Abstract
One of the most important functions of the clinical microbiology laboratory is the identification of the etiology of sepsis. For this study, aliquots from 405 positive blood cultures were tested against a unique array of DNA probes directed against rRNA subsequences of bacteria and fungi for identification. Another 280 samples that were negative after 5 days of incubation were also tested. Blood culture bottles were incubated in a BacT/Alert3D instrument. For the rRNA assay, a 0.4-ml aliquot was removed, and the cells were pelleted by centrifugation. The pellet was washed and frozen at −70°C. Analysis of the pellet involved a lysis step and then the addition of samples to the reaction wells containing the probes in a microtiter plate format. Analysis was performed by using a hybridization protection assay. Results were taken through a series of deductive steps to obtain species, or in some cases genus, identification. Batch sample preparation required approximately 15 min, and sample analysis required another 60 min. Probe results were compared to conventional biochemical identifications. The probe test was negative for the 280 samples that were negative by the BacT/Alert 3D system and for another 21 samples that were false positive (the instrument signaled, but there was no growth). Microorganisms from the remaining 384 signal-positive samples included 60 Enterobacteriaceae, 10 Pseudomonas aeruginosa, 10 other gram-negative bacteria, 40 Staphylococcus aureus, 152 coagulase-negative staphylococci, 28 streptococci, 22 enterococci, 21 other gram-positive bacteria, 8 anaerobes, and 16 yeast organisms. Seventeen cultures were polymicrobial, and one was gram positive and culture negative. Discordance between probe and conventional identification results was noted for only 12 (1.75%) samples. This novel rapid molecular approach to the identification of bacteria and yeast in blood cultures was highly sensitive (100%) and specific (96%).
Traditional culture and biochemical identification (ID) of microorganisms that cause bloodstream infections is currently the only routine method for detecting the etiological agents of sepsis. In the past 2 decades, techniques for culturing blood have gone from manual and visual methods to the use of automated continuous monitoring blood culture instruments, such as the Bactec (Becton-Dickinson), BacT/Alert3D (Organon Teknika, Durham, N.C.), and ESP (Trek Diagnostic Systems) systems. These instruments detect CO2 gas production or consumption when organisms grow in a broth medium above a certain growth threshold. Typically, the average time to detection of a positive culture ranges from 6 to 60 h, depending on the organism group. After the instrument signals positive, a Gram stain and subculture are performed. From the time the blood culture is noted to be positive, ID and susceptibility testing of the isolate(s) can take an additional 24 to 72 h. Due to the high morbidity and mortality associated with sepsis, there remains a need for rapid organism ID and susceptibility testing.
With the advent of molecular techniques, the goal of such rapid testing has become a closer reality for routine testing in the clinical laboratory. Numerous studies have documented the use of nucleic acid tests for rapid ID of positive blood cultures. Direct DNA probing of blood cultures using chemiluminescent labels has been used in several studies (4, 7; D. A. Bruckner, L. Gibson, J. Hindler, J. Hogan, I. Andruszkiewicz, K. Clark-Dickey, and W. Weisberg, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. C-1, 2001; D. Fuller and T. Davis, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1558, 1999; J. F. Hindler, S. Kozen, and D. A. Bruckner, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1557, 1999). Other rapid DNA detection systems have used amplified formats (5, 15, 17) and in situ approaches (8, 13, 14, 16). Vibrational spectroscopic approaches have also been able to identify pathogens in positive blood cultures (9). Chemiluminescent DNA probes without amplification have been used to detect bacteria that are growing in blood products (2, 3). Many of these techniques have shown value but have also proven to be labor intensive, expensive, or at risk for a high rate of false positives.
The objective of this study was to examine the use of a Gen-Probe (San Diego, Calif.) research prototype assay to rapidly identify organisms directly from positive blood cultures. This system consists of a DNA probe matrix directed towards rRNA targets which utilizes seven different probe cocktails (Fig. 1). The application of this probe matrix directly to positive blood cultures allows rapid ID of a majority of blood pathogens to a level that would answer most clinical questions related to ID of the causative agent(s) of the patient's septicemia. For this study, a total of 685 blood cultures, 405 positive and 280 negative, were examined to determine the ability of the DNA probe matrix to detect and identify the organism(s) present.
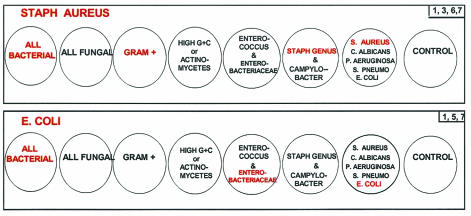
FIG. 1.
Examples of probe cocktails used in the probe matrix ID system. The probe cocktails are specifically arranged in the 8 wells of a column in a 96-well plate. The top column is an example of a specimen positive for S. aureus. As a result, wells 1, 3, 6, and 7 would produce a signal. The bottom column is an example of a specimen positive for E. coli. In this case, wells 1, 5, and 7 would produce a signal.
MATERIALS AND METHODS
Study design.
Specimens were obtained from routine blood cultures collected in standard aerobic and anaerobic BacT/Alert bottles (Organon Teknika Corp.) and submitted to the UCLA Medical Center Clinical Microbiology Laboratory. All bottles were incubated in a BacT/Alert3D continuous monitoring system (Organon Teknika Corp.). Both standard biochemical and probe matrix ID tests were performed on each specimen. Standard biochemical testing was not performed on isolates that, on the basis of Gram stain colony morphology and a rapid coagulase test, were probable skin contaminants (e.g., Corynebacterium and coagulase-negative staphylococci [CoNS]). Phenotypic ID of organisms was accomplished by use of conventional test systems (e.g., Vitek and API strips) routinely employed at the UCLA Medical Center laboratory. A total of 405 positive blood cultures were tested. Set A (n = 103) comprised samples that were removed and processed within minutes of signaling positive by the BacT/Alert3D system. Set B (n = 302) comprised samples that were removed and processed within 8 h after signaling positive. Set B represented those specimens that signaled positive between 12 a.m. and 7 a.m., when the laboratory was closed. A total of 280 bottles were sampled after 5 days of incubation following a final negative report.
Specimen processing.
The samples were collected at the same time as Gram stain samples, with the standard airway vent needle used for piercing the septum on the blood culture bottle. For specimen processing, a 0.4-ml aliquot of sample was added to a microcentrifuge tube containing 1.1 ml of a saponin-based wash solution and was mixed with a vortex machine for 1 s. The sample was then centrifuged for 1 min at 10,000 × g to pellet the organism. The supernatant was immediately aspirated, and another 1.1 ml of wash solution was added and mixed briefly with a vortex machine. The specimen was then centrifuged for 1 min, the supernatant was aspirated, and pellets were frozen at −70°C until further analysis.
Probe matrix detection.
DNA probe assays were performed at Gen-Probe Incorporated by use of a probe matrix directed towards rRNA targets. Probe analysis was performed by using a hybridization protection assay (HPA) as described previously (3). Briefly, the HPA employs chemiluminescent acridinium ester-labeled single-stranded DNA probes complementary to a highly conserved bacterial or fungal rRNA region. The labeled DNA probe combines with the complementary rRNA to form a stable DNA-RNA hybrid, and unhybridized probe is selectively hydrolyzed. A chemiluminescent reaction is then initiated in the presence of base and hydrogen peroxide, and the amount of luminescence is quantified in relative light units (RLU). Gen-Probe Incorporated supplied all proprietary reagents employed in the assay.
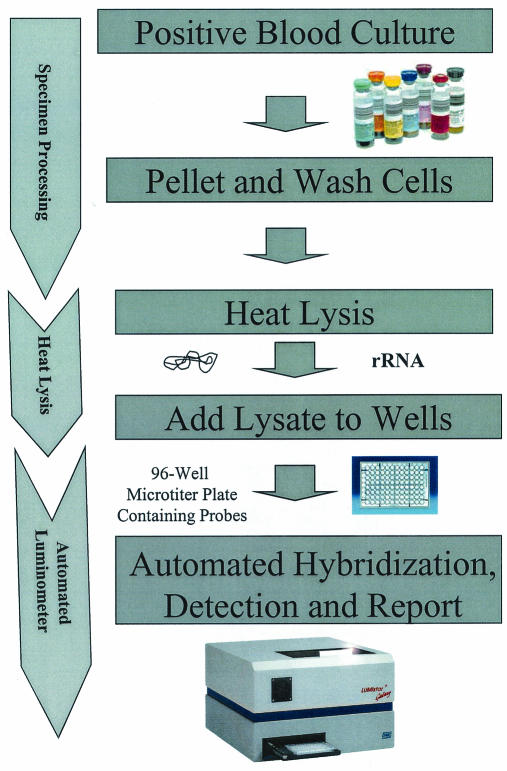
The probe cocktails were placed into white 96-well microtiter plates (Whatman, Clifton, N.J.) and dried until use. A single column of the microtiter plate contained seven individual probe cocktails, with the remaining well used as a sample background control (Fig. 1). The frozen blood culture test pellets were thawed and lysed at 100°C in 250 μl of a succinate-buffered detergent cocktail for 10 min. After the addition of an equal volume of hybridization buffer, 50-μl aliquots of the sample were placed into each of the eight wells in an individual column on the reaction plate. The reactions then had a 50-μl overlay of mineral oil added to prevent evaporation. The reactions were hybridized at 60°C and run through the HPA steps automatically by a luminometer (Lumistar 500; BMG Technologies, Offenburg, Germany) (1, 12). When the RLU levels from the HPA results were outside of the linear range of the luminometer, the lysed specimens were diluted 10-fold and then retested. Through a series of deductive reasoning steps based upon known taxonomic relationships, ID of the organism in the sample falls into categories, as shown in Table 1. A flow chart summarizing the probe matrix ID system is shown in Fig. 2.
TABLE 1.
Probe matrix identification key
| Well reactions | ID |
|---|---|
| 2 | Fungal, not C. albicans |
| 2, 7 | C. albicans |
| 1 | Gram negative, not Enterobacteriaceae, P. aeruginosa, or Campylobacter |
| 1, 7 | P. aeruginosa |
| 1, 6 | Campylobacter subgroup |
| 1, 5 | Enterobacteriaceae, not E. coli |
| 1, 5, 7 | E. coli |
| 1, 3, 4 | Gram positive, Actinomycetes (high G+C content) |
| 1, 3 | Gram positive, low G+C content |
| 1, 3, 5 | Enterococcus |
| 1, 3, 6, 7 | S. aureus |
| 1, 3, 6 | Staphylococcus, not S. aureus |
| 1, 3, 7 | S. pneumoniae |
FIG. 2.
Flow chart showing specimen processing, probe hybridization, and detection for the probe matrix ID system for positive blood cultures.
RESULTS
The overall correlation between conventional and DNA probe matrix ID was excellent, with a sensitivity and specificity of 100 and 96.8%, respectively. A total of 685 blood culture samples were screened (Table 2). Of these, 280 blood bottles were negative after 5 days of incubation in the BacT/Alert3D instrument and were included as negative controls. None of these negative bottles displayed growth upon blind subculturing, and none were positive by the DNA probe matrix assay.
TABLE 2.
Summary of total positive and negative blood culture bottles
| BacT/Alert3D signal | No. of specimens | No. with growth on subculture | No. with same probe matrix result |
|---|---|---|---|
| Positive | 384 | 384 | 384 |
| False positive | 21b | 0 | 0 |
| Negativea | 280 | 280 | 280 |
Sampled after 5 days of incubation and a negative report.
One specimen revealed gram-negative rods by Gram stain and no growth on subculture.
Four hundred five bottles were signal positive by the BacT/Alert3D instrument, with 384 of 405 samples demonstrating growth when the sample was subcultured on agar medium (Table 2). Examples of reaction patterns seen for some of the more commonly found organisms are shown in Fig. 1. There were 21 bottles that were signaled positive by the instrument but demonstrated no growth upon subculture. All 21 of these samples were negative by the DNA probe matrix test. False-positive cultures are occasionally encountered with automated blood culture systems and are usually attributed to respiration by the eukaryotic cells in the blood samples. The probe matrix test can offer a significant advantage for identifying these cultures, due to the lack of reactivity of the all-bacterial and all-fungal probes. Microorganisms isolated from the 385 signal-positive cultures included 60 (15.6%) Enterobacteriacae isolates, 10 (2.6%) Pseudomonas aeruginosa isolates, 10 (2.6%) other gram-negative bacteria, 40 (10.4%) Staphylococcus aureus isolates, 152 (39.5%) CoNS, 28 (7.3%) Streptococcus isolates, 22 (5.7%) Enterococcus isolates, 21 (5.5%) other gram-positive bacteria, 8 (2.1%) anaerobes, and 16 (4.2%) yeast organisms (Tables 3, 4, 5, and 6).Polymicrobial growth was found in 17 (4.4%) of the cultures (Tables 7 and 8). Discordance was noted for 12 (1.8%) samples, 7 of which were polymicrobial, 2 of which were gram positive, and 3 of which were yeast organisms (Tables 6 and 8).
TABLE 3.
Monomicrobial positive blood cultures: gram-negative organismsa
| Conventional ID | No. of specimens | Probe matrix ID |
|---|---|---|
| Escherichia coli | 39 | E. coli |
| Pseudomonas aeruginosa | 10 | P. aeruginosa |
| Citrobacter freundii | 1 | Enterobacteriaceae, not E. coli |
| Enterobacter aerogenes | 3 | Enterobacteriaceae, not E. coli |
| Enterobacter cloacae | 2 | Enterobacteriaceae, not E. coli |
| Flexispira sp | 2 | Enterobacteriaceae, not E. coli |
| Klebsiella oxytoca | 1 | Enterobacteriaceae, not E. coli |
| Klebsiella pneumoniae | 9 | Enterobacteriaceae, not E. coli |
| Salmonella typhi | 1 | Enterobacteriaceae, not E. coli |
| Serratia marcescens | 2 | Enterobacteriaceae, not E. coli |
| Achromobacter xylosoxidans | 1 | Gram-negative, not Enterobacteriaceae, Campylobacter, or P. aeruginosa |
| Acinetobacter lwoffii | 2 | Gram-negative, not Enterobacteriaceae, Campylobacter, or P. aeruginosa |
| Agrobacterium radiobacter | 1 | Gram-negative, not Enterobacteriaceae, Campylobacter, or P. aeruginosa |
| Bacteroides fragilis | 4 | Gram-negative, not Enterobacteriaceae, Campylobacter, or P. aeruginosa |
| Pasteurella multocida | 1 | Gram-negative, not Enterobacteriaceae, Campylobacter, or P. aeruginosa |
| Pseudomonas putida | 1 | Gram-negative, not Enterobacteriaceae, Campylobacter, or P. aeruginosa |
| Stenotrophomonas maltophilia | 4 | Gram-negative, not Enterobacteriaceae, Campylobacter, or P. aeruginosa |
There were no discrepancies between conventional and probe matrix IDs for gram-negative organisms.
TABLE 4.
Monomicrobial positive blood cultures: gram-positive organisms
| Conventional ID | No. of specimens | Probe matrix ID | No. of discrepancies |
|---|---|---|---|
| Staphylococcus aureus | 40 | S. aureus | 0 |
| CoNS | 152 | Staphylococcus, not S. aureus | 0 |
| Streptococcus pneumoniae | 7 | S. pneumoniae | 0 |
| Enterococcus faecalis | 10 | Enterococcus | 0 |
| Enterococcus faecium | 11 | Enterococcus | 0 |
| Enterococcus sp. | 1 | Enterococcus | 0 |
| Corynebacterium sp. | 8 | Actinomycetes (high G+C) | 2 |
| Propionibacterium acnes | 1 | Actinomycetes (high G+C) | |
| Bacillus sp. | 4 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Clostridium sp. | 1 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Fusobacterium sp. | 1 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Group B Streptococcus | 4 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Lactobacillus sp. | 3 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Micrococcus sp. | 5 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Peptostreptococcus magnus | 1 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Stomatococcus sp. | 1 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Streptococcus bovis | 1 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Streptococcus mitis | 1 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
| Viridans group Streptococcus | 15 | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | 0 |
TABLE 5.
Monomicrobial positive blood cultures: fungal organisms
| Conventional ID | No. of specimens | Probe matrix ID | No. of discrepancies |
|---|---|---|---|
| Candida albicans | 5 | C. albicans | 2 |
| Candida glabrata | 4 | Fungi, not C. albicans | 0 |
| Candida krusei | 1 | Fungi, not C. albicans | 0 |
| Candida parapsilosis | 3 | Fungi, not C. albicans | 0 |
| Candida tropicalis | 2 | Fungi, not C. albicans | 0 |
| Cryptococcus neoformans | 1 | Fungi, not C. albicans | 0 |
TABLE 6.
Monomicrobial discrepancies between conventional ID and probe matrix ID
| Specimen no. | Conventional ID | Probe matrix ID | Comment(s) |
|---|---|---|---|
| 52 | Corynebacterium sp. | Staphylococcus, not S. aureus | Conventional ID based on colony and Gram stain morphology |
| 62 | C. albicans | Fungi, not C. albicans | Germ tube positive, chlamydospores and blastoconidia in clusters, API ID = C. albicans |
| 201 | C. parapsilosis | Fungi, not C. albicans; Staphylococcus, not S. aureus | Probe result repeated with archived specimens |
| 242 | Corynebacterium sp. | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes | Conventional ID based on colony and Gram stain morphology |
| 265 | C. albicans | Fungi, Not C. albicans | Not viable at time of reevaluation |
TABLE 7.
Polymicrobial positive blood cultures: both organisms identified
| Specimen no. | Conventional ID | Probe matrix ID |
|---|---|---|
| 6 | E. faecalis, P. aeruginosa | Enterococcus, small amount of P. aeruginosa |
| 12D | P. aeruginosa, CoNS | Staphylococcus, not S. aureus, small amount of P. aeruginosa |
| 19D | E. coli, viridans group Streptococcus | E. coli, small amount of gram-positive (low G+C) organism |
| 51 | Enterobacter cloacae, Enterobacter sp. | Enterobacteriaceae, not E. coli |
| 56D | Fusobacterium mortiforum, CoNS | Gram-positive (low G+C),a small amount of Staphylococcus, not S. aureus |
| 60 | Clostridium perfringens, CoNS | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes; small amount of Staphylococcus, not S. aureus |
| 127 | Klebsiella oxytoca, Bacillus sp. | Enterobacteriaceae, not E. coli; small amounts of gram-positive (low G+C) organism, not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes |
| 182 | P. aeruginosa, E. faecium | P. aeruginosa, Enterococcus |
| 245 | Klebsiella pneumoniae, Enterobacter sp. | Enterobacteriaceae, not E. coli |
Fusobacterium mortiforum is a gram-negative rod, but phylogenetically, based on rRNA sequencing, falls within the gram-positive lineage (Sydney Finegold, personal communication).
TABLE 8.
Polymicrobial positive blood cultures: probe matrix was unable to discriminate multiple organisms present
| Specimen no. | Conventional ID | Probe matrix ID |
|---|---|---|
| 96 | Enterobacter cloacae, P. aeruginosa | Enterobacteriaceae, small amounts of either P. aeruginosa or E. coli |
| 111 | Viridans group Streptococcus, P. aeruginosa | Gram-positive (low G+C), not Staphylococcus, Enterococcus, S. pneumoniae, or Actinomycetes |
| 165 | S. pneumoniae, CoNS | Staphylococcus, not S. aureus; either S. pneumoniae or P. aeruginosa |
| 203 | P. aeruginosa, CoNS | P. aeruginosa, small amount of S. aureus or Staphylococcus, not S. aureus |
| 227 | Serratia marcescens, P. aeruginosa | Enterobacteriaceae, not E. coli; small amounts of either P. aeruginosa or E. coli |
| 223 | C. glabrata, CoNS, pleomorphic gram-positive rod | Fungi, not C. albicans; Staphylococcus, not S. aureus |
| 253 | Corynebacterium sp., CoNS | Actinomycetes (high G+C); either Campylobacter or Staphylococcus, not S. aureus |
There was complete agreement for all 60 of the Enterobacteriacae. Of the gram-positive organisms, 2 of 267 demonstrated discordant results (Table 6). Specimen 52 was identified conventionally as a Corynebacterium sp., whereas the probe matrix repeatedly identified the isolate as belonging to the Staphylococcus genus. The conventional result was only based on colony and Gram stain morphology. Similarly, specimen 242 was conventionally identified as a Corynebacterium sp., compared to the probe matrix ID of a low-G+C, gram-positive organism that was not a Staphylococcus, Enterococcus, or Streptococcus pneumoniae.
There were 2 of 16 specimens with discrepant fungal IDs for which the probe matrix ID was fungal, but not Candida albicans, while the conventional ID for both was C. albicans (Table 6). Repeat microscopic morphology, API 20C AUX, and germ tube testing gave the same C. albicans results for specimen 62. No further culture-based work could be done with specimen 265, since it was no longer viable.
There were a total of 16 polymicrobial positive blood cultures identified by culture (Tables 7 and 8). Two organisms were correctly identified for 9 of 16 of these cultures (Table 7). For the seven samples listed in Table 8, the polymicrobial specimens 51 and 245 had two different Enterobacteriaceae identified in each sample by conventional methods. Although the probe matrix test correctly identified the sample as containing Enterobacteriaceae, not Escherichia coli, due to the limited number of specific Enterobacteriaceae probes in this version of the matrix, the probe assay was not able to identify the components more precisely. The probe assay was not able to indicate that these specimens were polymicrobial, since the magnitude of the Enterobacteriaceae probe signal was comparable to that of the all-bacterial probe signal. If one of the Enterobacteriaceae had been E. coli, then the difference in magnitude of this species probe versus the all-bacterial and Enterobacteriaceae signals might have indicated a polymicrobial composition. For five of nine of the polymicrobial specimens, the relative strength of the probe signals in different wells indicated that more than 95% of the all-bacterial signal accounted for one unique ID. The remaining 5% of the all-bacterial signal could be correlated to the low-level RLU signal seen in one of the other reactive wells (two of these, specimens 96 and 227, had low levels of P. aeruginosa). After entering a normalization value in the luminometer interpretation software for each probe's RLU value relative to that of the all-bacterial probe (established with pure cultures), the relative amount of each species can be estimated in many cases. The research software package was not used for this study; rather, the interpretation of the results was done manually. Because the routine workup and reporting of blood cultures are generally not done on a semiquantitative basis (i.e., few, moderate, or many), the relative amounts of each organism present in the polymicrobial cultures could not be determined by the culture reports for this study.
For one polymicrobial specimen, the probe matrix was able to identify a mixed culture that standard culturing missed (Table 6). Specimen 201 had a probe matrix ID of Candida, not C. albicans, and a Staphylococcus, not S. aureus, while culturing only identified the Candida parapsilosis component. This result was confirmed with follow-up testing of archived lysed samples.
DISCUSSION
This study evaluated the performance of a DNA probe matrix for rapidly identifying microorganisms directly from positive blood cultures. The two discrepant gram-positive specimens, 52 and 242 (Table 6), were reported by conventional methods to be Corynebacterium sp. These conventional ID results were based on colony and Gram stain morphology only. Corynebacterium is a member of the high-G+C or Actinomycetes grouping of gram-positive bacteria. Other clinically relevant bacteria in this group include Rhodococcus, Nocardia, and Mycobacterium (6). Conversely, gram-positive bacteria commonly found in positive blood cultures that fall into the low-G+C group include organisms such as Staphylococcus, Streptococcus, and Enterococcus. The high-G+C-Actinomycetes well was nonreactive for both specimens, suggesting questionable results. For sample 52, the magnitude of the chemiluminescent signal for the Staphylococcus genus probe was equivalent to the signals for the all-bacterial and gram-positive probes, strongly supporting a monobacterial culture and a proper probe ID. These probe matrix results were repeated on duplicate pellets that had been archived for discrepancy testing. Retrospective checking of this patient's record for results with other blood culture bottles indicated that Staphylococcus was also recorded. Sequencing of the 16S ribosomal DNA was done for these two patient samples, which were grown in subsequent broth cultures to attempt further clarification of the ID of these organisms. Sample 52 showed homology in 16S ribosomal sequence to Staphylococcus hominis. Sequencing of sample 242 showed that the culture was a mix of more than one organism, with one of the partial ribosomal DNA sequences showing possible similarity, but no exact matches, to a Corynebacterium. More extensive sequencing of this isolate and other clinically relevant Corynebacterium isolates is under way to better resolve the identity of this isolate.
Resolution of the two fungal discrepant specimens, samples 62 and 265 (Table 6) required modification of the C. albicans probe in the matrix. We expanded the specificity from a reaction with only C. albicans to a reaction with all members of the C. albicans/Candida dubliniensis group (10). Follow-up testing of archived lysed samples gave a proper probe matrix ID of C. albicans/C. dubliniensis group for both of these specimens.
Mixtures of two different organisms have been accurately identified, and the relative amounts have been accurately estimated, in other research studies in which known amounts of two different organisms were mixed together and subjected to the probe matrix test (data not shown). This feature of the probe matrix assay may not have practical clinical applications since the amount of each organism in the patient sample is not necessarily reflected in the amounts of each organism present when the culture is signaled positive. In 7 of 16 polymicrobial specimens (Table 8), the probe matrix system indicated that a culture was mixed, but it was only able to definitively ID one of the organisms, while the other IDs were inconclusive.
Two of 16 of these polymicrobial specimens were discerned with the Gram stain. Specimen 165 had a conventional ID of S. pneumoniae and CoNS and a probe matrix ID of Staphylococcus, not S. aureus, and either S. pneumoniae or P. aeruginosa. The Gram stain of this specimen showed gram-positive cocci in pairs and clusters, with no indication of a gram-negative rod (P. aeruginosa). Therefore, the combination of the Gram stain result from the initial positive blood culture and the probe matrix result enabled a more definitive rapid ID result for sample 165. Specimen 253 had a conventional ID of Corynebacterium sp. and CoNS, while the probe matrix ID was a member of Actinomycetes and either Campylobacter or Staphylococcus, not S. aureus. The Gram stain result from the initial positive blood culture of this specimen was gram-positive cocci in clusters, with no indication of curved gram-negative rods. In this case, even though the Gram stain did not indicate any pleomorphic gram-positive rods (Corynebacterium), it still enabled a more rapid definitive result for sample 253 because of the absence of a curved gram-negative rod (Campylobacter).
In this study, 1 of 21 of the false positives did show a positive Gram stain result, implying that it was not a false positive result, but rather an organism which was nonviable upon subculture. It was also probe negative at the time of testing. The rapid loss of a viable organism in the late stages of cultivation is possible with certain fragile species, such as S. pneumoniae. If an organism were to autolyse, nothing would be seen on subculture, the rRNA would degrade quickly, and no probe signal would result. The pattern of test results seen with this sample could represent either an autolysis type of event or possibly an error in the interpretation or transcription of the Gram stain. Retrospective checking of this patient's record found no other positive cultures.
The blood culture system used for the probe matrix test does not affect the results. For this study, we used standard BacT/Alert bottles. However, in other studies, BACTEC plus aerobic and anaerobic blood culture bottles, which contain resin, have been used for the probe matrix test (Fuller and Davis, 39th ICAAC). Initial studies evaluating the effect of various blood culture media on the probe system demonstrated that even bottles containing activated charcoal did not interfere with the results (data not shown).
The probe matrix sample acquisition procedure utilizes the same blood bottle access system as that used for preparing the Gram stain and subculture plates. Specimen processing does not require nucleic acid extraction and purification. With batch testing, approximately 12 to 24 specimens can be processed in 15 min. The remaining DNA probe assay is automated, and final results are available 40 min later. The probe matrix result could be available within 1 h of the Gram stain result. As pointed out above, the combination of the Gram stain and probe matrix results in some cases improves the informational content for polymicrobial samples. In a recent study, Munson et al. (11) found that the most important information provided by the clinical microbiology laboratory in respect to antimicrobial management of bloodstream infections was the initial phone call reporting the positive blood culture and Gram stain. Giving a probe matrix result at the initial physician contact could result in prudent therapy. For example, the physician would be able to implement therapy for either a Pseudomonas or Enterobacteriaceae infection much sooner with the probe matrix result than with a Gram stain result that would only indicate gram-negative bacilli.
Workflow is always an issue when new tests are considered for the clinical laboratory. Because these specimens were processed at UCLA and later tested at Gen-Probe Incorporated, the issue of total workflow has not been fully examined in this particular study. Other research studies involving automated blood culture systems with immediate testing of positive blood cultures ascertained that the probe matrix was amenable to the standard workflow, and additional studies are planned to further evaluate cost savings, ergonomics and clinical outcome (7; Fuller and Davis, 39th ICAAC).
Overall, the rapid DNA probe matrix assay was able to provide ID information for nearly all of the positive blood cultures at a level sufficient to enhance clinical decisions. Further improvements have been made to the probe matrix system, including the addition of more species-level probes. Other versions of the probe matrix could be tailored to include previously developed probes for organisms contained in other specimen types, such as sputum, urine, throat swabs, or cerebrospinal fluid. Additional probes may include those for group A Streptococcus, group B Streptococcus, Haemophilus influenzae, Proteus mirabilis, Cryptococcus neoformans, Histoplasma capsulatum, Mycobacterium avium, Mycobacterium tuberculosis, and other Candida species or groups. In addition to the use of the probe matrix assay on positive blood cultures, we applied it to isolated colonies with equal success. Investigations are currently under way to optimize its utility for reliable ID of organisms present in positive blood cultures. A reflex test for methicillin-resistant Staphylococcus and vancomycin-resistant Enterococcus found in positive blood cultures or from isolated colonies is also currently being evaluated (J. J. Hogan, S. K. Kaplan, E. M. Marlowe, and D. A. Bruckner, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. C-084, 2003). This would allow important antibiotic susceptibility results to also be included in the initial communication with the physician.
REFERENCES
- 1.Arnold, L. J., P. W. Hammond, W. A. Wiese, and N. C. Nelson. 1989. Assay formats involving acridinium-ester-labeled DNA probes. Clin. Chem. 35:1588-1594. [PubMed] [Google Scholar]
- 2.Brecher, M. E., D. G. Heath, S. N. Hay, S. J. Rothenberg, and L. C. Stutzman. 2002. Evaluation of a new generation of culture bottle using an automated bacterial culture system for detecting nine common contaminating organisms found in platelet components. Transfusion 42:774-779. [DOI] [PubMed] [Google Scholar]
- 3.Brecher, M. E., J. J. Hogan, G. Boothe, A. Kerr, L. McClannnan, M. R. Jacobs, R. Yomtovian, V. Chongokolwatana, G. Tegtmeier, S. Henderson, A. Pineda, V. Halling, M. Kemper, K. Kuramato, P. V. Holland, and M. Longiaru. 1994. Platelet bacterial contamination and the use of a chemiluminescence-linked universal bacterial ribosomal RNA gene probe. Transfusion 34:750-755. [DOI] [PubMed] [Google Scholar]
- 4.Davis, T. E., and D. D. Fuller. 1991. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J. Clin. Microbiol. 29:2193-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Brik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox, G. E., K. R. Pechman, and C. R. Woese. 1977. Comparative cataloguing of 16S ribosomal ribonucleic acid: molecular approach to prokaryotic systematics. Int. J. Syst. Bacteriol. 27:44-57. [Google Scholar]
- 7.Hertz, D., D. Fuller, T. Davis, L. Papile, D. Stevenson, and J. Lemmon. 1999. Comparison of DNA probe technology and automated continuous-monitoring blood culture systems in the detection of neonatal bacteremia. J. Perinatol. 19:290-293. [DOI] [PubMed] [Google Scholar]
- 8.Jansen, G. J., M. Mooibroek, J. Idema, H. J. M. Harmsen, G. W. Welling, and J. E. Degener. 2000. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J. Clin. Microbiol. 38:814-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maquelin, K., C. Kirschner, L.-P. Choo-Smith, N. A. Ngo-Thi, T. van Vreeswijk, M. Stämmler, H. P. Endtz, H. A. Bruining, D. Naumann, and G. Puppels. 2003. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 41:324-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milliman, C. L., G. Bee, and J. J. Hogan. December2002. Polynucleotide probes for detection and quantitation of Candida albicans and Candida dubliniensis. U.S. patent 6,495,327.
- 11.Munson, E. L., D. J. Diekema, S. E. Beekmann, K. C. Chapin, and G. V. Doern. 2003. Detection and treatment of blood stream infection: laboratory reporting and antimicrobial management. J. Clin. Microbiol. 41:495-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson, N. C., M. A. Reynolds, and L. J. Arnold, Jr. 1995. Detection of acridinium esters by chemiluminescence, p. 392-429. In L. J. Kricka (ed.), Nonisotopic probing, blotting and sequencing. Academic Press, New York, N.Y.
- 13.Oliveira, K., G. W. Procop, D. Wilson, J. Coull, and H. Stender. 2002. Rapid identification of Staphyloccocus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 40:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigby, S., G. W. Procop, G. Haase, D. Wilson, G. Hall, C. Kurtzman, K. Oliveria, S. Von Oy, J. J. Hyldig-Nielson, J. Coull, and H. Stender. 2002. Fluorescence in situ hybridization with peptide nucleic acid probes for rapid identification of Candida albicans directly from blood culture bottles. J. Clin. Microbiol. 40:2182-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sen, Y., and D. M. Asher. 2001. Multiplex PCR for detection of Enterobacteriaceae in blood. Transfusion 41:1356-1364. [DOI] [PubMed] [Google Scholar]
- 16.Volkhard, A., J. Kempf, K. Trebesius, and I. B. Autenrienth. 2000. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 38:830-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen Tan, T., S. Corden, R. Barnes, and B. Cookson. 2001. Rapid identification of methicillin-resistant Staphylococcus aureus from positive blood cultures by real-time fluorescence PCR. J. Clin. Microbiol. 39:4529-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]