Abstract
Pathogenic strains of Clostridium difficile commonly produce two large clostridial toxins (LCTs), A and B, virulence factors responsible for C. difficile disease. Some strains have been reported to produce an additional toxin, a binary toxin designated CDT. Binary toxin has cytotoxic effects on cells in culture, but its role in human disease is not yet defined. In this study we examined the frequency of binary toxin genes (cdtB and cdtA) among C. difficile isolates that do not produce LCTs (A− B−) from a large United States-based collection organized by restriction endonuclease analysis (REA) typing. Of 58 strains tested, 9 (15.5%) were cdtB and cdtA positive, including 4 of 46 (8.7%) non-LCT-producing REA groups, with an estimated prevalence of at least 2% of all non-LCT-producing isolates within the collection. Five of the binary toxin-positive strains belonged to toxinotype XI, which does not produce LCTs but has minor parts of the LCT coding region or pathogenicity locus (PaLoc). We describe two new binary toxin-positive variants, one without any remnant of the LCT genes. This previously unknown variation was found in three isolates that were unrelated by REA typing. LCT-negative, binary toxin-positive strains were isolated from symptomatic and asymptomatic patients and from the hospital environment.
Clostridium difficile is a causative agent of antibiotic-associated diarrhea and potentially lethal pseudomembranous colitis (4, 11, 15). Toxigenic strains of C. difficile commonly produce two large clostridial toxins (LCTs), toxin A (TcdA) and toxin B (TcdB), to which disease symptoms are attributed.
Some C. difficile strains have been reported to produce an additional toxin, a binary toxin designated CDT (6, 19, 27). As is typical for binary toxins found in other clostridial species, CDT is composed of two unlinked components, which are both needed for toxic activity (5, 18). The CDTb component is responsible for binding to an as yet unknown cell receptor and for translocation of the enzymatic component, CDTa, into the cell. CDT is an actin-specific ADP-ribosyltransferase and is cytotoxic for cultured eukaryotic cells (18). However, determination of the cytotoxicity effect due to binary toxin in C. difficile has been difficult for three reasons: (i) the amount of binary toxin produced in vitro is low and culture supernatants need to be concentrated (18), (ii) binary toxin needs to be activated with trypsin, and (iii) most binary toxin-producing strains also produce other cytotoxins (TcdA and/or TcdB). The arborizing effect of CDT on cultured cells is similar to the characteristic cytopathic effect due to TcdB of reference strain VPI 10463 (18). Therefore, distinguishing this effect from that due to the LCTs in patient stool specimens would be difficult, and there are no commercial immunoassays for CDT available at this time.
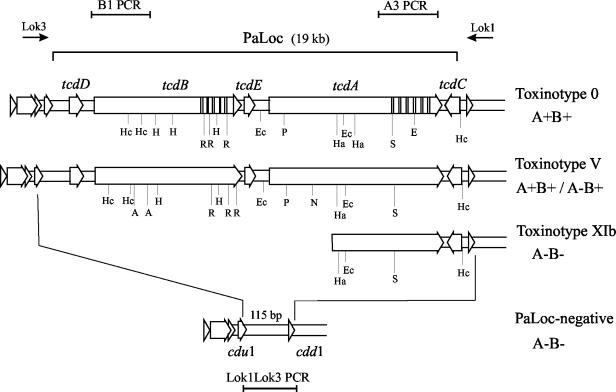
Genes for all three toxins produced by C. difficile have been sequenced. Genes coding for the two components of CDT, cdtA and cdtB, are reported to be chromosomal (18, 19). The LCTs, TcdA and TcdB, are encoded by genes tcdA and tcdB, which together with three additional genes (tcdC, tcdD, and tcdE; Fig. 1) form the well-characterized 19.6-kb pathogenicity locus (PaLoc) (7, 12). The PaLoc is also chromosomal but is not obviously linked to cdtA and cdtB. Variant C. difficile strains with deletions, insertions, or polymorphic restriction sites within the PaLoc have been described and categorized on this basis into 20 different toxinotypes (I to XX) (20, 24). Some variability has also been detected in the binary toxin genes by partial sequencing (27), but it is less extensive than that seen in the PaLoc. Binary toxin genes are found only in certain variant toxinotypes and not in toxinotype 0 strains (including the reference strain VPI 10463) or in toxinotypes with minor changes of toxin genes tcdA and tcdB (24, 27).
FIG. 1.
Schematic representation of the C. difficile PaLoc region coding for the LCTs TcdA and TcdB in reference toxinotype 0 and toxinotypes V and XI. One binary toxin gene-positive isolate was defined as toxinotype V. Whereas previously described toxinotype V isolates produce both LCTs (A+ B+), this isolate (isolate 4380, REA type AA2) was shown to represent a new type of A− B+ C. difficile variant. Three A− B− isolates belonged to toxinotype XIb, and two additional A− B− isolates were typed as toxinotype XIa (not shown), which differs from type XIb within the A3 PCR fragment, where a deletion is present in XIa but not in XIb. Note that toxinotypes XI and V have similar restriction sites. The upstream deletion in toxinotype XI is not yet completely defined, but the remainder of the PaLoc cannot be detected with specific PCRs. PCR results from three of the binary toxin gene-positive strains were consistent with typical “PaLoc-negative” (A− B−) strains that contain a 115-bp segment in place of the PaLoc. The 115-bp segment was detected with the Lok1-Lok3 PCR (115-bp segment is not drawn to scale). The positions of primers Lok3 and Lok1 and PCR fragments B1, A3, and Lok1-Lok3 are shown on top of and below the figure. Restriction sites are indicated below each PaLoc schematic: A, AccI; Ec, EcoRV; E, EcoRI; Ha, HaeIII; H, HindIII; Hc, HincII; N, NsiI; P, PstI; R, RsaI; S, SpeI.
Most of the binary toxin-producing C. difficile strains reported to date also produce TcdA and/or TcdB (19, 27). Only three isolates, all of the same toxinotype (toxinotype XI), have been reported as TcdA negative, TcdB negative, and binary toxin CDT positive (A− B− CDT+) (27). Two of the isolates were recovered from humans, and the third was isolated from a diseased foal that eventually died. Since the effect of binary toxin is not seen on standard cytotoxicity testing, A− B− CDT+ strains would be likely reported as nontoxigenic isolates when tested in a clinical laboratory.
Because these unique C. difficile strains which produce only binary toxin may help define the role of binary toxin in the pathogenesis of C. difficile disease, we screened a large U.S. clinical isolate collection for the prevalence of A− B− C. difficile strains that have binary toxin genes.
(This work has previously been presented in part [B. Geric, M. Rupnik, M. Grabnar, S. Sambol, D. Gerding, and S. Johnson, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. K-1236, 2001.])
Selection of non-LCT-producing strains and detection of the binary toxin genes.
A large collection of C. difficile isolates obtained from multiple U.S. and other sources over a 20-year period has been maintained in the research laboratory of Dale Gerding and colleagues. This collection of primarily clinical isolates has been catalogued by HindIII restriction endonuclease analysis (REA) of whole-cell genomic DNA (9, 13). Isolates showing six or fewer visible restriction band differences (a similarity index of >90%) are placed within the same REA group and designated by letter. Isolates with identical restriction patterns are assigned a specific REA type designated by number (Table 1). Currently, this collection includes over 5,000 isolates, representing 108 REA groups and 436 REA types (D. Gerding, personal communication). LCT production (TcdA and TcdB) is highly correlated with REA typing designations (9).
TABLE 1.
Strains from the non-LCT-producing REA groups tested for the binary toxin (CDT) genes cdtB and cdtAa
| REA type | No. of isolates in collection | Presence of cdt genes |
|---|---|---|
| AA | ||
| AA1 | 5 | + |
| AA2b | 1 | + |
| AD1 | 13 | − |
| AE1 | 1 | − |
| AI1 | 1 | − |
| AK1 | 7 | − |
| AM1 | 1 | + |
| AP1 | 20 | − |
| AQ1 | 1 | − |
| AR1 | 1 | − |
| AU1 | 1 | − |
| AX1 | 1 | − |
| AY1 | 1 | − |
| AZ1 | 2 | − |
| BB1 | 4 | − |
| BC1 | 1 | − |
| BD1 | 1 | − |
| BF1 | 1 | − |
| BG1 | 2 | − |
| BJ1 | 1 | − |
| BO1 | 1 | − |
| BP1 | 3 | − |
| BQ1 | 4 | − |
| BS1 | 5 | − |
| BU1 | 1 | − |
| BY1 | 2 | − |
| C1 | 19 | − |
| CA1 | 1 | − |
| CC1 | 1 | − |
| CD1 | 3 | − |
| CE1 | 1 | − |
| CH1 | 1 | − |
| CI1 | 1 | − |
| CM1 | 2 | − |
| CQ | ||
| CQ1c | 2 | +/− |
| CQ2 | 1 | − |
| CQ3 | 1 | − |
| CQ4 | 1 | − |
| CR1 | 1 | − |
| CS1 | 1 | + |
| CV1 | 1 | − |
| CZ1 | 1 | − |
| DE1 | 2 | − |
| DC1 | 1 | − |
| H1 | 4 | − |
| M | ||
| M1 | 6 | − |
| M23 | 58 | − |
| M3 | 109 | − |
| P1 | 4 | − |
| Q1 | 2 | − |
| S1 | 27 | − |
| T | ||
| T1 | 23 | − |
| T7 | 47 | − |
One representative isolate from each non-LCT-producing REA group was tested, except for REA groups AA and CQ, where all isolates were tested, and REA groups M and T, where three additional REA type isolates were tested, as these types (M3, M23, and T7) were the most prevalent non-LCT-producing REA types in the collection (9). All together 58 isolates representing 53 REA types were screened for cdtB. The total number of isolates was 403, and nine isolates were cdt+.
AA2 produced TcdB and was identified as a new A− B+ C. difficile variant despite relatedness to other non-LCT-producing (A− B−) isolates (REA type AA1 strains) within the same REA group.
Only one of the two REA type CQ1 isolates was positive for binary toxin genes.
Within this collection, 46 REA groups (and 146 unique REA types) have been recognized that contain non-LCT-producing strains (A− B−) as determined by lack of in vitro cytotoxin (TcdB) production (9) in at least one isolate from each unique REA type within each non-LCT-producing REA group. These strains are usually referred to as “nontoxigenic” as they do not react in the standard cytotoxin assay for TcdB. Only one REA group (AA) contains strains that are heterogenous for LCT production.
We have screened 58 strains from this collection for the presence of binary toxin gene cdtB. Included within this screen were representative isolates from each of the 45 exclusively non-LCT-producing (A− B−) REA groups and REA group AA. All isolates from REA groups with a binary toxin-positive representative strain were subsequently tested (nine strains; see below and Table 1). Additionally, three strains from the most prevalent non-LCT-producing (A− B−) C. difficile REA types in this collection, M3, M23, and T7, were also tested (9).
Crude DNA was prepared using 5% Chelex-100 resin (Bio-Rad Laboratories, Hercules, Calif.) as described by O'Neill et al. (17), and the presence of the binary toxin gene cdtB was determined by PCR (27). In the case of a positive result, PCR to detect the cdtA gene was also performed, to confirm that the entire binary toxin could be produced by the strain. Perelle et al. (18) have described strains with cdtB but not cdtA. In our studies we have found only strains with both genes (27).
Nine of 58 isolates tested (15.5%) were positive for binary toxin genes, and both cdtB and cdtA genes were detected in all nine (Table 2). Binary toxin genes were not found in the most prevalent non-LCT-producing (A− B−) REA types in this collection (M3, M23, and T7). Isolates from 4 of the 46 non-LCT-producing (A− B−) REA groups (including REA group AA) were cdtB positive and cdtA positive (groups AA, AM, CQ, and CS). Two isolates belonged to REA groups with a single known isolate (AM and CS; Table 1), whereas the other isolates belonged to REA groups with more than one known isolate (AA and CQ). We tested all isolates from REA groups AA (REA types AA1 and AA2) and CQ (REA types CQ1, CQ2, CQ3, and CQ4), to look for a correlation between REA types and REA groups and the presence of binary toxin genes. All REA type AA1 and AA2 isolates had binary toxin genes, whereas four of five REA group CQ isolates were negative for binary toxin genes. Only one of the two REA type CQ1 isolates was positive (Table 1). Both CQ1 isolates were typed again by HindIII REA, and identical restriction fragment patterns were obtained, confirming the original REA type designation. Therefore, REA type CQ1 represents an example of a lack of correlation between REA type and the presence of binary toxin genes.
TABLE 2.
Characterization of binary toxin-positive strainsa
| REA type | Isolate designation | LCT production
|
PCR
|
Toxinotyping
|
Patient data | ||||
|---|---|---|---|---|---|---|---|---|---|
| TcdA | TcdB | cdt genes | PaLocb | B1 HincII/AccIc | A3 EcoRIc | Toxinotype | |||
| AA2 | 4380 | − | + | + | + | 3 | 8 | V | Asymptomatic patient |
| AA1 | 5943 | − | − | + | + | − | 5 | XIa | No data |
| AA1 | 4680 | − | − | + | + | − | 5 | XIa | Symptomatic patient |
| AA1 | 3126 | − | − | + | + | − | 8 | XIb | Asymptomatic patientd |
| AA1 | 3133 | − | − | + | + | − | 8 | XIb | Asymptomatic patientd |
| AA1 | 3800 | − | − | + | + | − | 8 | XIb | Asymptomatic patient |
| AM1 | 3859 | − | − | + | − | − | − | Tox− | Hospital environment |
| CQ1 | 3376 | − | − | + | − | − | − | Tox− | Hospital environment |
| CS1 | 6009 | − | − | + | − | − | − | Tox− | Patient with diarrhea |
+, amplified or present; −, not amplified or not present.
Presence or absence of LCT genes (PaLoc) detected with Lok3-Lok1 PCR.
Restriction pattern of tcdB fragment B1 and tcdA fragment A3 (type according to the work of Rupnik et al.) (22).
Isolates 3126 and 3133 were recovered from the same patient.
PaLoc characterization of binary toxin-positive strains.
Toxinotype XI is the only known C. difficile variant that is positive for binary toxin CDT but negative for LCT production (A− B− CDT+) and is characterized by a truncated PaLoc with only a portion of the 3′ end still present (Fig. 1) (21, 24, 27).
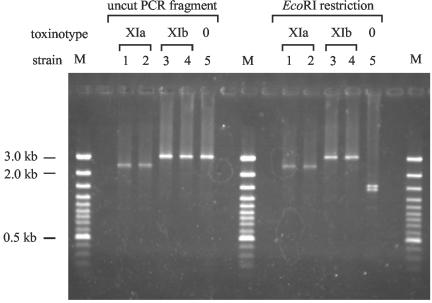
To confirm either an alteration in PaLoc structure or the complete absence of the PaLoc, all cdtB-positive strains were tested with PaLoc-specific PCRs and Lok1-Lok3 PCR (7) (Fig. 1). Ten fragments covering the whole PaLoc region (fragments PL1 to PL4, A1 to A3, and B1 to B3) were amplified in all nine strains. The primer sequences and amplification conditions were performed as previously described (22, 23) (http://www.uni-lj.si/∼bfbcdiff). PCR on REA types CQ1, CS1, and AM1 did not amplify any of the fragments, indicating the complete absence of the PaLoc (results for B1 and A3 PCR are shown in Table 2). PCR on the five REA type AA1 isolates amplified A2, A3, and PL4 fragments, indicating the presence of a portion of the 3′ end of the PaLoc (Fig. 1). Two AA1 isolates were assigned to the previously characterized toxinotype XIa. Toxinotype XIa is distinguished from toxinotype XIb, to which three other AA1 isolates were assigned, by a deletion within the A3 PCR fragment. Previously, only one toxinotype XIb isolate was known (21), and this isolate was incorrectly reported to have an A3 fragment that was smaller in size than that of toxinotype 0. In this study we compared A3 PCR fragments of the new toxinotype XI strains with those of previously characterized toxinotype XI strains. Three new XIb strains (Table 2) as well as the previously described strain R11402 (XIb) do not have a deletion within the A3 fragment (Fig. 2). Instead, the A3 PCR fragment from these AA1 isolates is similar to the A3 fragment found in toxinotype V (A3 restriction pattern 8, Table 2).
FIG. 2.
Comparison of length and restriction site polymorphisms found in PCR fragment A3 of toxinotype XI strains. Toxinotypes XIa and XIb show differences in A3 fragment length (a deletion is present in toxinotype XIa), but neither XIa nor XIb has the EcoRI restriction sites typical for toxinotype 0. Lanes 1, isolate 5943; lanes 2, isolate 4680; lanes 3, isolate 3126 (all three isolates are described in this study); lanes 4, control strain R11402 (type strain of toxinotype XIb); lanes 5, control strain VPI 10463 (type strain of toxinotype 0); lanes M, DNA ladder, 100 bp.
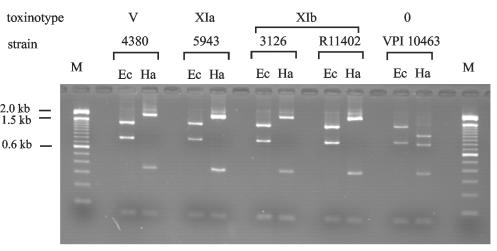
Unexpectedly, PCR fragments from both LCT genes (B1 and A3) were amplified in REA type AA2 isolate 4380 (Table 2), which was originally thought to be a non-LCT-producing strain (based on the REA typing result and the otherwise complete correlation of REA typing and LCT production). REA typing was repeated, and the restriction pattern confirmed the REA group AA designation for this isolate despite the marked PaLoc differences from the other REA group AA isolates (Table 2). When all 10 PaLoc-specific PCRs were performed, isolate 4380 was identical to toxinotype V. However, previously identified toxinotype V strains produce both LCTs (A+ B+) (22). Toxin testing of isolate 4380 (described below) showed this strain to be TcdA negative and TcdB positive (A− B+). Isolate 4380 therefore represents a new type of A− B+ variant. A− B+ C. difficile strains are increasingly recognized among clinical patient isolates. First described as nonvirulent (10), they have since been recovered from patients with C. difficile diarrhea and pseudomembranous colitis and have been associated with outbreaks of C. difficile diarrhea and colitis (1, 2, 3, 14, 16, 25). With respect to PaLoc composition, two A− B+ groups have been recognized: toxinotype X, with a single representative isolate (8864), and toxinotype VIII, which includes isolates recovered from patients worldwide (24). Toxinotype VIII strains, however, do not contain binary toxin genes (27). Recently, two new A− B+ isolates from Japan and Korea were described, representing two new toxinotypes, XVI and XVII (20). Despite major differences in PaLoc composition, the relatedness of the toxinotype XIa/b and V isolates in this study (REA types AA1 and AA2) is supported by REA typing and identical restriction patterns of PCR fragment A2 (Fig. 3). Toxinotypes V, VI, VII, and XI were reported previously to be a genetically related subgroup of C. difficile variants (21, 28). Additionally, results presented here indicate that some A− B+ strains are indistinguishable by PaLoc composition from A+ B+ strains of the same toxinotype.
FIG. 3.
Restriction patterns of PCR fragment A2 showing similarities between toxinotypes V and XI. Isolates 4380, 5943, and 3126 are described in this study. Strains R11402 and VPI 10463 are the previously described representative strains for toxinotypes XIb and 0, respectively. Ec, EcoRV; Ha, HaeIII; M, DNA ladder, 100 bp.
Primers Lok1 and Lok3 are positioned outside the PaLoc (Fig. 1), and Lok1-Lok3 PCR is used for confirmation of absence of the PaLoc (7). In toxigenic strains (A+ B+) the expected product would be 20 kb, but the product is not amplified under the selected conditions of amplification, whereas in typical nontoxigenic strains (A− B−, in which the PaLoc is replaced by a 115-bp segment) an amplified fragment of 700 bp is obtained (Fig. 1). The three isolates which were negative by PaLoc-specific PCR amplification had amplification of a 700-bp fragment by Lok1-Lok3 PCR. In distinction from toxinotype XI strains, which are also A− B− but which contain a portion of the 3′ end of the PaLoc, these isolates were typical nontoxigenic strains (A− B−) (Table 2). This particular genetic variation of C. difficile in which the binary toxin genes are present without any remnant of the LCT genes has not been reported previously. The three such isolates in this study do not appear to be closely related by REA typing.
Detection of toxin production.
All nine strains were retested for the production of TcdB and TcdA. One strain was sent to M. Popoff (Pasteur Institute, Paris, France) to test for the production of binary toxin.
Culture supernatants were assayed for TcdB production in a cell culture assay (cytotoxicity assay for C. difficile toxins; Bartels, Inc., Issaquah, Wash.) and TcdA production by immunoassay (C. difficile Tox-A test; TechLab, Blacksburg, Va.). Tests were performed according to manufacturers' instructions. This standard cytotoxicity assay will not detect binary toxin. All tested strains (except isolate 4380) were negative for TcdA and TcdB (Table 2). The cytopathic effect of supernatant from strain 4380 (A− B+) on this cell line was indistinguishable from that due to strain VPI 10463 (A+ B+).
Production of binary toxin components CDTa and CDTb in strain 6009 (REA type CS1) was confirmed by Western blotting as described previously (18, 27).
Prevalence of A− B− strains with binary toxin genes and clinical associations.
Excluding isolate 4380, eight strains that possessed genes for binary toxin were identified in this survey of non-LCT-producing (A− B−) C. difficile isolates. To estimate the prevalence of A− B− strains with binary toxin genes, we used, as a denominator, all isolates in the collection for the REA types screened (n = 402, excluding isolate 4380; Table 1) and made the assumption that the presence of binary toxin correlated with REA typing designations, since our previous studies have shown a good correlation between REA types and toxinotypes and between toxinotypes and the presence of binary toxin genes (25, 27). Binary toxin-positive strains were found mainly among REA types with only one known isolate but were also found among REA types with more than one isolate (AA1 and CQ1). Therefore, we estimate a prevalence of at least 2.0% (8 of 402) for binary toxin-positive strains among C. difficile isolates that do not produce LCT. This may be a conservative estimate, because at least one REA type (CQ1) was heterogenous for the presence of binary toxin.
This prevalence of binary toxin-only-producing strains in the studied collection is low but comparable to the prevalence of binary toxin among A+ B+ strains found in other studies: 1.6% (20), 4.6% (B. Geric, M. Rupnik, M. Grabnar, D. Gerding, and S. Johnson, Abstr. Anaerobe Olympiad 2002, abstr. SP-6, 2002), and 6.4% (27). In contrast, some small surveys have reported a prevalence of binary toxin-positive strains of 20 to 21% in A+ B+ strains (18, 26).
Two of the eight binary toxin-positive, A− B− strains were isolated from symptomatic patients. This finding does not, however, prove a role for binary toxin as a virulence factor, as C. difficile colonization among hospitalized patients is common and diarrhea may be a spurious association. The role of the binary toxin in C. difficile pathogenesis is unknown, but the highly related binary toxin in Clostridium spiroforme has been implicated as a virulence factor and involved in the pathogenesis of intestinal disease in rabbits (8). We hypothesize that binary toxin may act as an virulence factor in non-LCT-producing (A− B−) C. difficile strains or as an adjunctive virulence factor in A+ B+ strains.
In summary, we have surveyed a large U.S. collection of non-LCT-producing C. difficile strains for the presence of binary toxin genes. We have found nine binary toxin-positive strains, one of which was subsequently shown to produce TcdB and represents a new A− B+ C. difficile variant, though it is indistinguishable from toxinotype V. Five strains had a truncated PaLoc typical of toxinotype XI but did not produce LCTs. Three isolates with binary toxin genes did not contain any PaLoc sequences and were the first such variants described. Binary toxin-positive strains were isolated from the hospital environment and symptomatic and asymptomatic patients. We are currently testing these LCT-negative, binary toxin gene-positive isolates for virulence potential in different animal models. If CDT is shown to be an additional virulence factor for C. difficile, further clinical studies would be warranted to determine the role of binary toxin gene-positive strains in C. difficile-associated disease and the need for specific diagnostic testing.
Acknowledgments
This work was supported by the Ministry of Education, Science and Sport of Republic of Slovenia grants no. S1-487-002/20069/99 and P0-0523-0481 and by grants from the U.S. Department of Veterans Affairs Research Service and Pharmacia & Upjohn, USA.
We thank Michel Popoff (Institut Pasteur, Paris, France) for testing the production of binary toxin and Susan Sambol for testing the production of TcdA and TcdB.
REFERENCES
- 1.Al-Barrak, A., J. Embil, B. Dyck, K. Olekson, D. Nicoll, M. Alfa, and A. Kabani. 1999. An outbreak of toxin A negative, toxin B positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can. Commun. Dis. Rep. 25:65-69. [PubMed] [Google Scholar]
- 2.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barral, G. H. K. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbut, F., V. Lalande, B. Burghoffer, H. V. Thien, E. Grimprel, and J.-C. Petit. 2002. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J. Clin. Microbiol. 40:2079-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett, J. G., N. Moon, T. W. Chang, N. Taylor, and A. B. Onderdonk. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75:778-782. [PubMed] [Google Scholar]
- 5.Boquet, P., P. Munro, C. Fiorentini, and I. Just. 1998. Toxins from anaerobic bacteria: specificity and molecular mechanisms of action. Curr. Opin. Microbiol. 1:66-74. [DOI] [PubMed] [Google Scholar]
- 6.Braun, M., C. Herholz, R. Straub, B. Choisat, J. Frey, J. Nicolet, and P. Kuhnert. 2000. Detection of the ADP-ribosyltransferase toxin gene (cdtA) and its activity in Clostridium difficile isolates from Equidae. FEMS Microbiol. Lett. 184:29-33. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V., T. Hundesberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. [DOI] [PubMed] [Google Scholar]
- 8.Carman, R. J., S. Perelle, and M. R. Popoff. 1997. Binary toxins from Clostridium spiroforme and Clostridium perfringens, p. 359-367. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, London, United Kingdom.
- 9.Clabots, C. R., S. Johnson, K. M. Bettin, P. A. Mathie, M. E. Mulligan, D. R. Schaberg, L. R. Peterson, and D. N. Gerding. 1993. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J. Clin. Microbiol. 31:1870-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Depitre, C., M. Delmée, V. Avesani, R. L'Haridon, A. Roels, M. Popoff, and G. Corthier. 1993. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J. Med. Microbiol. 38:434-441. [DOI] [PubMed] [Google Scholar]
- 11.George, R. H., J. M. Symonds, F. Dimock, J. D. Brown, Y. Arabi, N. Shinagawa, M. R. Keighley, J. Alexander-Williams, and D. W. Burdon. 1978. Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br. Med. J. 1:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond, G. A., and J. L. Johnson. 1995. The toxigenic element of Clostridium difficile strain VPI 10463. Microb. Pathog. 19:203-213. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, S., S. P. Sambol, J. S. Brazier, M. Delmée, V. Avesani, M. M. Merrigan, and D. N. Gerding. 2003. International typing study of toxin A-negative, toxin B-positive Clostridium difficile variants. J. Clin. Microbiol. 41:1543-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, H., H. Kita, T. Karasawa, T. Maegawa, Y. Koino, H. Takakuwa, T. Saikai, K. Kobayashi, T. Yamagishi, and S. Nakamura. 2001. Colonization and transmission of Clostridium difficile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J. Med. Microbiol. 50:720-727. [DOI] [PubMed] [Google Scholar]
- 15.Knoop, F. C., M. Owens, and I. A. Crocker. 1993. Clostridium difficile: clinical disease and diagnosis. Clin. Microbiol. Rev. 6:251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuijper, E. J., J. de Weerdt, H. Kato, N. Kato, A. P. van Dam, E. R. van der Vorm, J. Weel, D. van Rheenen, and J. Dankert. 2001. Nosocomial outbreak of Clostridium difficile-associated diarrhoea due to a clindamycin-resistant enterotoxin A-negative strain. Eur. J. Clin. Microbiol. Infect. Dis. 20:528-534. [DOI] [PubMed] [Google Scholar]
- 17.O'Neill, G. L., F. T. Ogunsola, J. S. Brazier, and B. I. Duerden. 1996. Modification of a PCR-ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe 2:205-209. [Google Scholar]
- 18.Perelle, S., M. Gibert, P. Bourlioux, G. Corthier, and M. R. Popoff. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect. Immun. 65:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popoff, M. R., E. J. Rubin, D. M. Gill, and P. Boquet. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 56:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupnik, M., N. Kato, M. Grabnar, and H. Kato. 2003. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J. Clin. Microbiol. 41:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupnik, M., J. Brazier, B. Duerden, M. Grabnar, and S. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 22.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmée. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rupnik, M., V. Braun, F. Soehn, M. Janc, M. Hofstetter, R. Laufenberg-Feldmann, and C. von Eichel-Streiber. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148:197-202. [DOI] [PubMed] [Google Scholar]
- 24.Rupnik, M. 2001. How to detect Clostridium difficile variant strains in a routine laboratory. Clin. Microbiol. Infect. 7:417-420. [DOI] [PubMed] [Google Scholar]
- 25.Sambol, S. P., M. M. Merrigan, D. Lyerly, D. N. Gerding, and S. Johnson. 2000. Toxin gene analysis of a variant strain of Clostridium difficile that causes human clinical disease. Infect. Immun. 68:5480-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 28.Wozniak, G., P. Trontelj, and M. Rupnik. 2000. Genomic relatedness of Clostridium difficile strains from different toxinotypes and serogroups. Anaerobe 6:261-267. [Google Scholar]