Abstract
Six Cryptosporidium spp. were found in 50 of 179 Milwaukee wastewater samples collected weekly over a year. Of the eight subtypes of Cryptosporidium hominis and Cryptosporidium parvum present, allele Ib was found in 14 of 16 samples, and its sequence was identical to that of the subtype in human samples from the 1993 Milwaukee outbreak of cryptosporidiosis.
Two Cryptosporidium spp., Cryptosporidium hominis (previously known as the Cryptosporidium parvum human genotype or genotype 1) and C. parvum (previously know as C. parvum bovine genotype or genotype 2), are responsible for cryptosporidiosis outbreaks (3, 5). C. hominis is found almost exclusively in humans, while C. parvum is found in humans and domestic ruminants (5). C. hominis has been responsible for more outbreaks than C. parvum, even in countries where C. parvum is the predominant Cryptosporidium parasite in humans (3, 5). Recently, DNA sequence characterizations of several genes have identified intragenetic variations in C. hominis and C. parvum. These tools have been used effectively in the investigation of several outbreaks. One such subtyping tool is based on sequence analysis of the 60-kDa glycoprotein (GP60), which divides C. hominis and C. parvum into several allelic groups, each of which consists of multiple subtypes (1, 2, 4, 6, 7). One of the allele families, Ib, was responsible for the waterborne outbreaks of cryptosporidiosis in Milwaukee, Wis., which caused illness in over 400,000 people (6).
The distribution of Cryptosporidium species was previously characterized in 49 wastewater samples in Milwaukee (9). Cryptosporidium andersoni was identified as the most common parasite in wastewater, followed by Cryptosporidium muris, C. hominis, C. parvum, Cryptosporidium canis, Cryptosporidium felis, and the Cryptosporidium cervid genotype (W4). In the present study, we examined the seasonal distribution of Cryptosporidium species in raw Milwaukee wastewater and subtyped C. hominis and C. parvum isolates from wastewater to assess current cryptosporidiosis transmission in Milwaukee and its relationship to the 1993 outbreak.
Wastewater sample collection and processing.
A total of 179 raw wastewater samples were obtained from the Jones Island Wastewater Treatment Plant in Milwaukee from August 2000 to July 2001 with an average of two to four samples per week (no more than one sample per day). Fifteen more samples were also obtained in March 2002. Each sample was a composite from the three separate siphons that delivered influent to the treatment plant: domestic sewage, combined domestic and industrial sewage, and wastewater from the deep tunnel, which was comprised of infiltration and inflow during dry-weather periods and storm water-diluted sewage during periods of precipitation. For each siphon, a small quantity (10 to 15 ml) was automatically drawn every 15 min for 24 h, and the three collections were mixed together in proportion to the volumes delivered by the three systems. Only 50 ml of the 24-h composite wastewater was analyzed for each sample. Samples were concentrated by centrifuging at 1,000 × g for 10 min. Cryptosporidium oocysts were further purified by immunomagnetic separation, using magnetic beads coated with an anti-Cryptosporidium monoclonal antibody (Dynal, Inc., Lake Success, N.Y.).
PCR-RFLP analysis.
DNA was extracted from the immunomagnetic separation concentrates obtained above were used in DNA extraction without detachment of Cryptosporidium oocysts from beads (8, 9). Cryptosporidium species in DNA from wastewater were determined by a previously described small-subunit (SSU)-rRNA-based PCR-restriction fragment length polymorphism (PCR-RFLP) (8, 9). Each sample was analyzed at least three times by nested PCR using 0.5, 1.0, or 2.0 μl of DNA as the template. Positive (Cryptosporidium serpentis DNA) and negative controls were included in each PCR run. For the differentiation of Cryptosporidium species or genotypes, 10 μl of the secondary PCR product was subjected to restriction digestions by SspI (New England BioLabs, Beverly, Mass.) and VspI (GIBCO BRL, Grand Island, N.Y.). If C. muris or C. andersoni was present, they were differentiated from each other by restriction digestion with DdeI (New England BioLabs). Species and genotypes were determined by banding patterns in electrophoresis with 2% agarose gels (8, 9). Unusual Cryptosporidium parasites were confirmed by DNA sequencing of the PCR products.
Subtyping.
For subtyping, the GP60 gene was amplified by nested PCR with the primer sets 5′-ATAGTCTCGCTGTATTC-3′ and 5′-GCAGAGGAACCAGCATC-3′ in the primary PCR and 5′-TCCGCTGTATTCTCAGCC-3′ and 5′-GAGATATATCTTGGTGCG-3′ in the secondary PCR (1, 7). The secondary PCR amplicons were sequenced in both directions on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). Two PCR products per sample were sequenced to confirm the accuracy of diagnosis. Nucleotide sequences were aligned with known Cryptosporidium GP60 sequences, and a neighbor-joining tree was constructed based on evolutionary distances calculated by the Kimura two-parameter model with 1,000 bootstrap samplings (8, 9).
Cryptosporidium species or genotypes in wastewater.
A total of 179 wastewater samples were examined in this study, of which 50 were positive for Cryptosporidium by the SSU-rRNA-based PCR-RFLP technique. Restriction analysis of PCR products revealed the presence of six types of Cryptosporidium parasites in these wastewater samples (Table 1): C. hominis, C. parvum, C. andersoni, C. muris, the Cryptosporidium mouse genotype, and the Cryptosporidium cervid genotype. C. hominis was the most commonly detected parasite overall (24 samples, or 13.4% of the total samples). The order of prevalence for the remaining Cryptosporidium parasites was C. andersoni (12.8%), the Cryptosporidium cervid genotype (3.3%), C. parvum (2.8%), C. muris (2.2%), and the Cryptosporidium mouse genotype (0.6%). Among the 50 positive samples, 5 showed the concurrent presence of two or more Cryptosporidium parasites: 4 of these samples had two types of Cryptosporidium (samples 2466 and 2474 had C. hominis and C. andersoni, 2623 had C. muris and C. hominis, and 2683 had the Cryptosporidium cervid genotype and C. muris), and 1 (sample 4229) had three types of Cryptosporidium (C. hominis, C. parvum, and the Cryptosporidium cervid genotype).
TABLE 1.
Cryptosporidium species and subtypes in raw wastewater in Milwaukee
| Month | No. of samples | No. (%) positive | Species or genotypea | GP60 allelea |
|---|---|---|---|---|
| July 2000 | 12 | 4 (33.3) | C. andersoni (1), C. hominis (2), cervid genotype (1), C. parvum (1) | Ia (1), Ie (1) |
| Aug. | 8 | 0 (0) | ||
| Sept. | 12 | 4 (33.3) | C. hominis (3), C. muris (1), cervid genotype (1) | Ib (1) |
| Oct. | 15 | 5 (33.3) | C. andersoni (4), C. hominis (2), C. parvum (1) | Ia (1), Ib (1), Ie (1) |
| Nov. | 12 | 2 (16.7) | C. andersoni (2), C. hominis (1) | Ib (1) |
| Dec. | 13 | 8 (61.5) | C. andersoni (4), C. hominis (2), C. muris (2), cervid genotype (W4), (2), C. parvum (1) | Ia (1), Ib (2), IIa (1) |
| Jan. 2001 | 14 | 2 (14.3) | C. andersoni (1), C. hominis (1) | Ib (1) |
| Feb. | 12 | 2 (16.7) | C. andersoni (1), mouse genotype (1) | |
| Mar. | 12 | 2 (16.7) | C. andersoni (1), C. hominis (1) | Ib (1) |
| Apr. | 15 | 6 (40) | C. andersoni (4), C. hominis (2) | Ib (2) |
| May | 12 | 3 (25) | C. andersoni (2), C. hominis (1), C. parvum (1) | Ia (1), Ib (1), IIa (1) |
| June | 12 | 1 (8.3) | C. andersoni (1) | |
| July | 15 | 4 (26.7) | C. andersoni (1), C. hominis (2), C. parvum (1), cervid genotype (2) | Ib (2) |
| March 2002 | 15 | 7 (46.7) | C. hominis (7), C. muris (1), C. andersoni (1) | Ib (2) |
| Total | 179 | 50 (27.9) | 6 species/genotypes | 4 alleles |
Numbers in parentheses are numbers of samples positive for each species or subtype allele.
Subtypes of C. hominis and C. parvum in wastewater.
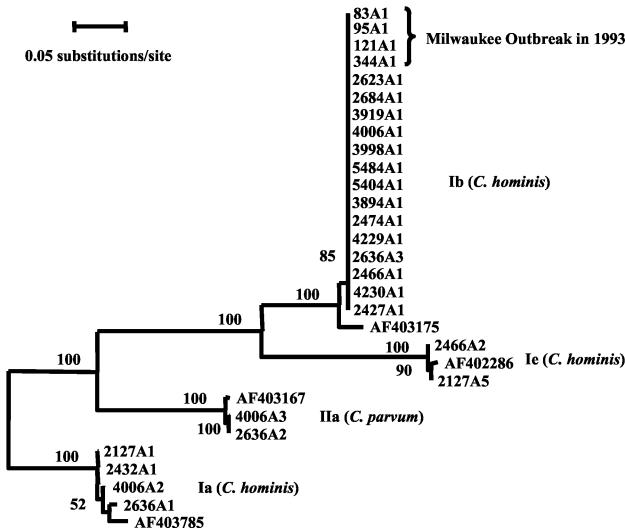
Wastewater samples positive for C. hominis (24 isolates) and C. parvum (5 isolates) were subtyped by GP60 locus analysis, and 16 isolates were positive for GP60 amplification. The reduced detection rate by GP60 PCR was probably due to the single-copy nature of the target gene (instead of five copies for the SSU-rRNA gene) and low numbers of oocysts in water samples. The obtained nucleotide sequences were compared with the known subtype sequences and those present in four stool samples from the 1993 Milwaukee outbreak. Phylogenetic analysis of GP60 sequences obtained suggested the presence of four allelic groups (eight subtypes) in 16 wastewater samples: Ia (four subtypes), Ib (one subtype), Ie (two subtypes), and IIa (one subtype) (Fig. 1). In contrast, there was only one subtype allele (Ib) in human stool samples from the 1993 Milwaukee outbreak. Fourteen of the wastewater samples had allele Ib, and all allele Ib sequences were identical to sequences obtained from four stool samples collected during the 1993 outbreak (Fig. 1). Most of the samples (i.e., 12) had only one subtype allele: Ib (11 samples) or Ia (1 sample). However, two samples had two subtypes (Ia and Ie in sample 2127; Ib and Ie in sample 2466), and two samples had three subtypes (Ia, Ib, and IIa in both 2636 and 4006) (Fig. 1).
FIG. 1.
Relationship among C. parvum and C. hominis subtypes inferred by neighbor-joining analysis of GP60 nucleotide sequences. The Kimura two-parameter model was used in distance calculation. Numbers on branches are percent bootstrap values from 1,000 resamplings.
Public health significance.
Results of this study support a previous finding of the complexity of Cryptosporidium in raw urban wastewater (9). Six Cryptosporidium species or types were found in raw wastewater samples from Milwaukee, with C. hominis and C. andersoni as the most common. Because these host-adapted parasites occur in humans (C. hominis and C. parvum), farm animals (C. andersoni and C. parvum), rodents (C. muris and the Cryptosporidium mouse genotype), and deer (the Cryptosporidium cervid genotype), these results confirmed the previous conclusion that humans, slaughtered farm animals, rodents, and deer all contributed to Cryptosporidium contamination in wastewater. In the previous study conducted in Milwaukee, a few samples were found to be positive for C. canis and C. felis, which were not found in this study.
Results of subtype analysis further support the complexity of human-pathogenic Cryptosporidium in wastewater. Four subtype alleles of C. hominis and C. parvum were found in Milwaukee wastewater: Ia, Ib, Ie, and IIa. Subtype alleles Ia, Ib, and Ie belonged to C. hominis and thus were likely of human origin, whereas IIa belonged to C. parvum and probably originated from cattle as well as humans. Nevertheless, allele Ib was the predominant subtype of C. hominis in Milwaukee wastewater samples. All parasites in this group in the wastewater had identical GP60 sequences and belonged to the subtype involved in the 1993 Milwaukee outbreak.
The frequent detection of C. hominis in Milwaukee wastewater all year round indicates that there is stable transmission of human cryptosporidiosis in Milwaukee in the absence of an outbreak. The high-frequency detection of the allele Ib parasite, which was probably responsible for the 1993 outbreak, suggests that this parasite is probably quite virulent or infectious and has been circulating in humans in the Milwaukee area at least since the outbreak. Even though there are many subtypes of C. hominis and C. parvum in humans, quite a few food-borne and waterborne outbreaks of cryptosporidiosis in the United States and Northern Ireland are caused by allele Ib parasites (1, 6).
In summary, results of this study suggest that molecular tools can be very useful in assessing the human-infective potential of Cryptosporidium oocysts in wastewater and the source of Cryptosporidium oocyst contamination. The distribution of Cryptosporidium species indicates that even in urban areas, a significant proportion of Cryptosporidium oocysts in wastewater are probably not from humans and are of no significant public health concern. Continuous monitoring of Cryptosporidium species and subtypes in urban wastewater can be an effective tool in postoutbreak surveillance and may provide useful information for understanding Cryptosporidium transmission in humans and the occurrence of waterborne outbreaks of cryptosporidiosis.
Nucleotide sequence accession numbers.
Nucleotide sequences were submitted to the GenBank database under accession numbers AY262027 to AY262034.
Acknowledgments
This study was supported in part by a research grant from the AWWA Research Foundation.
We thank Jeff MacDonald of the Milwaukee Metropolitan Sewerage District and Birhane Dashew for their assistance in sample collection.
REFERENCES
- 1.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. J. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng, M. M., O. Matos, W. Gatei, P. Das, M. Stantic-Pavlinic, C. Bern, I. M. Sulaiman, S. Glaberman, A. A. Lal, and L. Xiao. 2001. A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. 2001(Suppl.):28S-31S. [DOI] [PubMed] [Google Scholar]
- 5.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulaiman, I. M., A. A. Lal, and L Xiao. 2001. A population genetic study of the Cryptosporidium parvum human genotype parasites. J. Eukaryot. Microbiol. 2001(Suppl.):24S-27S. [DOI] [PubMed] [Google Scholar]
- 7.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]