 44 | 706 | AAG AAT ACT CGT TGT AGC |
| CTC ACT GTC TTA TTA TCG | ||||
| cpa | A95/443 | 48 | 1,400 | CTC AAA ATG CTA TTT GGT AT |
| ATT TCC CAT CTT TAG CTA CT | ||||
| cpa-1 | A93/195 | 48 | 1,000 | TGT GAA CTT CCA TTT TTA TT |
| AGA GTA GCA CAC GAT TTA AG | ||||
| fba | A92/039 | 46 | 587 | GGT GAT TCA ACA TCA GTT AC |
| CGT TTT GTG ACT AAA AGA CT | ||||
| fbp-54 | 05A338 | 48 | 795 | CTT CAG AAT CTG TTT CTT TG |
| AGT TCA CAG GTT GTC TAT TG | ||||
| pfbp | A94/131 | 48 | 597 | CTG AAT ATG CTG CTT TTA CT |
| TTA TCC TTC GTT ACT TCT TG | ||||
| prtf-1 | A93/121 | 46 | 806 | CCT TTG TAG ATT ATG CTC AC |
| TTC TGT CTC AAC CAT ATT TC | ||||
| prtf-2 | A95/443 | 47 | 767 | AAA GCA ATT ATA TTA CTA ATG |
| TTT TGT TTC ATA CAG GTC | ||||
| prtf-15 | A93/190 | 48 | 399 | TGG GAG TAC AGA AAC TTT TA |
| ATC AGG TAC ATA TTC AGC AC | ||||
| sciA | 01A188 | 47 | 267 | TGA CAT CAA AGG AGA GAC AA |
| CAC GAG CAC CAG CTT TAC | ||||
| sciB | 05A633 | 47 | 592 | TGA CAA ACA AAC AAA CTC ACT |
| ATA AAC TGC AAA ATC CCA AA | ||||
| sfb | A95/721 | 46 | 1,001 | CAT ATC AGG CTT ATT GTT TT |
| TTC TGT CTC AAC CAT ATT TC | ||||
| sfb-2 | 05A633 | 42 | 399 | ATG ACA AAA GAG AAT TTT GA |
| TGT GAT ATT TTC ATT TAC CC |
Typing by PFGE.
For pulsed-field gel electrophoresis (PFGE), the bacteria were suspended to a density of a McFarland no. 3 standard, and bacterial plugs were made in 0.75% pulsed-field-certified agarose (Bio-Rad, Hercules, Calif.) with Tris-HCl and NaCl (pH 8.0) at final concentrations of 10 mM and 1 M, respectively. After solidification, the plugs were treated with lysis buffer (6 mM Tris-HCl, 1 M NaCl, 100 mM EDTA [pH 8.0], 0.2% sodium desoxycholate, 0.5% sodium laurylsarcosine, 0.5% Brij 58) containing freshly added lysozyme (2.88 g/liter; Sigma, St. Louis, Mo.) for 48 h at 37°C. After 48 h of incubation at 56°C in 0.5 M EDTA (pH 9.0)-1.0 g of proteinase K per liter, the plugs were washed in TE (Tris-EDTA) buffer and stored at 4°C until use. Digestion of the bacterial DNA was performed with restriction endonuclease SmaI (Roche, Almere, The Netherlands) according to the guidelines of the manufacturer. Restriction fragments were then separated by PFGE in a 1% pulsed-field-certified agarose gel (Bio-Rad) with 1 ml of thiourea per liter with a CHEF-DRII drive module (Bio-Rad). The initial pulse time of 1.0 s was increased linearly to 50.0 s over 24 h at a voltage of 6 V/cm and a temperature of 11.3°C. The gels were then stained with ethidium bromide for 15 min, and the restriction fragments were visualized under UV light. The images of the gels were imported into Gelcompar software (version 4.1; Applied Maths, Kortrijk, Belgium) by using band-based clustering, and the banding patterns were evaluated for each individual M type. By use of the dendrogram, isolates with a genetic relatedness of >80% were considered to represent the same PFGE type. Confirmation of genetic similarity or difference was performed by visual interpretation of the gels.
Statistical analysis.
Chi-square tests were performed to compare data from patients with different site-specific infections. P values <0.05 were considered significant. To show more complex relationships between some virulence genes and site-specific disease independent of the M genotype, logistic regression was performed by using SPSS (version 10) software.
RESULTS
Surveillance.
In our nationwide surveillance system, a total of 170 isolates were obtained between 1992 and 1996 from patients with microbiologically defined cases of invasive GAS disease causing meningitis (27 of 170), arthritis (41 of 170), necrotizing fasciitis (61 of 170), and puerperal sepsis (41 of 170). Twenty-eight percent (47 of 170) of these cases of invasive GAS disease were complicated by the occurrence of TSS. The isolates were derived from 170 different patients and represented 26% (170 of 650) of a collection of all isolates from patients infected with invasive GAS isolates that were sent to the national public health laboratory during the time period.
Typing.
In order to establish whether there is a relationship between genetic profile and disease type, all isolates were subjected to M genotyping and PFGE analysis. Table 2 shows the results of M genotyping of the isolates involved in the four different site- or tissue-specific infections. A total of 20 different M types were detected; however, 30 isolates could not be typed because of a lack of hybridization with any of the oligonucleotides used. Predominant M types were defined as those that contained five or more isolates that could be typed. These predominant M types were M1 (25% of isolates), M2 (3%), M3 (14%), M4 (6%), M6 (6%), M9 (4%), M12 (6%), and M28 (7%). They comprised a total of 122 (72%) of all M types involved and were nonuniformly distributed among the patients with different site- or tissue-specific infections. Remarkably, M1 predominated in TSS cases but was not as prevalent in non-TSS cases (21 of 47 and 22 of 123, respectively [P < 0.001]). TSS was seen as a complication in all groups with invasive GAS diseases but significantly less often in patients with puerperal sepsis (3 of 41 versus 44 of 129 [P < 0.001]). M3 also tended to be overrepresented among TSS cases, but this was not supported by statistical significance.
TABLE 2.
Association of M genotypes with GAS disease and TSS
| Disease or TSS occurrence | No. of cases | No. (%) of TSS cases | No. (%) of the following M types:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M6 | M9 | M12 | M28 | Othera | |||
| Meningitis | 27 | 6 (22) | 8 (30) | 0 (0) | 4 (15) | 2 (7) | 5b (19) | 1 (4) | 1 (4) | 0 (0) | 6 (22) |
| Arthritis | 41 | 8 (20) | 7 (17) | 0 (0) | 6 (15) | 3 (7) | 2 (5) | 1 (2) | 6c (15) | 4 (10) | 12 (29) |
| Necrotizing fasciitis | 61 | 30 (49) | 19 (31) | 0 (0) | 12 (20) | 3 (5) | 1 (2) | 4 (7) | 3 (5) | 4 (7) | 15 (25) |
| Puerperal sepsis | 41 | 3d (7) | 9 (22) | 5d (12) | 1c (2) | 3 (7) | 2 (5) | 0 (0) | 1 (2) | 5 (12) | 15 (35) |
| Non-TSS | 123 | 22 (13) | 4 (2) | 14 (8) | 9 (5) | 10 (6) | 5 (4) | 9 (5) | 12 (7) | 38 (22) | |
| TSS | 47 | 21d (45) | 1 (2) | 9 (19) | 2 (4) | 0 (0) | 1 (2) | 2 (4) | 1 (2) | 10 (21) | |
Single M types representing less then 4% of all isolates (except M nontypeable).
P < 0.005.
P < 0.025.
P < 0.001.
PFGE and identification of toxin and fibronectin binding protein genes.
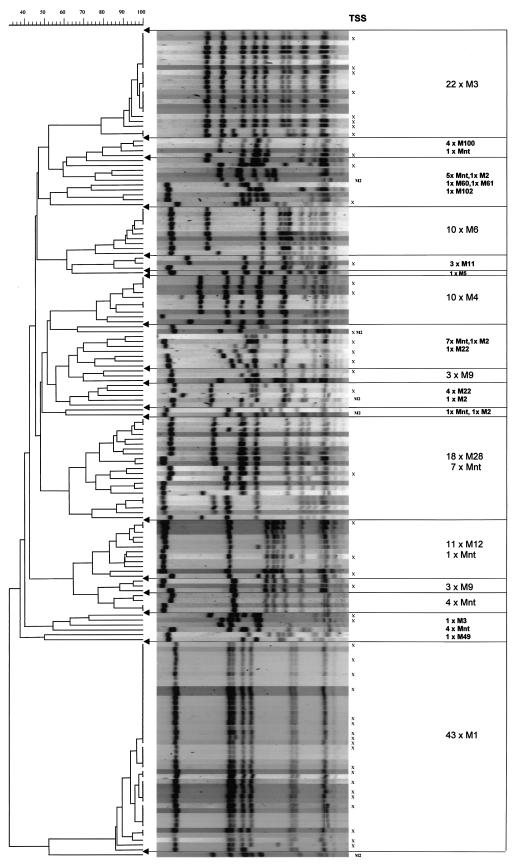
Figure 1 shows the PFGE patterns and their relation to the M types. Among the eight predominant M types (i.e., M1, M2, M3, M4, M6, M9, M12, and M28), we found 23 different patterns with differences of more than three bands (Fig. 1 and Table 3). Clonal lineages were demonstrated among M1, M3, M6, and M12 isolates, whereas M2 isolates were genetically very different.
FIG. 1.
PFGE patterns of SmaI-digested chromosomal DNA and their association with M type and TSS. The numbers before the multiplication signs indicate the numbers of isolates of the indicated M type. nt, nontypeable.
TABLE 3.
Distribution of virulence genes among predominant M and PFGE types
| M type | Pulsotype | Presence of toxin genes
|
Presence of fibronectin binding protein genes
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| speA | speB | speC | speF | speG | speH | speJ | smeZ | ssa | cpa | cpa-1 | fba | fbp-54 | pfbp | prtf-1 | prtf-2 | prtf-15 | sciA | sciB | sfb | sfb-2 | ||
| M1 | 1A (n = 43) | + | + | + | + | + | + | + | + | + | + | + | ||||||||||
| M2 | 2A (n = 1) | + | + | + | + | + | + | + | + | + | + | |||||||||||
| 2B (n = 1) | + | + | + | + | + | + | + | |||||||||||||||
| 2C (n = 1) | + | + | + | + | + | + | + | |||||||||||||||
| 2D (n = 1) | + | + | + | + | + | + | + | |||||||||||||||
| 2E (n = 1) | + | + | + | + | + | + | + | + | ||||||||||||||
| M3 | 3A (n = 20) | + | + | + | + | + | + | + | ||||||||||||||
| 3A (n = 1) | + | + | + | + | + | + | ||||||||||||||||
| 3A (n = 1) | + | + | + | + | + | |||||||||||||||||
| 3B (n = 1) | + | + | + | + | + | |||||||||||||||||
| M4 | 4A (n = 4) | + | + | + | + | + | + | + | ||||||||||||||
| 4A (n = 2) | + | + | + | + | + | + | + | + | ||||||||||||||
| 4B (n = 2) | + | + | + | + | + | + | + | + | ||||||||||||||
| 4B (n = 1) | + | + | + | + | + | + | + | |||||||||||||||
| 4C (n = 2) | + | + | + | + | + | + | + | |||||||||||||||
| M6 | 6A (n = 6) | + | + | + | + | + | + | + | ||||||||||||||
| 6A (n = 2) | + | + | + | + | + | + | + | + | ||||||||||||||
| 6A (n = 2) | + | + | + | + | + | + | ||||||||||||||||
| 6A (n = 1) | + | + | + | + | + | + | + | + | ||||||||||||||
| M9 | 9A (n = 3) | + | + | + | + | + | + | + | ||||||||||||||
| 9B (n = 1) | + | + | + | + | + | + | ||||||||||||||||
| 9C (n = 1) | + | + | + | + | + | |||||||||||||||||
| 9D (n = 1) | + | + | + | + | + | + | + | + | + | |||||||||||||
| M12 | 12A (n = 4) | + | + | + | + | + | + | + | + | |||||||||||||
| 12A (n = 3) | + | + | + | + | + | + | + | + | + | |||||||||||||
| 12A (n = 2) | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| 12B (n = 1) | + | + | + | + | + | + | + | + | + | |||||||||||||
| 12B (n = 1) | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| M28 | 28A (n = 5) | + | + | + | + | + | + | + | + | + | + | |||||||||||
| 28A (n = 1) | + | + | + | + | + | + | + | + | ||||||||||||||
| 28B (n = 1) | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| 28B (n = 1) | + | + | + | + | + | + | + | + | ||||||||||||||
| 28C (n = 2) | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| 28D (n = 2) | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| 28E (n = 1) | + | + | + | + | + | + | + | |||||||||||||||
The relationship between the distributions of the 21 genes for which the isolates were tested and the genetic backgrounds of the isolates, as defined by M genotyping and PFGE analysis, is shown in Table 3. M1 and M3 strains shared a number of striking similarities. They represented 39% of all isolates tested, and all of these isolates except for one M3 isolate carried an speA allele and were lacking the speC gene. cpa-1 and fbp-54 were found exclusively in all M1 strains. Furthermore, the smeZ gene was found in all M1 isolates, a feature shared only with all M4 strains. In M3 strains, the existence of the ssa allele stood out. The M2 group of strains showed a large degree of genetic variability in terms of both their PFGE types and their gene profiles. M4 strains were characterized by the absence of speG and the presence of ssa, smeZ, and fba. Size variations were observed for the fba gene, as well as for the prtf and sfb genes. M6 isolates were found to be positive for speC, prtf-1, and, in most cases, speA. In general, PFGE allowed a better distinction of the different gene profiles than M genotyping, although some slight variations were observed among isolates indistinguishable by PFGE (i.e., pulsotypes 3A, 4A, 4B, 6A, 12A, 12B, 28A, and 28B) as well as identical patterns among different PFGE types (i.e., pulsotypes 4B and 4C and pulsotypes 12A and 12B).
To uncover potential associations of virulence genes and types of GAS disease, the rate of occurrence of these genes in isolates that had given rise to the different manifestations of invasive streptococcal disease was analyzed. Some toxin and matrix binding protein genes were nonuniformly distributed among the GAS isolates causing different types of disease (Table 4). However, most of these correlations reflect the M-type distributions among the various site-specific infections (Table 2) and the association of these M types with certain genes (Table 3). For instance, within the group of isolates causing meningitis, speA and prtf-1 predominated, a representation of the genetic makeup of meningitis-specific genotype M6. In isolates that had given rise to GAS arthritis, speH and pfbp were predominant (M12), as was the case for speC, prtf-15, and sfb among isolates causing puerperal sepsis, in which M2 was exclusively found. The overrepresentation of the fba gene among the isolates causing puerperal sepsis, however, could not be attributed to an association of one M type with puerperal sepsis. In this case, different strains shared one gene that gave rise to a similar clinical syndrome. Remarkably, no specific toxin genes or genes encoding matrix binding proteins were associated with necrotizing fasciitis.
TABLE 4.
Association of gene profile and type of invasive GAS disease
| Disease | No. of cases | No. (%) of isolates with the following genes:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| speA | speC | speH | fba | pfbp | prtf-1 | prtf-15 | sfb | ||
| Meningitis | 27 | 17a (63) | 11 (41) | 1 (4) | 16 (59) | 1 (4) | 5b (19) | 1 (4) | 3 (11) |
| Arthritis | 41 | 15 (37) | 19 (46) | 12c (29) | 24 (59) | 10b (24) | 2 (5) | 7 (17) | 9 (22) |
| Necrotizing fasciitis | 61 | 31 (51) | 16 (26) | 8 (13) | 45 (74) | 8 (13) | 1 (2) | 9 (15) | 9 (15) |
| Puerperal sepsis | 41 | 14 (34) | 25c (61) | 2 (5) | 35a (85) | 2 (5) | 3 (7) | 15c (37) | 15b (37) |
P < 0.05.
P < 0.01.
P < 0.001.
The invasive isolates were scrutinized for the existence of a toxin gene profile associated with severe disease, as defined by TSS. The distribution of genes significantly associated with a severe clinical outcome is illustrated in Table 5. The toxin genes significantly overrepresented among isolates causing TSS were smeZ and speA, whereas the presence of speC appeared to be associated with a more favorable course of invasive GAS disease. This is again a reflection of the genetic makeup of those M genotypes associated with the development of TSS (i.e., M1 and M3).
TABLE 5.
Association of gene profile and development of TSS
| TSS occurrence | No. of cases | No. (%) of isolates with the following genes:
|
||||||
|---|---|---|---|---|---|---|---|---|
| speAa | speCb | smeZa | cpa-1b | fbp-54b | prtf-1c | sfbc | ||
| TSS | 47 | 29 (62) | 10 (21) | 22 (47) | 20 (43) | 20 (43) | 0 (0) | 3 (6) |
| Non-TSS | 123 | 48 (39) | 61 (50) | 32 (26) | 23 (19) | 23 (19) | 11 (9) | 33 (27) |
P < 0.01.
P < 0.001.
P < 0.05.
DISCUSSION
The reemergence of severe invasive GAS infections provided impetus to scientists and epidemiologists to define the factors responsible for disease manifestations and clinical outcomes. Traditionally, M1 and M3 serotypes that contain the speA and speB genes have been particularly implicated in the pathogenesis of invasive GAS disease (10, 26, 27, 37, 45, 52, 62, 73). More recently, the importance of other GAS virulence factors has been underscored in several studies (11, 47, 50, 51, 76). In this study we have focused on GAS strain characteristics and their correlation to clinical syndromes. To this end we analyzed all microbiologically defined cases of meningitis, arthritis, necrotizing fasciitis, and puerperal sepsis in a national surveillance study conducted in The Netherlands between 1992 and 1996. Twenty-eight percent of these infections were complicated by the occurrence of TSS. To address the question of the intrinsic virulence in GAS, the strains were characterized for their M types, PFGE types, and the presence of genes encoding toxins and matrix binding proteins.
In accordance with other studies (5, 15, 38), M1, M3, M12, and M28 isolates were the most frequently present. Interestingly, the different manifestations of invasive GAS disease (meningitis, arthritis, necrotizing fasciitis, and puerperal sepsis) appeared to be associated with certain M types (Table 2). M2 was found exclusively in puerperal sepsis cases, whereas M3 was a rare causative agent of this disease. M6 was associated with meningitis, and M12 was associated with arthritis. M1 was distributed equally among cases of these four different manifestations of invasive GAS disease, while no particular M type was associated with necrotizing fasciitis. A preferential distribution of different M genotypes in skin and throat isolates has been well described (7, 8), but this is the first report of a nonrandom distribution of M genotypes among cases of the different clinical manifestations of invasive GAS disease.
Among all invasive isolates, the development of TSS was caused primarily by M1 types. The overrepresentation of M1 isolates among isolates responsible for TSS implies an increased intrinsic virulence for isolates of this M type. In the past, a large number of different studies that corroborate this notion have been published (13, 19, 20, 26, 30, 45, 60, 65, 71). A recent publication by Johnson et al. (31) could not identify a virulent clone among isolates of serotypes M1, M3, and M28 but suggested that the prevalence of the “virulent” clones reflected their normal prevalence in the population. Nonetheless, those investigators “still favor the hypothesis that GAS strains of M-types 1 and 3 have an increased association with invasive infections.” On the basis of an increased propensity of the M1 and M3 types among all invasive GAS isolates in our study to cause TSS, we agree with the notion that these strains have increased intrinsic virulence.
All strains with the same PFGE pattern belonged to the same M type. As expected, PFGE proved to have a discriminatory power superior to that of M genotyping (68). Clonality was observed among isolates of the M1, M3, M6 and M12 types (Fig. 1 and Table 3). In contrast, a larger degree of heterogeneity was found among M4, M9, M12, and M28 isolates; and no homology was observed among the M2 types. The M1 isolates constituted the largest cluster of strains (n = 43).
In order to study the strain characteristics contributing to aggressive clinical behavior, the isolates were evaluated for the presence of different toxin genes and other presumptive virulence factors encoded by chromosomal as well as plasmid-located genes. We showed that the patterns of the protein binding factor and toxin genes were exclusively associated with particular M types. Similarly, Natanson et al. (46) reported on the detection of the prtf protein in M6 and M28 strains and found that it was absent from M1, M3, and M18 isolates. Also, the presence of pfbp in M12 strains and its association with prtf-2 have been reported previously (29, 61). We noted size variations in the sfb, prtf, and fba genes. This has been reported to be the result of intragenic repeats (46, 72, 74). The size of the fba gene in M1 isolates was unique to this genotype.
Some of the observed associations between M types and toxin genes have been reported by others. Murakami et al. (44) detected the ssa gene in the majority of M2 and M4 strains. Furthermore, they detected speC, speG, and speH in all M12 isolates as well as the absence of the speG gene in M4 isolates. These findings, with the exception of the presence of ssa in M2 strains, are completely in accordance with those of our study. Descheemaeker et al. (17) described the exotoxin gene profile of their M1 genotype as speA, speB, and speF positive and speC and ssa negative. To this profile we would like to add the speG-, speJ-, and smeZ-positive and speH-negative genotype as well as the exclusive presence of cpa-1 and fbp-54. In M1, the exclusive combination of speA and smeZ, which both encode proteins known to be major immunoactive agents, might contribute to increased intrinsic virulence. Although it is speculative, the link between M types and the presence of virulence genes appears to be best conserved for the most virulent M types, such as M1 and M3. Their specific set of virulence genes might render them more successful as pathogens.
Since we do not know if particular virulence factors are involved in different types of invasive GAS disease, we searched for a particular site-specific profile. Different repertoires of adhesins and the interactions of different adhesins with each other may determine tissue tropism and the pathogenicity of S. pyogenes (16). Therefore, isolates with differences in tropism (i.e., isolates causing meningitis, necrotizing fasciitis, arthritis, and puerperal sepsis) were differentiated as non-TSS and TSS isolates and were analyzed for the presence of microbial surface adhesins as well as superantigens. We sought to define a “pathogenic” toxin gene profile that resulted in the development of severe complications such as TSS. Although the presence of some factors such as speA or smeZ, as opposed to speC, appears to be positively correlated with the development of severe complications, the development of severe complications in patients infected with strains with these genes is a reflection of the association of these genes with the M1 and M3 types. The same applies to genes found significantly more often in association with GAS strains responsible for infections at the different sites. Most of these associations mirror the differences in M-type distributions seen in different disease manifestations and the correlation of these M types to various virulence genes. However, the overrepresentation of the fba gene among isolates causing puerperal sepsis could not be attributed to the association of one M type with puerperal sepsis. This was the only gene shared by different strains that gave rise to a similar clinical syndrome.
In conclusion, the profiles of exotoxin and protein binding factor genes were closely associated with M types. Each M type was characterized by one or two dominant gene profiles that were exclusive for a given type, whether or not these profiles also included a variable number of less frequently occurring patterns. How can the linkage between M types and gene profile be explained? Most genes analyzed in this study are located on bacteriophages. A phenotypic correlation between resistance to bacteriophage infection and M-protein surface expression has been described (12, 69). Thus, the nonrandom association between M types and gene distribution that was observed could suggest a direct or indirect biological interaction between M-protein surface structures and bacteriophages. In our study, toxin and protein binding factor gene profiling revealed for each M type a number of less frequent profiles, in addition to one or two dominant toxin profiles. Remarkably, all gene profiles were exclusive for their respective M types. Although this illustrates the fundamental nature of the lines of division drawn by M typing, the different genotypes were distinguished at a higher resolution by PFGE.
Toxin profiles are highly conserved among the most virulent M types (M1 and M3). These strains do not seem to easily incorporate or release any virulence genes, suggesting that an optimal composition for their pathogenetic potential has been achieved. In this respect, it is noteworthy that in a study in Japan (44) the genotype profiles of M1 isolates changed from the lack of speA in the early 1980s to a speA-positive genotype in the 1990s. Furthermore, most associations between the clinical manifestations of invasive GAS disease and virulence genes could be attributed to the nonrandom distribution patterns of M types among the isolates that cause these clinical manifestations. However, this reasoning could well be reversed: the M type associated with the presence of genes encoding exotoxins and extracellular matrix binding proteins, which enable the streptococcus to selectively interact with certain extracellular matrix components of different tissues and organs, might explain the observed tropism of particular M types for certain organs. This view is further supported by the observation that one matrix binding gene (fba) was more strongly correlated with a particular GAS disease manifestation than one particular M type. Nonetheless, in this geographically and temporally narrowly defined study, a diversity of clones displaying specific genetic patterns proved to have the capacity to cause similar types of invasive disease. This underscores the complexity of the interplay between bacterial virulence factors, bacterial gene regulation, and host factors.
REFERENCES
- 1.Abe, J., J. Forrester, T. Nakahara, J. A. Lafferty, B. L. Kotzin, and D. Y. Leung. 1991. Selective stimulation of human T cells with streptococcal erythrogenic toxins A and B. J. Immunol. 146:3747-3750. [PubMed] [Google Scholar]
- 2.Anonymous. 1993. Defining the group A streptococcal toxic shock syndrome. Rationale and consensus definition. JAMA 269:390-391. [PubMed] [Google Scholar]
- 3.Banks, D. J., S. B. Beres, and J. M. Musser. 2002. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10:515-521. [DOI] [PubMed] [Google Scholar]
- 4.Basma, H., A. Norrby-Teglund, Y. Guedez, A. McGeer, D. E. Low, O. El Ahmedy, B. Schwartz, and M. Kotb. 1999. Risk factors in the pathogenesis of invasive group A streptococcal infections: role of protective humoral immunity. Infect. Immun. 67:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., R. Facklam, T. Hoenes, and B. Schwartz. 1997. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J. Clin. Microbiol. 35:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bessen, D. E., T. R. Fiorentino, and S. K. Hollingshead. 1997. Molecular markers for throat and skin isolates of group A streptococci. Adv. Exp. Med. Biol. 418:537-543. [DOI] [PubMed] [Google Scholar]
- 8.Bessen, D. E., M. W. Izzo, T. R. Fiorentino, R. M. Caringal, S. K. Hollingshead, and B. Beall. 1999. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J. Infect. Dis. 179:627-636. [DOI] [PubMed] [Google Scholar]
- 9.Bisno, A. L. 1991. Group A streptococcal infections and acute rheumatic fever. N. Engl. J. Med. 325:783-793. [DOI] [PubMed] [Google Scholar]
- 10.Carapetis, J., R. Robins-Browne, D. Martin, T. Shelby-James, and G. Hogg. 1995. Increasing severity of invasive group A streptococcal disease in Australia: clinical and molecular epidemiological features and identification of a new virulent M-nontypeable clone. Clin. Infect. Dis. 21:1220-1227. [DOI] [PubMed] [Google Scholar]
- 11.Chatellier, S., N. Ihendyane, R. G. Kansal, F. Khambaty, H. Basma, A. Norrby-Teglund, D. E. Low, A. McGeer, and M. Kotb. 2000. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68:3523-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleary, P. P., and Z. Johnson. 1977. Possible dual function of M protein: resistance to bacteriophage A25 and resistance to phagocytosis by human leukocytes. Infect. Immun. 16:280-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colman, G., A. Tanna, A. Efstratiou, and E. T. Gaworzewska. 1993. The serotypes of Streptococcus pyogenes present in Britain during 1980-1990 and their association with disease. J. Med. Microbiol. 39:165-178. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, H. D., A. McGeer, B. Schwartz, K. Green, D. Cann, A. E. Simor, D. E. Low, et al. 1996. Invasive group A streptococcal infections in Ontario, Canada. N. Engl. J. Med. 335:547-554. [DOI] [PubMed] [Google Scholar]
- 16.Delvecchio, A., B. J. Currie, J. D. McArthur, M. J. Walker, and K. S. Sriprakash. 2002. Streptococcus pyogenes prtFII, but not sfbI, sfbII or fbp54, is represented more frequently among invasive-disease isolates of tropical Australia. Epidemiol. Infect. 128:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Descheemaeker, P., F. Van Loock, M. Hauchecorne, P. Vandamme, and H. Goossens. 2000. Molecular characterisation of group A streptococci from invasive and non-invasive disease episodes in Belgium during 1993-1994. J. Med. Microbiol. 49:467-471. [DOI] [PubMed] [Google Scholar]
- 18.Efstratiou, A. 2000. Group A streptococci in the 1990s. J. Antimicrob. Chemother. 45(Suppl.):3-12. [DOI] [PubMed] [Google Scholar]
- 19.Ferrieri, P. 1991. Microbiological features of current virulent strains of group A streptococci. Pediatr. Infect. Dis. J. 10:S20-S24. [DOI] [PubMed] [Google Scholar]
- 20.Gaworzewska, E., and G. Colman. 1988. Changes in the pattern of infection caused by Streptococcus pyogenes. Epidemiol. Infect. 100:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlach, D., B. Fleischer, M. Wagner, K. Schmidt, S. Vettermann, and W. Reichardt. 2000. Purification and biochemical characterization of a basic superantigen (SPEX/SMEZ3) from Streptococcus pyogenes. FEMS Microbiol. Lett. 188:153-163. [DOI] [PubMed] [Google Scholar]
- 22.Goodfellow, A. M., M. Hibble, S. R. Talay, B. Kreikemeyer, B. J. Currie, K. S. Sriprakash, and G. S. Chhatwal. 2000. Distribution and antigenicity of fibronectin binding proteins (SfbI and SfbII) of Streptococcus pyogenes clinical isolates from the Northern Territory, Australia. J. Clin. Microbiol. 38:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goshorn, S. C., G. A. Bohach, and P. M. Schlievert. 1988. Cloning and characterization of the gene, speC, for pyrogenic exotoxin type C from Streptococcus pyogenes. Mol. Gen. Genet. 212:66-70. [DOI] [PubMed] [Google Scholar]
- 24.Goshorn, S. C., and P. M. Schlievert. 1988. Nucleotide sequence of streptococcal pyrogenic exotoxin type C. Infect. Immun. 56:2518-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauser, A. R., D. L. Stevens, E. L. Kaplan, and P. M. Schlievert. 1991. Molecular analysis of pyrogenic exotoxins from Streptococcus pyogenes isolates associated with toxic shock-like syndrome. J. Clin. Microbiol. 29:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm, S. E., A. Norrby, A. M. Bergholm, and M. Norgren. 1992. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988-1989. J. Infect. Dis. 166:31-37. [DOI] [PubMed] [Google Scholar]
- 28.Iwasaki, M., H. Igarashi, Y. Hinuma, and T. Yutsudo. 1993. Cloning, characterization and overexpression of a Streptococcus pyogenes gene encoding a new type of mitogenic factor. FEBS Lett. 331:187-192. [DOI] [PubMed] [Google Scholar]
- 29.Jaffe, J., S. Natanson-Yaron, M. G. Caparon, and E. Hanski. 1996. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two binding domains. Mol. Microbiol. 21:373-384. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, D. R., D. L. Stevens, and E. L. Kaplan. 1992. Epidemiologic analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever, or uncomplicated pharyngitis. J. Infect. Dis. 166:374-382. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, D. R., J. T. Wotton, A. Shet, and E. L. Kaplan. 2002. A comparison of group A streptococci from invasive and uncomplicated infections: are virulent clones responsible for serious streptococcal infections? J. Infect. Dis. 185:1586-1595. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, L. P., J. J. L'Italien, and P. M. Schlievert. 1986. Streptococcal pyrogenic exotoxin type A (scarlet fever toxin) is related to Staphylococcus aureus enterotoxin B. Mol. Gen. Genet. 203:354-356. [DOI] [PubMed] [Google Scholar]
- 33.Kamezawa, Y., T. Nakahara, S. Nakano, Y. Abe, J. Nozaki-Renard, and T. Isono. 1997. Streptococcal mitogenic exotoxin Z, a novel acidic superantigenic toxin produced by a T1 strain of Streptococcus pyogenes. Infect. Immun. 65:3828-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapur, V., S. Topouzis, M. W. Majesky, L. L. Li, M. R. Hamrick, R. J. Hamill, J. M. Patti, and J. M. Musser. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 15:327-346. [DOI] [PubMed] [Google Scholar]
- 35.Katerov, V., A. Andreev, C. Schalen, and A. A. Totolian. 1998. Protein F, a fibronectin-binding protein of Streptococcus pyogenes, also binds human fibrinogen: isolation of the protein and mapping of the binding region. Microbiology 144(Pt 1):119-126. [DOI] [PubMed] [Google Scholar]
- 36.Kaufhold, A., A. Podbielski, G. Baumgarten, M. Blokpoel, J. Top, and L. Schouls. 1994. Rapid typing of group A streptococci by the use of DNA amplification and non-radioactive allele-specific oligonucleotide probes. FEMS Microbiol. Lett. 119:19-25. [DOI] [PubMed] [Google Scholar]
- 37.Kaul, R., A. McGeer, D. E. Low, K. Green, B. Schwartz, et al. 1997. Population-based surveillance for group A streptococcal necrotizing fasciitis: clinical features, prognostic indicators, and microbiologic analysis of seventy-seven cases. Am. J. Med. 103:18-24. [DOI] [PubMed] [Google Scholar]
- 38.Kiska, D. L., B. Thiede, J. Caracciolo, M. Jordan, D. Johnson, E. L. Kaplan, R. P. Gruninger, J. A. Lohr, P. H. Gilligan, and F. W. Denny, Jr. 1997. Invasive group A streptococcal infections in North Carolina: epidemiology, clinical features, and genetic and serotype analysis of causative organisms. J. Infect. Dis. 176:992-1000. [DOI] [PubMed] [Google Scholar]
- 39.Kotb, M., A. Norrby-Teglund, A. McGeer, H. El Sherbini, M. T. Dorak, A. Khurshid, K. Green, J. Peeples, J. Wade, G. Thomson, B. Schwartz, and D. E. Low. 2002. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat. Med. 8:1398-1404. [DOI] [PubMed] [Google Scholar]
- 40.Kreikemeyer, B., S. R. Talay, and G. S. Chhatwal. 1995. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol. Microbiol. 17:137-145. [DOI] [PubMed] [Google Scholar]
- 41.Mascini, E. M., M. Jansze, J. F. Schellekens, J. M. Musser, J. A. Faber, L. A. Verhoef-Verhage, L. Schouls, W. J. van Leeuwen, J. Verhoef, and H. van Dijk. 2000. Invasive group A streptococcal disease in The Netherlands: evidence for a protective role of anti-exotoxin A antibodies. J. Infect. Dis. 181:631-638. [DOI] [PubMed] [Google Scholar]
- 42.Molinari, G., S. R. Talay, P. Valentin-Weigand, M. Rohde, and G. S. Chhatwal. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 65:1357-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mollick, J. A., G. G. Miller, J. M. Musser, R. G. Cook, D. Grossman, and R. R. Rich. 1993. A novel superantigen isolated from pathogenic strains of Streptococcus pyogenes with aminoterminal homology to staphylococcal enterotoxins B and C. J. Clin. Investig. 92:710-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami, J., S. Kawabata, Y. Terao, K. Kikuchi, K. Totsuka, A. Tamaru, C. Katsukawa, K. Moriya, I. Nakagawa, I. Morisaki, and S. Hamada. 2002. Distribution of emm genotypes and superantigen genes of Streptococcus pyogenes isolated in Japan, 1994-9. Epidemiol. Infect. 128:397-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natanson, S., S. Sela, A. E. Moses, J. M. Musser, M. G. Caparon, and E. Hanski. 1995. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J. Infect. Dis. 171:871-878. [DOI] [PubMed] [Google Scholar]
- 47.Newton, D., A. Norrby-Teglund, A. McGeer, D. E. Low, P. M. Schlievert, and M. Kotb. 1997. Novel superantigens from streptococcal toxic shock syndrome Streptococcus pyogenes isolates. Adv. Exp. Med. Biol. 418:525-529. [DOI] [PubMed] [Google Scholar]
- 48.Norrby-Teglund, A., S. Chatellier, D. E. Low, A. McGeer, K. Green, and M. Kotb. 2000. Host variation in cytokine responses to superantigens determine the severity of invasive group A streptococcal infection. Eur. J. Immunol. 30:3247-3255. [DOI] [PubMed] [Google Scholar]
- 49.Norrby-Teglund, A., and M. Kotb. 2000. Host-microbe interactions in the pathogenesis of invasive group A streptococcal infections. J. Med. Microbiol. 49:849-852. [DOI] [PubMed] [Google Scholar]
- 50.Norrby-Teglund, A., D. Newton, M. Kotb, S. E. Holm, and M. Norgren. 1994. Superantigenic properties of the group A streptococcal exotoxin SpeF (MF). Infect. Immun. 62:5227-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norrby-Teglund, A., P. Thulin, B. S. Gan, M. Kotb, A. McGeer, J. Andersson, and D. E. Low. 2001. Evidence for superantigen involvement in severe group A streptococcal tissue infections. J. Infect. Dis. 184:853-860. [DOI] [PubMed] [Google Scholar]
- 52.Nunthapisud, P., S. Sirilertpanrana, S. Reinprayoon, and A. Tanna. 1997. Detection of the erythrogenic toxin A, B, and C genes in group A streptococci isolated from clinical specimens. Adv. Exp. Med. Biol. 418:729-731. [DOI] [PubMed] [Google Scholar]
- 53.O'Brien, K. L., B. Beall, N. L. Barrett, P. R. Cieslak, A. Reingold, M. M. Farley, R. Danila, E. R. Zell, R. Facklam, B. Schwartz, and A. Schuchat. 2002. Epidemiology of invasive group a streptococcus disease in the United States, 1995-1999. Clin. Infect. Dis. 35:268-276. [DOI] [PubMed] [Google Scholar]
- 54.Okada, N., A. P. Pentland, P. Falk, and M. G. Caparon. 1994. M protein and protein F act as important determinants of cell-specific tropism of Streptococcus pyogenes in skin tissue. J. Clin. Investig. 94:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 56.Proft, T., S. L. Moffatt, C. J. Berkahn, and J. D. Fraser. 1999. Identification and characterization of novel superantigens from Streptococcus pyogenes. J. Exp. Med. 189:89-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proft, T., S. L. Moffatt, K. D. Weller, A. Paterson, D. Martin, and J. D. Fraser. 2000. The streptococcal superantigen SMEZ exhibits wide allelic variation, mosaic structure, and significant antigenic variation. J. Exp. Med. 191:1765-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasmussen, M., A. Eden, and L. Bjorck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reda, K. B., V. Kapur, J. A. Mollick, J. G. Lamphear, J. M. Musser, and R. R. Rich. 1994. Molecular characterization and phylogenetic distribution of the streptococcal superantigen gene (ssa) from Streptococcus pyogenes. Infect. Immun. 62:1867-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichardt, W., H. Muller-Alouf, J. E. Alouf, and W. Kohler. 1992. Erythrogenic toxins A, B and C: occurrence of the genes and exotoxin formation from clinical Streptococcus pyogenes strains associated with streptococcal toxic shock-like syndrome. FEMS Microbiol. Lett. 79:313-322. [DOI] [PubMed] [Google Scholar]
- 61.Rocha, C. L., and V. A. Fischetti. 1999. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infect. Immun. 67:2720-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schellekens, J. F., L. M. Schouls, W. van Pelt, M. Esveld, and W. J. van Leeuwen. 1998. Group A streptococci: a change in virulence? Neth. J. Med. 52:209-217. [DOI] [PubMed] [Google Scholar]
- 63.Schellekens, J. F., L. M. Schouls, A. van Silfhout, K. Elzenaar, H. Brunings, H. ten Broek, J. Top, and W. J. van Leeuwen. 1995. The resurgence of group A streptococcal disease. Ned. Tijdschr. Med. Microbiol. 4:78-83. [Google Scholar]
- 64.Schmitz, F. J., A. Beyer, E. Charpentier, B. H. Normark, M. Schade, A. C. Fluit, D. Hafner, and R. Novak. Toxin gene profile heterogeneity among endemic invasive European group A streptococcal isolates. J. Infect. Dis., in press. [DOI] [PubMed]
- 65.Schwartz, B., R. R. Facklam, and R. F. Breiman. 1990. Changing epidemiology of group A streptococcal infection in the USA. Lancet 336:1167-1171. [DOI] [PubMed] [Google Scholar]
- 66.Sela, S., A. Aviv, A. Tovi, I. Burstein, M. G. Caparon, and E. Hanski. 1993. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10:1049-1055. [DOI] [PubMed] [Google Scholar]
- 67.Sharkawy, A., D. E. Low, R. Saginur, D. Gregson, B. Schwartz, P. Jessamine, K. Green, and A. McGeer. 2002. Severe group A streptococcal soft-tissue infections in Ontario: 1992-1996. Clin. Infect. Dis. 34:454-460. [DOI] [PubMed] [Google Scholar]
- 68.Single, L. A., and D. R. Martin. 1992. Clonal differences within M-types of the group A Streptococcus revealed by pulsed field gel electrophoresis. FEMS Microbiol. Lett. 70:85-89. [DOI] [PubMed] [Google Scholar]
- 69.Spanier, J. G., and P. P. Cleary. 1980. Bacteriophage control of antiphagocytic determinants in group A streptococci. J. Exp. Med. 152:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stevens, D. L. 1994. Invasive group A streptococcal infections: the past, present and future. Pediatr. Infect. Dis. J. 13:561-566. [DOI] [PubMed] [Google Scholar]
- 71.Stromberg, A., V. Romanus, and L. G. Burman. 1991. Outbreak of group A streptococcal bacteremia in Sweden: an epidemiologic and clinical study. J. Infect. Dis. 164:595-598. [DOI] [PubMed] [Google Scholar]
- 72.Talay, S. R., P. Valentin-Weigand, K. N. Timmis, and G. S. Chhatwal. 1994. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol. Microbiol. 13:531-539. [DOI] [PubMed] [Google Scholar]
- 73.Talkington, D. F., B. Schwartz, C. M. Black, J. K. Todd, J. Elliott, R. F. Breiman, and R. R. Facklam. 1993. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect. Immun. 61:3369-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed] [Google Scholar]
- 75.Terao, Y., S. Kawabata, M. Nakata, I. Nakagawa, and S. Hamada. 2002. Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 277:47428-47435. [DOI] [PubMed] [Google Scholar]
- 76.Unnikrishnan, M., D. M. Altmann, T. Proft, F. Wahid, J. Cohen, J. D. Fraser, and S. Sriskandan. 2002. The bacterial superantigen streptococcal mitogenic exotoxin Z is the major immunoactive agent of Streptococcus pyogenes. J. Immunol. 169:2561-2569. [DOI] [PubMed] [Google Scholar]
- 77.Valentin-Weigand, P., S. R. Talay, A. Kaufhold, K. N. Timmis, and G. S. Chhatwal. 1994. The fibronectin binding domain of the Sfb protein adhesin of Streptococcus pyogenes occurs in many group A streptococci and does not cross-react with heart myosin. Microb. Pathog. 17:111-120. [DOI] [PubMed] [Google Scholar]
- 78.Whatmore, A. M. 2001. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology 147:419-429. [DOI] [PubMed] [Google Scholar]