Abstract
Investigations of the population genetics of Bartonella henselae have demonstrated a high level of diversity among strains, and the delineation of isolates into one of two subtypes, type I (Houston) and type II (Marseille), represented by specific 16S ribosomal DNA (rDNA) sequences, has long been considered the most significant genotypic division within the species. This belief is challenged by recent work suggesting a role for horizontal gene exchange in generating intraspecies diversity. We attempted to resolve this issue and extend exploration of the population structure of B. henselae by using multilocus sequence typing (MLST) to examine the distribution of polymorphisms within nine different genes in a sample of 37 human and feline isolates. MLST distinguished seven sequence types (STs) that resolved into three distinct lineages, suggesting a clonal population structure for the species, and support for these divisions was obtained by macrorestriction analysis using pulsed-field gel electrophoresis. The distribution of STs among isolates recovered from human infections was not random, and such isolates were significantly more often associated with one particular ST, lending further support to the suggestion that specific genotypes contribute disproportionately to the disease burden in humans. All but one isolate lay on lineages that bore the representative strain of either the Houston or Marseille subtype. However, the distribution of the two 16S rDNA alleles among the isolates was not entirely congruent with their lineage allocations, indicating that this is not a sensitive marker of the clonal divisions within the species. The inheritances of several of the genes studied could not be reconciled with one another, providing further evidence of horizontal gene transfer among B. henselae strains and suggesting that recombination has a role in shaping the genetic character of bartonellae.
Bartonella henselae is now well established as a significant human pathogen and is possibly the agent of the world's most common bacterial zoonosis acquired from a companion animal. The bacterium is naturally maintained through persistent, subclinical infections in felids and is transmitted between reservoir hosts via arthropods (Ctenocephalides felis). The prevalence of B. henselae bacteremia in domestic cats ranges from about 10 to 40% (7, 17), and infections have been encountered virtually worldwide. Cat scratch disease (CSD) is the most commonly encountered B. henselae-induced syndrome in humans, with an estimated 25,000 cases and several thousand hospitalizations occurring each year in the United States alone (16, 19, 21, 34). B. henselae is also unique in its invasion mechanism (9) and capacity to drive angiogenesis in vitro and in vivo (20). Although the prevalence of reported CSD and other B. henselae infections is relatively high (10 cases per 100,000 population per annum in the United States), the frequency of human inoculation by B. henselae is likely to be considerably greater, given the high carriage rates in domestic cats and the proximity in which cats and humans live. A recent survey of Australian blood donors detected seropositivity in 5% of samples tested (7), and B. henselae was detected in the bloodstream of 10% of attendees at a South African AIDS clinic (14). These observations suggest that B. henselae infections are commonly subclinical and/or markedly underreported, as problems with current diagnostic approaches are well recognized (13, 24, 25, 27).
Genotypic analyses of B. henselae isolates using a variety of different pangenomic or locus-specific typing approaches have identified a number of genetic groupings. These analyses have been applied not only to epidemiological investigations of human B. henselae infections but also to broader surveys of human and feline isolate collections. Such surveys have revealed limited diversity among human-infecting isolates (2, 5, 10) and even some type-specific differences in virulence (8, 31, 35) but, as yet, no direct evidence for any particular hypervirulent genotypes. Several B. henselae genes and genetic loci have been subjected to comparative analysis for typing purposes, including the 16S and 23S ribosomal DNA (rDNA), 16S-23S rDNA intergenic spacer region, and fragments of protein-encoding genes such as gltA, ftsZ, and pap31 (6, 37, 38). Examination of 16S rRNA-encoding gene sequences has revealed that B. henselae isolates possess one of two different sequence types, with strains being designated as type I or type II on this basis (5). Although the true significance of this heterogeneity remains uncertain, representative members of each type are serologically distinguishable (11) and possess consistently different protein profiles (15, 23). The type I (Houston)-type II (Marseille) delineation of B. henselae has thus become widely accepted as a consistent and meaningful phylogenetic division within the species.
However, other approaches to B. henselae genotyping are not entirely supportive of this delineation. One study has reported that the distributions of pap31 and groEL alleles among a small panel of B. henselae isolates were the same as that of 16S rDNA alleles (36), whereas a separate investigation has revealed noncongruence between the distributions of alleles of 16S rDNA and the citrate synthase-encoding gene (gltA) among a larger sample of isolates (10). Furthermore, pangenomic fingerprinting using arbitrarily primed (AP)-PCR, enterobacterial repetitive intergenic consensus (ERIC)-PCR, and infrequent restriction site (IRS)-PCR as well as macrorestriction analysis using pulsed-field gel electrophoresis (PFGE) have also indicated imperfect correlation with 16S rDNA type (2, 10).
Multilocus sequence typing (MLST) is a relatively new typing method that groups bacteria based on comparison of nucleic acid sequences derived from the internal fragments of a number (typically seven) of housekeeping genes. This approach is particularly powerful as, unlike the raw data obtained by other approaches to bacterial strain typing, nucleic acid sequences are unambiguous and can be stored and transferred between laboratories electronically with complete reliability. MLST as a typing tool has been developed for a number of gram-negative (26) and gram-positive bacteria (29). In this paper, MLST is applied to the study of B. henselae population structure for the first time. The approach is used for the characterization of 37 previously described B. henselae isolates derived from humans and cats from Australia and other countries. MLST results are compared to those obtained using conventional DNA fingerprinting methods.
MATERIALS AND METHODS
B. henselae isolates and growth conditions.
The B. henselae isolates analyzed in this study are a subset of those described previously (10) and are listed in Table 1. Berlin-2 is a feline isolate grown from the pet cat of a patient with CSD in Germany (3). Isolates were grown on chocolate blood agar plates (Blood Agar Base No. 2; Oxoid) supplemented with 5% defibrinated horse blood (BioMerieux) at 37°C in a 5% CO2-enriched atmosphere.
TABLE 1.
Genotypic characteristics of the 37 B. henselae isolates studied
| Isolate | Host of origina | Distribution of alleles at the nine loci studied
|
ST | ERIC type | AP type | IRS type | PFGE type | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16S | batR | eno | gltA | ftsZ | groEL | nlpD | ribC | rpoB | |||||||
| 49882 (H-1) | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 1 |
| 49793 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 1 | 1 |
| BH2 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 1 | 1 |
| BH3 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | NTb |
| BH5 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 1 |
| HC54 | F | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 1 |
| JR1 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 3 |
| JR3 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 3 |
| JR5 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 3 |
| JR6 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 3 |
| JR7 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 3 |
| JR8 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 3 |
| JR9 | H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 3 |
| NU4695 | F | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 1 | 2 |
| RMC3 | F | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 2 | 2 |
| RMC10 | F | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | H | 1 | 2 |
| JR2 | H | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 4 | H | 2 | 1 |
| HC62 | F | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | H | 2 | 5 |
| HC35 | F | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 4 | 3 | H | 1 | 4 |
| HC60 | F | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 4 | 3 | H | 1 | NT |
| RMC1 | F | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 4 | 1 | H | 2 | 5 |
| RMC8 | F | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 4 | 3 | H | 2 | 5 |
| RMC12 | F | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 4 | 3 | M | 1 | 5 |
| NU4714 | F | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 | 4 | H | 1 | 5 |
| R987 | H | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 | 4 | H | 2 | 4 |
| R1073 | H | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 | 4 | H | 2 | 4 |
| BH4 | H | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 4 | M | 2 | 6 |
| HC48 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 2 | M | 2 | 6 |
| HC71 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 2 | M | 2 | NT |
| NU4423 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 4 | H | 2 | 6 |
| NU4713 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 3 | H | 2 | 6 |
| RMC2 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 6 | M | 2 | 6 |
| RMC5 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 1 | M | 2 | NT |
| RMC7 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 5 | M | 2 | 6 |
| RMC11 | F | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 7 | M | 1 | 6 |
| URLLY8 | H | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 | 4 | M | 2 | 6 |
| BERL-2 | F | 2 | 4 | 1 | 2 | 3 | 1 | 2 | 2 | 1 | 7 | 5 | M | 2 | 7 |
H, human infection; F, feline infection.
NT, not tested.
DNA fingerprinting methods.
Macrorestriction analysis using PFGE of SmaI-digested genomic DNA and computer-assisted designation and analysis of PFGE fingerprints were carried out as described previously (2). AP-PCR, ERIC-PCR, and IRS-PCR typing have been reported previously (10).
MLST.
Internal fragments of approximately 300 to 500 bp were amplified from each of nine genetic loci and evaluated for use in the MLST scheme (Table 2). The gltA sequence data for all isolates and the 16S rDNA sequence data for some of the isolates examined have been reported previously (10), and all were here sequenced and many resequenced on both strands for confirmation. DNA extracts were prepared by using a commercially available kit (Promega Corporation, Madison, Wis.) according to the manufacturer's instructions and stored at 4°C. Primers for amplification and sequencing were designed using Prime software (Genetics Computer Group) (ftsZ, groEL, and ribC) or Primer Designer 4 software (SciEd Central; Scientific and Educational Software) (batR, eno, nlpD, and rpoB) and are detailed in Table 2. Reaction mixtures (30 to 50 μl) contained the following (at the indicated final concentrations): 1× Mastermix (ABGene) or 1× NH4 buffer (Bioline) with 1 U of BioTaq DNA polymerase (Bioline), deoxynucleoside triphosphates (dATP, dCTP, dGTP, and dTTP) (200 μM each), MgCl2 (2.5 mM), 20 pmol or 200 nM concentrations of each primer (from MWG or GeneWorks), 1 to 2 μl of DNA template, and sterile distilled water. PCR amplifications varied slightly with the primers but were generally as follows: denaturation at 95 to 96°C for 5 min, followed by 35 to 40 cycles (94 to 96°C for 1 min, 55 to 60°C for 10 to 60 s, and 72°C for 45 to 50 s) and a final extension step at 72°C for 6 to 10 min.
TABLE 2.
Primers used for amplification and sequencing of the nine loci evaluated for the B. henselae MLST schemea
| Locus | Putative gene product | Product size (bp) | Position on gene (GenBank no.) | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Reference |
|---|---|---|---|---|---|---|
| 16S rDNA | RNA small subunit | 511 | 10-521 (M73229) | AGAGTTTGATCCTGGYTCAG | CTTTACGCCCARTAAWTCCG | 5 |
| batR | Two-component regulator | 487 | 69-555 (AJ300267) | GACCGCAATATTTTGACATC | GCATCCATCAAAGCATCACGACTT | This study |
| eno | Enolase | 472 | 378-849 (AY074768) | AGCAGCAGAGTCATTGTC | CTCAGCCATACCATCTTC | This study |
| ftsZ | Cell division protein | 522 | 1095-1616 (AF061746) | GCCTTCTCATCCTCAACTTC | CTTTGTTTTAAACGCTGCC | This study |
| gltA | Citrate synthase | 379 | 782-1160 (L38987) | GGGGACCAGCTCATGGTGG | AATGCAAAAAGAACAGTAAACA | 30 |
| groEL | Hsp60 chaperone | 405 | 1185-1589 (AF014829) | GTTGATGATGCCTTGAAC | TGGTGTGTCTTTCTTTGG | This study |
| nlpD | Cell surface glycoprotein | 494 | 1061-1554 (AF484425) | GGCGCTGGTATGATACAA | GACATCTGTGCGGAAGAA | This study |
| ribC | Riboflavin synthase | 321 | 1230-1550 (AJ132928) | AGCGAGGATCAAAACAAC | GCTCTTCAACACAATTAACG | This study |
| rpoB | RNA polymerase beta subunit | 471 | 1702-2172 (AF171070) | CGTGACGTACATCCTACA | AACAGCAGCTCCTGAATC | This study |
All reference sequences were obtained from B. henselae Houston-1T, with the exception of AJ300267 (batR), which was obtained from Bartonella bacilliformis KC583T. Y = C or T; R = A or G; W = A or T.
Amplification products were purified (GeneClean Spin kit from Bio101 or Qiaquick PCR purification kit from Qiagen) and then sequenced directly using an ABI PRISM Big Dye cycle sequencing ready reaction kit (Perkin-Elmer) and an ABI 373 or 377 DNA sequencer (Applied Biosystems). All DNA sequences were analyzed manually for polymorphism, which was confirmed by multiple sequence alignments by use of EclustalW (ANGIS), Pileup and Pretty software (Genetics Computer Group), or Align Plus 4 software (SciEd Central). For every locus, alleles were assigned a number according to the order in which they were encountered (although alleles possessed by the B. henselae type strain, Houston-1T, were always assigned allele number 1 for each locus examined). For each isolate, the combination of alleles at each of the loci examined (the allelic profile) defined the sequence type (ST), and these were also assigned arbitrary numbers in the order of encounter. The nucleotide numbering follows that for sequences assigned the following GenBank accession numbers: AJ300267, batR; AY074768, eno; AF061746, ftsZ; AF014829, groEL; AF484425, nlpD; AJ132928, ribC; and AF171070, rpoB (Table 2).
Analysis of MLST data.
Cluster analysis of the 37 isolates from a matrix of pairwise similarities between the allelic profiles was performed by the unweighted pair-group method with arithmetic averages (UPGMA). The definition of clonal complexes and the examination of relationships between STs within clonal complexes were carried out using BURST analysis (http://burst.mlst.net).
Nucleotide sequence accession numbers.
Newly encountered alleles have been submitted to GenBank under the following accession numbers: AY289790, batR allele 1 (Houston-1); AY289791, batR allele 2 (HC35); AY289792, batR allele 3 (URLLY8); AY289793, batR allele 4 (Berlin-2); and AY289794, nlpD allele 2 (Berlin-2).
RESULTS
Development of an MLST scheme for B. henselae.
Sequence data were obtained for the nine selected genetic loci from all 37 isolates included in the study. The amount of sequence data obtained at each locus ranged from 321 to 522 nucleotides (Table 2), with the proportion of sites at which nucleotide variation was observed within each locus examined ranging from 0 (eno) to 0.99% (groEL). The number of different alleles (those which differed in DNA sequence) which were encountered at each locus was low, ranging from 1 (eno) to 4 (batR) (Table 3). Nucleotide variations observed within the 16S rDNA, ftsZ, gltA, groEL, ribC, and rpoB fragments of the isolates examined were consistent with those reported in earlier studies (4, 12, 36), and no additional alleles of any of these genes were encountered. 16S sequences conflicted in three instances with those previously reported (10) due to disagreement with the primer-based method of assignment (5) we had originally used. This was resolved by the complete sequencing of all 16S loci.
TABLE 3.
Characteristics of the nine loci evaluated for the B. henselae MLST scheme
| Locus | No. (%) of variable sites | No. of alleles | Allelic polymorphism, by reference to Houston-1T (allele 1) |
|---|---|---|---|
| 16S rDNA | 4 (0.78) | 2 | 2: TTAG 143-146 TATTT |
| batR | 4 (0.82) | 4 | 2: T302C |
| 3: C65T, C68A, A218G, T302C | |||
| 4: C65T, A218G, T302C | |||
| eno | 0 | 1 | |
| ftsZ | 4 (0.77) | 3 | 2: A1185T, G1404A, G1467T |
| 3: A1185T, G1467T, C1537T | |||
| gltA | 2 (0.53) | 2 | 2: G648A, C1026T |
| groEL | 1 (0.25) | 2 | 2: G1343A |
| nlpD | 1 (0.20) | 2 | 2: G1453A |
| ribC | 4 (1.25) | 2 | 2: A1310G, G1360A, A1494G, G1499A |
| rpoB | 1 (0.21) | 2 | 2: G1779A |
Identity of B. henselae isolates at selected genetic loci.
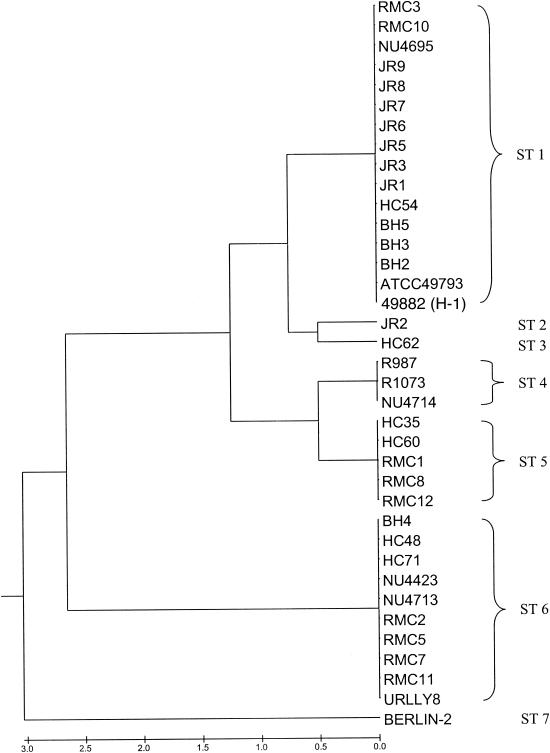
The dendrogram shown as Fig. 1 was constructed from a matrix of pairwise allelic differences between the STs of the 37 isolates studied. Seven STs were encountered among the isolates. ST1 was the most commonly encountered ST (16 isolates), while other STs contained between 1 and 10 members. On the basis of calculated linkage distances, the seven STs were assigned to three lineages. One of these lineages constituted a clonal complex (the ST1 complex), containing five STs and 26 isolates. BURST analysis supported the delineation of this complex and proposed ST1 as the founder ST within the complex. The remaining two lineages comprised single STs (ST6, 10 isolates; ST7, 1 isolate).
FIG. 1.
Dendrogram of 37 isolates of B. henselae constructed by UPGMA cluster analysis of MLST data. The seven STs to which the isolates were assigned are indicated at right.
That 36 of the 37 isolates examined lay in one of two lineages reinforces the notion of quite distinct Marseille (ST6) and Houston (ST1 complex) subtypes. Membership of the Houston subtype was characterized by distinct ftsZ and gltA alleles in this sample, whereas all Marseille subtypes shared specific batR, ftsZ, and rpoB alleles.
Distribution of 16S rDNA alleles does not reflect clonality.
Eleven of the 37 isolates possess STs different from that of the Houston-1 type strain and that of URLLY8, the representative strain of the Marseille subtype. Two of these 11 isolates were proven human pathogens from the same region of Australia. Significantly, the 16S rDNA type II (Marseille) allele (allele 2) was found to be present in lineages carrying both the Houston-1 type strain and the URLLY8 Marseille subtype. In both lineages, the 16S rDNA allele 2 was encountered in isolates recovered from humans and cats.
Evidence of recombination among B. henselae isolates.
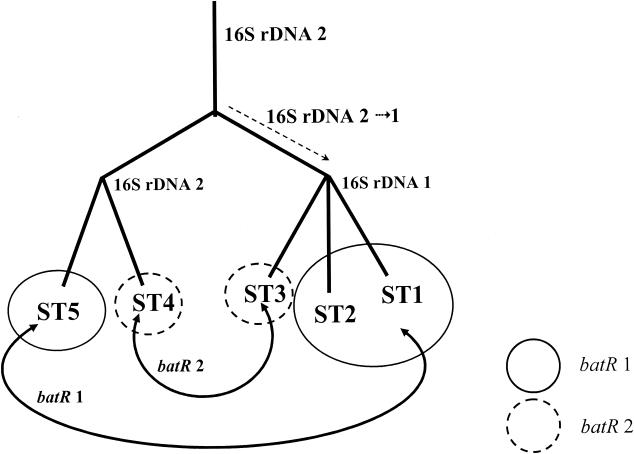
The distribution of alleles among the isolates examined suggested that, in general, loci had been inherited in a congruent manner. However, the observed distributions of batR and groEL alleles suggested that the inheritance of these loci could not be reconciled with those of other loci. As demonstrated in Table 1, only ST1 and ST7 strains possess groEL allele 1, a distribution that suggests a clustering of two STs that lie apart in relatedness assessments based on other loci. Furthermore, within the ST1 complex, the inheritance of 16S rDNA alleles (specifically the evolution of allele 1 by a common ancestor of STs 1 to 3) is not congruent with the inheritance inferred from distribution of batR alleles 1 and 2 among STs 1 to 5 (Table 1). These two observations can be reconciled by evoking horizontal genetic transfer among B. henselae strains, as demonstrated for batR alleles in Fig. 2.
FIG. 2.
Evidence for horizontal genetic transfer within the ST1 clonal complex: lack of congruence between 16S rDNA and batR allelic inheritance. The 16S rDNA allele 1 is fixed in a common ancestor of STs 1, 2, and 3 following its divergence from strains possessing the ancestral allele 2. Distribution of batR alleles among the STs within the complex suggests horizontal transfer of either allele 1 between an ST1-ST2 common ancestor and ST5 or allele 2 between ST3 and ST4.
Relationships between MLST and data derived from other approaches to genotyping.
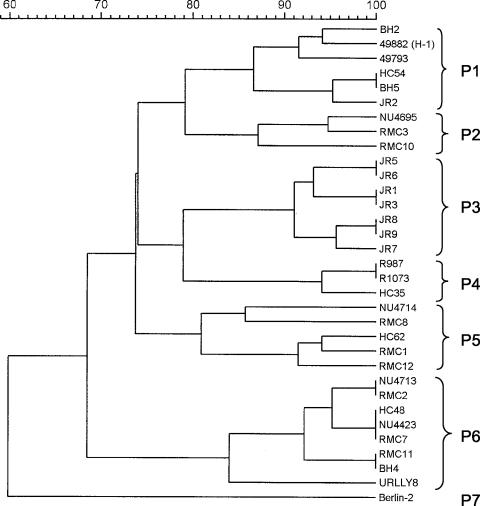
A dendrogram derived from UPGMA analysis of SmaI restriction digest PFGE profiles for 33 of the 37 isolates is presented in Fig. 3. Groupings within this dendrogram, as defined by a Dice coefficient of 81%, are presented in Table 1. Previously assigned groupings derived from AP-PCR, ERIC, and EagI-HhaI IRS-PCR analyses of all 37 isolates are also presented in Table 1.
FIG. 3.
Dendrogram of 33 isolates of B. henselae constructed by UPGMA cluster analysis of PFGE profiles. The seven PFGE types to which the isolates were assigned are indicated at right.
PFGE analysis groups the isolates into three lineages that are identical to those obtained by MLST (Fig. 3). The two MLST lineages that contain single STs (ST6 and ST7) each possess a single and unique PFGE type (P6 and P7). P6 includes human and feline isolates from temporally and geographically disparate sources, whereas P7 contains only one feline isolate. Within the ST1 clonal complex, five PFGE types, P1 to P5, were encountered. The three PFGE types noted among ST1 isolates were almost exclusive to the ST, with the exception being P1, which was possessed by the single ST2 isolate, JR2. The remaining eight isolates from the other three STs (ST3 to ST5) within the ST1 clonal complex yielded two PFGE types (P4 and P5), neither of which was ST specific but rather was shared by different STs.
ERIC typing proved to be less discriminating than MLST, and the two approaches to strain delineation are generally incongruent. However, although four different STs are encountered among the common E4 group, representing two of the three B. henselae lineages, all ST1, -2, and -5 isolates (within the ST1 clonal complex) were E4 members. Conversely, ST6 included strains belonging to all seven ERIC types. EagI-HhaI IRS-PCR fingerprinting did not correlate well with MLST, PFGE, or other typing methods.
Relationships between ST, isolate host, and country of isolation.
Of the 37 isolates examined, 17 were isolated from humans and 20 from cats. Four of the seven STs were found within the 17 human isolates and all except ST2 were found among the 20 feline isolates. However, the distribution of isolates from each host among the STs was not uniform. Of the 16 ST1 strains, 12 were of human origin, compared with 5 of the 21 non-ST1 strains. The overrepresentation of human isolates in ST1 was statistically significant (chi-squared, P < 0.05). As our strain collection was assembled opportunistically, we attempted to further test the validity of the association described above by using a subset of isolates originating within the state of New South Wales (strains with BH, HC, or RMC prefixes). Three of six ST1 isolates but only one of 13 non-ST1 isolates were of human origin. This difference has statistical support (chi-squared, P < 0.05). The feline isolates we examined were more evenly distributed among STs, with 11 of 19 belonging to the ST1 clonal complex and 8 of 19 belonging to ST6. No ST possessed a statistically significant over- or underrepresentation of feline isolates.
As 33 of the 37 isolates used in our study were of Australasian origin, exploration of a correlation between geography and ST variation was not possible. However, ST1, ST5, and ST6 comprise isolates from Australia and New Zealand, and ST1 and ST6 comprise isolates from Australasia and the United States or France. The Berlin-2 strain, isolated in Germany (3), is an outlier in both MLST and PFGE analyses. This strain possessed four alleles (batR 4, ftsZ 3, nlpD 2, and ribC 2) not found in the Australasian population.
DISCUSSION
The importance of a portable, reproducible, and discriminatory approach to delineating B. henselae strains is now more apparent than ever before. Although an increasing number of genotypic assessments of isolates are being published, broader insights into the population structure of the species are obscured by methodological differences between studies. Furthermore, the reliability of the pangenomic fingerprinting schemes that have been employed in the majority of these investigations must now be questioned following evidence for intragenomic rearrangement by B. henselae both in vivo and during in vitro passage. The introduction of strain typing based on the comparison of DNA sequences, reflecting genomic content rather than organization, circumvents this problem and therefore potentially offers a reliable indicator of the genetic identity of B. henselae strains. Furthermore, and of particular importance to fastidious microorganisms such as bartonellae that are notoriously difficult to isolate in most disease-associated situations, MLST can be applied directly to infected material, permitting the genotyping of uncultured strains.
Whereas most prior evaluations of MLST in other bacterial species have used large representative strain panels, no accessible collection of B. henselae strains representing the geographical and genetic diversity of the species has yet been assembled. We reasoned that a demonstration of the value of MLST in defining lineages among a limited set of well-characterized strains would provide us with an indication as to its value when applied to a larger, more representative collection. Our choice of loci included fragments of genes encoding well-characterized housekeeping functions (16S rDNA, eno, ftsZ, gltA, groEL, ribC, and rpoB) together with other less-well-defined proteins. BatR encodes a putative homologue of the Brucella abortus regulatory protein BvrR (33), whereas the product of nlpD encodes a putative cell surface glycoprotein of unknown function. While the ribC locus had the greatest number of variable positions, there were only two alleles observed. Thus, the number of variable positions within each locus is not a direct reflection of genetic divergence. Although alleles at all loci differed very little, some by as little as a single base change, we were confident of the accuracy of our observations because (i) all amplification products were sequenced directly rather than by subcloning of single amplicons, (ii) all loci were sequenced at least once on both strands and, (iii) most alleles were possessed by more than one isolate. The occurrence of the same one or two altered nucleotides in different strains as a result of coincident, independent mutation (homoplasy) is extremely unlikely. Among the protein-encoding loci, most alleles encountered (4) belonged to batR, and these varied at only four (synonymous) positions within almost 500 bp. A Marseille subtype-like strain (Fizz) has been reported to be distinguishable from both Marseille (URLLY-8) and Houston-1 (ATCC 49882) representative strains on the basis of groEL sequence comparison, although the extent of this variation was not described (38) and was not found in our sample. Comparison of five separate alleles of the pap31 sequence suggested that this strain (Fizz) was quite similar to another (CAL-1) but less similar to the Marseille representative strain. These Marseille subtype-like strains were in turn quite distinct from those of the Houston subtype, in which three apparently identical strains (90-615, SA-2, and Houston-1) were distinguishable from one other strain, ZF-1, by pap31 sequencing (36). Whether pap31 is a good indicator of genetic relatedness is not yet clear, but such diversity in a gene that may encode an essential hemin-binding protein (28) should prove useful in an evolved B. henselae MLST scheme. Although the uniqueness of the Berlin-2 isolate (ST7) illustrates that there are alleles of several loci that may be present in Europe and elsewhere that were not encountered in the smaller Australasian gene pool, previous studies have suggested that the number of genetic subgroups within the species is limited (10, 13, 36-38). Indeed, the remainder of the European and U.S. isolates included in our study correspond to the two lineages within the species that supported all of the Australasian isolates.
Our results demonstrate that allelic variation at the 16S rDNA locus is not a highly discriminatory indicator of the clonal structure of B. henselae. The type II 16S rDNA allele was encountered in all three lineages defined within the species and cannot, therefore, be relied upon for defining the Marseille (i.e., ST6) subtype. In our study, the type I 16S rDNA allele was confined to the ST1 clonal complex that includes the type I representative strain, Houston-1, but whether this would hold generally true is also unknown. Reconstructions of genetic relatedness among our isolates suggest that the type II 16S rDNA allele is ancestral, with the type I allele appearing later in the diversification of the species. Although present observations suggest that variation at other loci may serve as good predictors of clonal relationships within B. henselae, analysis of variation at multiple loci is clearly a far more reliable approach to obtaining an accurate, highly discriminate picture of population structure in this and other species
In demonstrating apparent noncongruent inheritance of alleles at two loci in some of the isolates studied, these data are consistent with a previously hypothesized role for horizontal gene transfer in shaping the genetic structure of the species (10). However, the small size of our survey and the lack of high levels of allelic diversity means that larger data sets are necessary before interclonal relationships can be reconstructed with a high degree of confidence. Nevertheless, the present study provides an invaluable B. henselae population framework and a convincing case for the future use of MLST in this species. This is illustrated by the fact that the major branches of the dendrogram are largely supported by other typing methods. The highest congruence was found between MLST and PFGE, an approach that has previously been found to have a very high discriminatory power relative to other methods applied to the molecular typing of B. henselae (32). In some cases, DNA fingerprinting by other typing methods, including AP, ERIC, IRS, and 16S-23S intergenic spacer region sequencing (10), discriminates effectively between members of our STs but correlates imperfectly with MLST-derived clonal complexes. Such noncongruence suggests that intragenomic rearrangement results from a process quite distinct from that leading to interstrain recombination.
Thus, one might conceive a B. henselae gene pool from which have emerged subgroups with enhanced virulence (at least in humans). These may not have the same phenotypic requirements and the inheritance of relevant traits is not necessarily linked; B. henselae carries a bacteriophage of as-yet-unknown significance (1) and appears to undergo intragenomic rearrangement which does not inevitably lead to loss of virulence potential nor irreversible change (18, 22), and the present study suggests the potential for horizontal genetic transfer in this species. Our study adds further weight to the hypothesis that human-infecting strains of B. henselae are more homogeneous than those associated with feline reservoirs. It is possible that this partly reflects a sampling bias as over half the human isolates we studied were obtained from a single region (Queensland) whereas analyzed feline isolates came from Europe (n = 1) and New Zealand (n = 4), but mostly from metropolitan Sydney (n = 6 from one collection and n = 9 from another). Despite this, there is good evidence for a lower level of diversity among human strains than in the feline reservoir. Three of four New South Wales human isolates belonged to ST1, whereas the 15 feline isolates from the same region were assigned to four different MLST groups (ST1, ST3, ST4, and ST6). The hypothesis that strains belonging to the Houston subtype are less tolerant of intragenomic rearrangement (as human-associated isolates do seem to be) than strains belonging to the Marseille subtype (when controlled for source of isolate) is worthy of further investigation.
B. henselae may cause subclinical or minor infection in both feline and human hosts and, adapted also to an invertebrate vector, is the paradigm par excellence for the versatile pathogen. MLST represents a discriminatory and portable typing scheme for the sensitive delineation of strains within B. henselae and should provide a practical basis for multicenter collaborative analyses in a way that was not previously feasible. Detailed analysis of the virulent subgroupings within this species and the evolutionary processes leading to the emergence of these clones may be facilitated by identification of loci with greater allelic variation and will require a larger and more representative strain collection to address the questions raised in this study.
Acknowledgments
This work was supported by grant RN084/99 from the Clive and Vera Ramaciotti Foundations and grants from the Westmead Millennium Institute to J.I. D.B. is the recipient of an Australian Postgraduate Research Award. M.A. is supported by grant 325-4471-02/05 from the German Ministry of Health. E.J.F. is supported by a Medical Research Council Career Development Award. R.J.B. is supported by a Wellcome Trust Medical Microbiology Fellowship.
We thank Belinda Dillon for help with repeat 16S rDNA sequencing and alignments and with preparation of the manuscript.
REFERENCES
- 1.Anderson, B. E., C. Goldsmith, A. Johnson, I. Padmalayam, and B. Baumstark. 1994. Bacteriophage-like particle of Rochalimaea henselae. Mol. Microbiol. 13:67-73. [DOI] [PubMed] [Google Scholar]
- 2.Arvand, M., A. J. Klose, D. Schwartz-Porsche, H. Hahn, and C. Wendt. 2001. Genetic variability and prevalence of Bartonella henselae in cats in Berlin, Germany, and analysis of its genetic relatedness to a strain from Berlin that is pathogenic for humans. J. Clin. Microbiol. 39:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvand, M., C. Wendt, T. Regnath, R. Ullrich, and H. Hahn. 1998. Characterisation of Bartonella henselae isolated from bacillary angiomatosis lesions in a human immunodeficiency virus-infected patient in Germany. Clin. Infect. Dis. 26:1296-1299. [DOI] [PubMed] [Google Scholar]
- 4.Bereswill, S., S. Hinkelmann, K. Manfred, and A. Sander. 1999. Molecular analysis of riboflavin synthesis genes in Bartonella henselae and use of the ribC gene for differentiation of Bartonella species by PCR. J. Clin. Microbiol. 37:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmans, A. M. C., J. F. P. Schellekens, J. D. A. van Embden, and L. M. Schouls. 1996. Predominance of two Bartonella henselae variants among cat-scratch disease patients in the Netherlands. J. Clin. Microbiol. 34:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birtles, R. J., and D. Raoult. 1996. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int. J. Syst. Bacteriol. 46:891-897. [DOI] [PubMed] [Google Scholar]
- 7.Branley, J., C. Wolfson, P. Waters, T. Gottlieb, and R. Bradbury. 1996. Prevalence of Bartonella henselae bacteraemia, the causative agent of cat-scratch disease, in an Australian cat population. Pathology 28:262-265. [DOI] [PubMed] [Google Scholar]
- 8.Chang, C. C., B. B. Chomel, R. W. Kasten, J. W. Tappero, M. A. Sanchez, and J. E. Koehler. 2002. Molecular epidemiology of Bartonella henselae infection in human immunodeficiency virus-infected patients and their cat contacts, using pulsed-field gel electrophoresis and genotyping. J. Infect. Dis. 186:1733-1739. [DOI] [PubMed] [Google Scholar]
- 9.Dehio, C., M. Meyer, J. Berger, H. Schwarz, and C. Lanz. 1997. Interaction of Bartonella henselae with endothelial cells results in bacterial aggregation on the cell surface and the subsequent engulfment and internalisation of the bacterial aggregate by a unique structure, the invasome. J. Cell Sci. 110:2141-2154. [DOI] [PubMed] [Google Scholar]
- 10.Dillon, B., J. Valenzuela, R. Don, D. Blanckenberg, D. I. Wigney, R. Malik, A. J. Morris, J. M. Robson, and J. Iredell. 2002. Limited diversity among human isolates of Bartonella henselae. J. Clin. Microbiol. 40:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drancourt, M., R. Birtles, G. Chaumentin, F. Vandenesch, J. Etienne, and D. Raoult. 1996. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet 347:441-443. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenborg, C., L. Wesslen, A. Jakobson, G. Friman, and M. Holmberg. 2000. Sequence variation in the ftsZ gene of Bartonella henselae isolates and clinical samples. J. Clin. Microbiol. 38:682-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier, P.-E., J. Robson, Z. Zeaiter, R. McDougall, S. Byrne, and D. Raoult. 2002. Improved culture from lymph nodes of patients with cat scratch disease and genotypic characterization of Bartonella henselae isolates in Australia. J. Clin. Microbiol. 40:3620-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frean, J., S. Arndt, and D. Spencer. 2002. High rate of Bartonella henselae infection in HIV-positive outpatients in Johannesburg, South Africa. Trans. R. Soc. Trop. Med. Hyg. 96:549-550. [DOI] [PubMed] [Google Scholar]
- 15.Iredell, J., J. McHattan, P. Kyme, B. Dillon, and D. Blanckenberg. 2002. Antigenic and genotypic relationships between Bartonella henselae strains. J. Clin. Microbiol. 40:4397-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, L. A., B. A. Perkins, and J. D. Wenger. 1993. Cat-scratch disease in the United States: an analysis of three national databases. Am. J. Pub. Health 83:1707-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph, A. K., C. W. Wood, J. M. Robson, S. L. Paul, and A. J. Morris. 1997. Bartonella henselae bacteraemia in domestic cats from Auckland, New Zealand. Vet. J. 45:85-187. [DOI] [PubMed] [Google Scholar]
- 18.Kabeya, H., S. Maruyama, M. Irei, R. Takahashi, M. Yamashita, and T. Mikami. 2002. Genomic variations among Bartonella henselae isolates derived from naturally infected cats. Vet. Microbiol. 89:211-221. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, S., J. Rawlings, C. Paddock, J. Childs, R. Regnery, and M. Reynolds. 2002. Cat-scratch disease in children—Texas, September 2000-August 2001. Morb. Mortal. Wkly. Rep. 51:212-214. [PubMed] [Google Scholar]
- 20.Kempf, V. A., B. Volkmann, M. Schaller, C. A. Sander, K. Alitalo, T. Riess, and I. B. Autenrieth. 2001. Evidence of a leading role for VEGF in Bartonella henselae-induced endothelial cell proliferations. Cell. Microbiol. 3:623-632. [DOI] [PubMed] [Google Scholar]
- 21.Koehler, J. E., C. A. Glaser, and J. W. Tappero. 1994. Rochalimaea henselae infection: a new zoonosis with the domestic cat as reservoir. JAMA 271:531-535. [DOI] [PubMed] [Google Scholar]
- 22.Kyme, P., Dillon, B., and J. Iredell. 2003. Phase variation in Bartonella henselae. Microbiology 149:621-629. [DOI] [PubMed] [Google Scholar]
- 23.La Scola, B., Z. Liang, Z. Zeaiter, P. Houpikian, P. A. D. Grimont, and D. Raoult. 2002. Genotypic characteristics of two serotypes of Bartonella henselae. J. Clin. Microbiol. 40:2002-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Scola, B., and D. Raoult. 1999. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998). J. Clin. Microbiol. 37:1899-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Scola, B., and D. Raoult. 1996. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J. Clin. Microbiol. 34:2270-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matar, G. M., J. E. Koehler, G. Malcolm, M. A. Lambert-Fair, J. Tappero, S. B. Hunter, and B. Swaminathan. 1999. Identification of Bartonella species directly in clinical specimens by PCR-restriction fragment length polymorphism analysis of a 16S rRNA gene fragment. J. Clin. Microbiol. 37:4045-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minnick, M. F., K. N. Sappington, L. S. Smitherman, S. G. E. Andersson, O. Karlberg, and J. A. Carroll. 2003. Five-member gene family of Bartonella quintana. Infect. Immun. 71:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nallapareddy, S. R., R.-W. Duh, K. V. Singh, and B. E. Murray. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norman, A. F., R. Regnery, P. Jameson, C. Greene, and D. C. Krause. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Reilly, K. L., R. W. Bauer, R. L. Freeland, L. D. Foil, K. J. Hughes, K. R. Rohde, A. F. Roy, R. W. Stout, and P. C. Triche. 1999. Acute clinical disease in cats following infection with a pathogenic strain of Bartonella henselae (LSU16). Infect. Immun. 67:3066-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sander, A., M. Ruess, S. Bereswill, M. Schuppler, and B. Steinbrueckner. 1998. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J. Clin. Microbiol. 36:2973-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sola-Landa, A., J. Pizarro-Cerdá, M.-J. Grilló, E. Moreno, I. Moriyón, J.-M. Blasco, J.-P. Gorvel, and I. López-Goñi. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 34.Tompkins, L. S. 1996. Bartonella species infections, including cat-scratch disease, trench fever, and bacillary angiomatosis—what molecular techniques have revealed. West. J. Med. 164:39-41. [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto, K., B. B. Chomel, R. W. Kasten, C. M. Hew, D. K. Weber, and W. I. Lee. 2002. Experimental infection of specific pathogen free (SPF) cats with two different strains of Bartonella henselae type I: a comparative study. Vet. Res. 33:669-684. [DOI] [PubMed] [Google Scholar]
- 36.Zeaiter, Z., P.-E. Fournier, and D. Raoult. 2002. Genomic variation of Bartonella henselae strains detected in lymph nodes of patients with cat scratch disease. J. Clin. Microbiol. 40:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeaiter, Z., Z. Liang, and D. Raoult. 2002. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J. Clin. Microbiol. 40:3641-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeaiter, Z., P. E. Fournier, H. Ogata, and D. Raoult. 2002. Phylogenetic classification of Bartonella species by comparing groEL sequences. Int. J. Syst. E vol. Microbiol. 52:165-171. [DOI] [PubMed] [Google Scholar]