Abstract
The distribution of EDL 933 O island 122 (OI-122) was investigated in 70 strains of Verocytotoxin-producing Escherichia coli (VTEC) of multiple serotypes that were classified into five “seropathotypes” (A through E) based on the reported occurrence of serotypes in human disease, in outbreaks, and/or in the hemolytic-uremic syndrome (HUS). Seropathotype A comprised 10 serotype O157:H7 and 3 serotype O157:NM strains. Seropathotype B (associated with outbreaks and HUS but less commonly than serotype O157:H7) comprised three strains each of serotypes O26:H11, O103:H2, O111:NM, O121:H19, and O145:NM. Seropathotype C comprised four strains each of serotypes O91:H21 and O113:H21 and eight strains of other serotypes that have been associated with sporadic HUS but not typically with outbreaks. Seropathotype D comprised 14 strains of serotypes that have been associated with diarrhea but not with outbreaks or HUS, and seropathotype E comprised animal VTEC strains of serotypes not implicated in human disease. All strains were tested for four EDL 933 OI-122 virulence genes (Z4321, Z4326, Z4332, and Z4333) by PCR. Negative PCRs were confirmed by Southern hybridization. Overall, 28 (40%) strains contained OI-122 (positive for all four virulence genes), 27 (38.6%) contained an “incomplete” OI-122 (positive for one to three genes), and 15 (21.4%) strains did not contain OI-122. The seropathotype distribution of complete OI-122 was as follows: 100% for seropathotype A, 60% for B, 36% for C, 15% for D, and 0% for E. The differences in the frequency of OI-122 between seropathotypes A, B, and C (associated with HUS) and seropathotypes D and E (not associated with HUS) and between seropathotypes A and B (associated with epidemic disease) and seropathotypes C, D, and E (not associated with epidemic disease) were highly significant (P < 0.0001).
Verocytotoxin (VT)-producing Escherichia coli (VTEC) (34), also referred to as Shiga toxin-producing E. coli (11), is the cause of a potentially fatal food- or waterborne illness whose clinical spectrum includes nonspecific diarrhea, hemorrhagic colitis, and the hemolytic-uremic syndrome (HUS) (19, 30, 31). The serious public health concern about VTEC infection is due to the risks of massive outbreaks (1, 12, 13, 22, 41, 54) and of HUS (30, 31), the leading cause of acute renal failure in children (30). Ruminants, especially cattle, are the main reservoir of VTEC, which is transmitted to humans primarily via contaminated foods and water (19, 28, 30, 45). Although serotype O157:H7 has been implicated in most outbreaks and in most cases of HUS (19, 28, 30, 45), there is growing concern about the risk to human health associated with non-O157 VTEC serotypes (3, 25, 61), more than 200 of which have now been associated with human illness (70). First reported in association with HUS in Canada (31, 32), non-O157 serotypes have since been more commonly implicated in HUS than serotype O157:H7 in Latin America (37) and Australia (15), and their frequency may be rising in Europe (18, 66). As many as 20% of HUS cases in North America may be associated with non-O157 VTEC (2). Some non-O157 VTEC serotypes (e.g., serotypes O26:H11, O103:H2, O111:NM, O121:H19, and O145:NM) are associated with outbreaks and HUS, but less commonly than serotype O157:H7 (3, 19, 25, 39, 70). Others (e.g., O113:H21 and O91:H21) generally do not cause outbreaks but are associated with sporadic episodes of HUS (3, 25, 45). A large number of VTEC serotypes have been isolated from patients with diarrhea but have not been associated with outbreaks or HUS (3, 25, 45, 70), and yet others, isolated from cattle, have never been associated with human disease (69, 70). Thus, VTEC serotypes appear to differ in pathogenic potential, but the scientific basis for this is not known. Consequently, assessment of clinical and public health risks is greatly compromised when non-O157 VTEC strains are found in humans, foods, animals, and the environment.
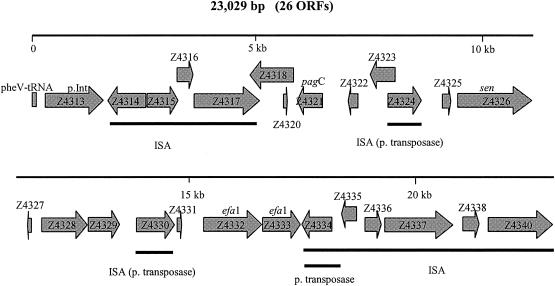
Increasing evidence suggests that major differences in virulence between groups of strains within, for example, E. coli, Salmonella spp., Shigella spp., and Helicobacter pylori are due to specific virulence characteristics encoded on large, horizontally acquired “gene cassettes” referred to as pathogenicity islands (PAIs) (26). An example of this in enteropathogenic E. coli (EPEC), E. coli O157:H7, and some non-O157 VTEC strains is the locus of enterocyte effacement (LEE) (27), which is a ca. 43-kb chromosomal PAI (in E. coli serotype O157:H7) containing genes that encode all the virulence factors necessary for forming the characteristic attaching and effacing (AE) lesions on enterocytes. LEE-positive VTEC serotypes are commonly referred to as enterohemorrhagic E. coli (EHEC) (27). LEE appears to confer enhanced virulence, since LEE-positive VTEC serotypes (such as O157:H7, O26:H11, O103:H2, O111:NM, O121:H19, and O145:NM) are much more commonly associated with HUS and with epidemic diseases than are LEE-negative serotypes (5, 27, 45). On the other hand, serotype O157:H7 is associated with outbreaks and HUS much more commonly than other LEE-positive serotypes (27, 45), and some LEE-positive serotypes from bovines have never been associated with human disease (69). Furthermore, LEE-negative serotypes are also associated with serious human disease (45). These observations suggest that, in addition to LEE, other hitherto unknown factors, possibly PAIs, may also enhance the virulence potential of VTEC strains. Genome sequencing of two epidemic strains of E. coli O157:H7, EDL 933 (50) and the Sakai strain (21), has revealed several additional candidate PAIs which, in the EDL 933 genome, include O island 1 (OI-1), OI-43, OI-48, OI-115, OI-122, OI-140, OI-141, and OI-154. Nothing is known about the role of these newly discovered potential PAIs in the virulence of E. coli O157:H7 or about their presence and role in non-O157 VTEC. Our hypothesis is that, like LEE, some of these newly recognized O islands might contribute to virulence differences between VTEC serotypes. The results of preliminary pilot studies looking at the distribution and potential significance of different O islands in VTEC seropathotypes suggested that EDL 933 OI-122 (50), referred to as SpLE3 in the Sakai strain (21), was a promising candidate for more detailed investigation. Hence the objective of this study was to investigate the distribution and possible public health significance of this PAI in VTEC seropathotypes. OI-122 (Fig. 1) is a 23,029-bp genomic island that is composed of 26 open reading frames (ORFs) including those that show significant homology to virulence genes, namely, Salmonella enterica serovar Typhimurium pagC (51), Shigella flexneri enterotoxin 2 (sen) (46), and the EHEC factor for adherence (efa1) (47), which is also referred to as lymphocyte inhibition factor (lifA) (33). OI-122 is adjacent to a pheV tRNA locus. The terminus closest to the pheV locus consists of a P4 integrase gene and four sequences that show homology to E. coli ISEc8. The downstream terminal region consists of six sequences homologous to E. coli ISEC8 and IS629. Apart from four putative virulence genes, OI-122 also contains three putative transposases and nine genes of unknown function.
FIG. 1.
OI-122 in the genome of VTEC strain EDL 933 (O157:H7). p.Int, putative pathogenicity island integrase; ISA, insertion sequence-associated protein; p. transposase, putative transposase.
MATERIALS AND METHODS
Seropathotype classification.
A “seropathotype” classification was developed for VTEC serotypes based on their reported frequencies in human illness (in qualitative terms such as “high,” “moderate,” or “rare”), and their known associations with outbreaks and with severe disease, such as HUS and hemorrhagic colitis (3, 25, 45, 70), as shown in Table 1. Assignment of serotypes to seropathotype groups was based on published references (3, 25, 45, 70) and on two large Internet databases of non-O157 VTEC serotypes (available at http://www.microbionet.com.au/frames /feature/vtec/brief01.html and http://www.lugo.usc.es/∼ecoli/SEROTIPOSHUM.htm).
TABLE 1.
Classification of VTEC serotypes into seropathotypes
| Seropathotype | Relative incidence | Frequency of involvement in outbreaks | Association with severe diseasea | Serotypes |
|---|---|---|---|---|
| A | High | Common | Yes | O157:H7, O157:NM |
| B | Moderate | Uncommon | Yes | O26:H11, O103:H2, O111:NM, O121:H19, O145:NM |
| C | Low | Rare | Yes | O91:H21, O104:H21, O113:H21; others |
| D | Low | Rare | No | Multiple |
| E | Nonhuman only | NAb | NA | Multiple |
HUS or hemorrhagic colitis.
NA, not applicable.
Bacterial strains.
The 70 study strains, sorted by seropathotype, are listed in Table 2. Positive-control strains were EDL 933 (50) and the Sakai strain (21) of serotype O157:H7. The negative-control strain was the E. coli K-12 strain MG 1655 (50). Selection of strains was, in general, random but was influenced by the following criteria: all strains belonging to the same serotype were selected to ensure that they were isolates from different patients or animals that were not linked temporally and that they gave distinct macrorestriction enzyme digest patterns by pulsed-field gel electrophoresis (PFGE) (6, 58). The bacterial strain typing criteria of Tenover et al. (64) were used as a general guide in interpreting differences in PFGE patterns. Five main serotypes have been reported with clinical and epidemiological features consistent with the criteria for seropathotype B (serotypes O26:H11, O103:H2, O111:NM, O121:H19, and O145:NM) (3, 25, 39, 45, 70). We selected three strains for each of these serotypes. The best-known serotypes that conform to the features of seropathotype C are O91:H21 and O113:H21 (3, 25, 45, 70). We included four strains for each of these two serotypes, and the remainder of the seropathotype C strains were selected randomly. Given that multiple serotypes have been reported to conform to seropathotypes D and E (3, 25, 45, 52, 53, 69, 70), study strains were selected randomly, although no more than two strains belonging to the same serotype were included.
TABLE 2.
List of study strains
| Pathotype | Serotype | Strain | Host | Sourcea | Presence or absenceb of virulence gene:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| VT1 | VT2 | eae | hlyA | espP | katP | |||||
| Control | O157:H7 | EDL 933 | J. Kaper, Baltimore, Md. | + | + | + | + | + | + | |
| Control | O157:H7 | Sakai | T. Honda, Osaka, Japan | + | + | + | + | + | + | |
| Control | (K-12) | MG 1655 | ATCC 700926 | − | − | − | − | − | − | |
| A | O157:H7 | 93111 | Human | T. Whittam, East Lansing, Mich. | + | + | + | + | + | + |
| O157:H7 | OK1 | Human | T. Whittam | + | + | + | + | + | + | |
| O157:H7 | 278F1 | Human | LFZ | − | + | + | + | + | + | |
| O157:H7 | 279F1 | Human | LFZ | + | + | + | + | + | + | |
| O157:H7 | 237F1 | Human | LFZ | + | + | + | + | + | + | |
| O157:H7 | 235F1 | Human | LFZ | + | + | + | + | + | + | |
| O157:H7 | D103F5 | Human | LFZ | + | + | + | + | + | + | |
| O157:H7 | 254 | Human | LFZ | − | + | + | + | + | + | |
| O157:H7 | E48F9 | Human | LFZ | + | + | + | + | + | + | |
| O157:H7 | 157F1 | Human | LFZ | − | + | + | + | + | + | |
| O157:NM | 158F2 | Human | LFZ | + | + | + | + | + | + | |
| O157:NM | E32511 | Human | H. Smith, London, United Kingdom | − | + | + | + | + | + | |
| O157:NM | ER63-94 | Human | F. Jamieson, OPHL | + | + | + | + | + | + | |
| B | O26:H11 | CL1 | Human | LFZ | + | − | + | + | + | + |
| O26:H11 | CL4 | Human | LFZ | + | − | + | + | + | + | |
| O26:H11 | CL9 | Human | LFZ | + | − | + | + | + | + | |
| O103:H2 | N01-2454 | Human | BCCDC, NLEP | + | − | + | + | + | − | |
| O103:H2 | N01-7015 | Human | BCCDC, NLEP | + | − | + | + | − | − | |
| O103:H2 | N02-1626 | Human | BCCDC, NLEP | + | − | + | + | − | + | |
| O111:NM | R82F2 | Human | LFZ | + | − | + | − | − | − | |
| O111:NM | C69F1 | Human | LFZ | + | − | + | + | − | − | |
| O111:NM | CL101 | Human | LFZ | + | − | + | + | − | − | |
| O121:H19 | CL106 | Human | LFZ | − | + | + | − | + | − | |
| O121:H19 | Z3F1 | Human | LFZ | − | + | + | − | + | − | |
| O121:H19 | 274F4 | Human | LFZ | − | + | + | − | + | − | |
| O145:NM | N01-2051 | Human | BCCDC, NLEP | − | + | + | + | + | + | |
| O145:NM | N00-6496 | Human | BCCDC, NLEP | + | + | + | + | + | + | |
| O145:NM | N02-5149 | Human | BCCDC, NLEP | + | − | + | + | + | + | |
| C | O5:NM | N00-4067 | Human | BCCDC, NLEP | + | − | + | + | + | + |
| O5:NM | N00-4541 | Human | BCCDC, NLEP | + | − | + | + | + | − | |
| O91:H21 | B2F1 | Human | LFZ | − | + | − | − | − | − | |
| O91:H21 | EC7-181 | Human | LFZ | − | + | − | − | − | − | |
| O91:H21 | EC6-990 | Human | S. Aleksic, Munich, Germany | − | + | − | − | − | − | |
| O91:H21 | EC6-936 | Human | L. Beutin, Berlin, Germany | + | + | − | − | − | − | |
| O104:H21 | G5506 | Human | T. Whittam | − | + | − | + | + | − | |
| O113:H21 | CL3 | Human | LFZ | − | + | − | + | + | − | |
| O113:H21 | N99-3504 | Human | BCCDC, NLEP | − | + | − | − | + | − | |
| O113:H21 | N89-0541 | Human | J. Preiksaitis, APLPH | + | + | − | − | + | − | |
| O113:H21 | N90-0657 | Human | P. VanCaeseele, CPL | − | + | − | − | + | − | |
| O121:NM | N99-4390 | Human | BCCDC, NLEP | + | + | + | + | + | − | |
| O121:NM | N99-4389 | Human | BCCDC, NLEP | − | + | + | + | + | − | |
| O165:H25 | N00-4540 | Human | BCCDC, NLEP | − | + | + | + | + | + | |
| D | O7:I14 | EC3-480 | Bovine | LFZ | − | + | − | − | − | − |
| O69:H11 | EC7-821 | Human | LFZ | + | − | + | + | + | + | |
| O103:H25 | N00-4859 | Human | BCCDC, NLEP | + | − | + | + | − | − | |
| O103:H25 | N02-2616 | Human | BCCDC, NLEP | + | − | + | + | − | − | |
| O113:H4 | EC6-371 | Bovine | LFZ | + | + | − | − | − | − | |
| O117:H7 | N02-4495 | Human | BCCDC, NLEP | + | − | − | − | − | − | |
| O117:H7 | N02-0035 | Human | BCCDC, NLEP | + | − | − | − | − | − | |
| O119:H25 | EC2-267 | Human | LFZ | + | − | + | + | + | − | |
| O132:NM | EC2-051 | Human | LFZ | − | + | − | − | − | − | |
| O146:H21 | N02-1625 | Human | BCCDC, NLEP | + | + | + | + | − | − | |
| O171:H2 | EC2-032 | Bovine | LFZ | − | + | − | − | − | − | |
| O172:NM | EC6-484 | Bovine | LFZ | − | + | + | + | + | + | |
| O174:H8 | A2EV659 | Human | BCCDC | + | + | − | − | − | − | |
| Orough:H2 | N00-3105 | Human | G. Horsman, SHLDC | + | − | + | + | − | − | |
| E | O6:H34 | EC6-626 | Bovine | J. Preiksaitis, APLPH | − | + | − | − | − | − |
| O8:H19 | EC6-448 | Bovine | LFZ | + | + | − | + | + | − | |
| O39:H49 | EC2-293 | Bovine | LFZ | + | + | − | + | + | − | |
| O46:H38 | EC7-451 | Bovine | LFZ | + | + | − | + | + | − | |
| O76:H7 | EC9-333 | Ovine | P. Desmarchelier, Brisbane, Australia | + | − | + | + | − | + | |
| O84:NM | EC2-044 | Bovine | LFZ | + | − | + | + | + | − | |
| O88:H25 | EC4-453 | Bovine | LFZ | − | + | − | + | − | − | |
| O98:H25 | EC3-377 | Bovine | LFZ | + | − | + | + | + | − | |
| O113:NM | EC2-211 | Bovine | LFZ | − | + | − | − | − | − | |
| O136:H12 | EC3-208 | Bovine | LFZ | + | − | − | + | − | − | |
| O136:NM | EC2-258 | Bovine | LFZ | + | − | − | + | − | − | |
| O153:H31 | EC2-104 | Bovine | LFZ | + | − | − | − | − | − | |
| O156:NM | EC2-020 | Bovine | LFZ | − | + | − | − | + | − | |
| O163:NM | EC2-459 | Bovine | LFZ | + | + | − | + | + | − | |
LFZ, Laboratory for Foodborne Zoonoses; OPHL, Ontario Public Health Laboratory; BCCDC, British Columbia Centre for Disease Control; NLEP, National Laboratory for Enteric Pathogens; APLPH, Alberta Provincial Laboratory for Public Health; CPL, Cadham Provincial Laboratory, Winnipeg, Manitoba, Canada; SHLDC, Saskatchewan Health Laboratory and Disease Control Services Branch.
+, presence; −, absence.
Bacterial strain characterization.
All strains were serotyped by using reference O- and H-specific antisera by the method of Edwards and Ewing (16). They were tested for the presence of the VT1, VT2 (including VT2 variants), eaeA, and hlyA genes by multiplex PCR using the method and primers reported by Paton and Paton (48). The presence of espP and katP was detected by using the methods and primers reported by Brunder et al. (7-9). All test strains were tested for XbaI macrorestriction enzyme digest patterns by PFGE (6, 58). This was done to ensure that all strains, especially those belonging to the same serotype, were distinct.
Investigation of strains for the presence of OI-122. (i) PCR.
Strains were screened for the presence of OI-122 by testing for four EDL 933 OI-122 putative virulence genes (Z4321, Z4326, Z4332, and Z4333) (Fig. 1) by PCR using the primers listed in Table 3. All PCR amplifications were carried out in 50-μl reaction mixtures containing 1× PCR buffer (Perkin-Elmer Applied Biosystems, Foster City, Calif.), 250 μM concentrations of deoxynucleoside triphosphates, 1 mM MgCl2, 25 pmol of each primer, and 2 U of Taq DNA polymerase (AmpliTaq; Applied Biosystems). Cycling conditions for all OI-122 genes consisted of an initial denaturation step at 94°C for 5 min, followed by 30 cycles, each consisting of denaturation at 94°C for 30 s, annealing at 56°C for 1 min, and elongation at 72°C for 2.5 min. There was a final elongation step at 72°C for 5 min.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′-to-3′ direction) | Target gene | Amplicon size (bp) | Location within gene | GenBank accession no. |
|---|---|---|---|---|---|
| Z4321-a | ATGAGTGGTTCAAGACTGG | pagC | 521 | 14-33 | NP_289546 |
| Z4321-b | CCAACTCCAACAGTAAATCC | 534-517 | |||
| Z4326-a | GGATGGAACCATACCTGG | sen | 551 | 927-944 | NP_289551 |
| Z4326-b | CGCAATCAATTGCTAATGC | 1477-1459 | |||
| Z4332-a | CTCCCAGAGATAATTTTGAGG | efa1 | 504 | 428-448 | NP_289557 |
| Z4332-b | CAACTGTATGCGAATAGTACTC | 931-910 | |||
| Z4333-a | CTGTCAGACGATGACATTGG | efa1 | 547 | 103-122 | NP_289558 |
| Z4333-b | GAAGGATGGGCATTGTGTC | 649-631 |
(ii) Southern hybridization.
Strains that were negative for any of the four genes by PCR were retested by Southern hybridization (55, 56) for the presence of that gene by using digoxigenin (DIG)-labeled probes generated by PCR (36) using the primers shown in Table 3. Genomic DNA was isolated by using the DNeasy Tissue kit (Qiagen, Hilden, Germany). Approximately 2 μg of DNA digested with an excess of EcoRI and run on a 0.6% agarose gel was transferred to a nylon membrane (Roche Diagnostics, Mannheim, Germany). DIG-labeled probes were synthesized by using a PCR DIG Probe Synthesis kit (Roche Diagnostics, Germany) according to the manufacturer's manual. Hybridizations were carried out overnight at 42°C. The DIG Nucleic Acid Detection kit (Roche Diagnostics) was used to detect hybridized bands. EDL 933 and the Sakai strain of serotype O157:H7 were used as positive controls, and the E. coli K-12 strain MG 1655 was used as a negative control.
The presence of all four putative virulence genes was taken as evidence for the presence of a complete OI-122 (COI-122). The absence of one or more of the genes was considered to indicate an “incomplete” OI-122, whereas the absence of all four genes indicated an absent OI-122.
Statistical analysis.
Statistical analyses were performed with SAS for Windows (version 8.01; SAS Institute, Cary, N.C.). Associations between seropathotypes and the presence of COI-122 and eae were analyzed using Fisher's exact test (17). The ability of COI-122 and eae to identify the seropathotype was assessed by calculating the sensitivity, specificity, and predictive value.
RESULTS
Strain characterization.
All 70 strains had distinct macrorestriction enzyme digest patterns by PFGE (data not shown). The VT genotypes and the genotypes for putative plasmid virulence factors (hlyA, katP, and espP) and eae are listed for each strain in Table 2. In cases where serotypes were represented by more than one strain (serotypes O157:H7, O157:NM, O26:H11, O103:H2, O111:NM, O121:H19, O145:NM, O5:NM, O91:H21, O113:H21, O121:NM, O103:H25, and O117:H7), strains with the same serotype were either all positive or all negative for eae. In contrast, strain-to-strain variation in individual serotypes was evident for VT genotypes and/or for the plasmid genes (hlyA, katP, and espP).
Detection and frequency distribution of OI-122 in different serotypes and seropathotypes.
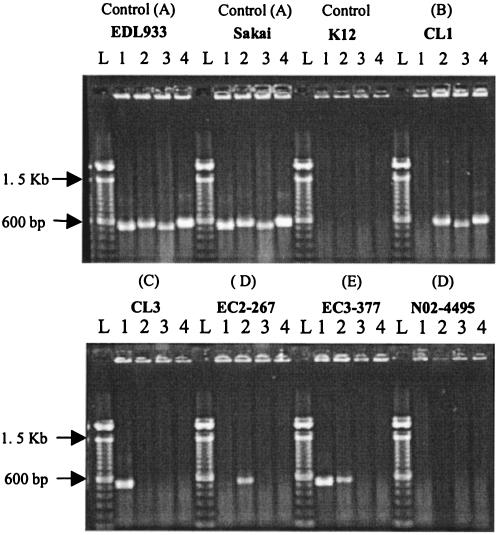
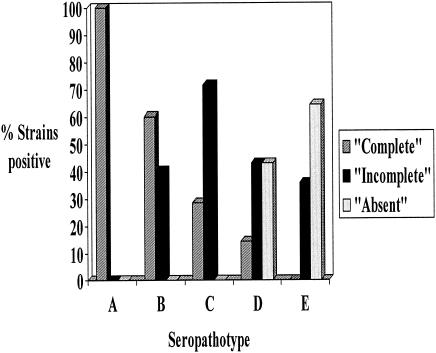
Figure 2 shows the PCR amplification products of the four OI-122 genes that were investigated (Z4321, Z4326, Z4332, and Z4333) in control strains and in representative strains from different seropathotypes. All strains negative by PCR were confirmed as negative by Southern hybridization. The distribution of OI-122 genes and eae in different serotypes and seropathotypes is shown in Table 4. Overall, 28 of 70 (40%) strains had a COI-122, 27 (38.6%) had an incomplete OI-122, and OI-122 was absent in 15 (21.4%) strains. There was a progressive decrease in the frequency of COI-122 from seropathotype A to seropathotype E and a concomitant increase in the frequencies of incomplete and “absent” OI-122s (Fig. 3). The difference in the frequency of COI-122 between seropathotypes A and B was not significant (P = 0.2). However, the difference in the frequency of COI-122 between seropathotype A and each of seropathotypes C (P = 0.0002), D (P < 0.0001), and E (P < 0.0001) was highly significant. There were no significant differences in the frequency of COI-122 between seropathotypes B and C, C and D, C and E, or D and E, but there was a significant difference in the frequency of COI-122 between seropathotypes B and D (P = 0.02) and B and E (P < 0.001). The difference in the frequency of COI-122 between seropathotypes A and B (which are associated with epidemic disease), on the one hand, and serotypes C, D, and E (which are not associated with epidemic disease), on the other, was highly significant (P < 0.0001; odds ratio [OR] = 22.0; 95% confidence interval [95% CI], 6.3 to 76). The difference in the frequency of COI-122 between seropathotypes A, B, and C (which are all associated with HUS), and seropathotypes D and E (which are not) was also highly significant (P < 0.0001; OR = 21.1; 95% CI, 4.4 to 101.3).
FIG. 2.
Distribution of OI-122 genes in representative strains of different seropathotypes (A to E) and controls. Lanes: L, 100-bp ladder; 1, pagC (Z4321); 2, sen (Z4326); 3, efa1 (Z4332); 4, efa1 (Z4333).
TABLE 4.
Serotype distribution of OI-122 virulence genes and eae
| Seropathotype | Serotype | No. of strains | No. of strains positive by PCR for:
|
||||
|---|---|---|---|---|---|---|---|
| Z4321 | Z4326 | Z4332 | Z4333 | eae | |||
| Control (EDL 933) | O157:H7 | 1 | 1 | 1 | 1 | 1 | 1 |
| Control (Sakai) | O157:H7 | 1 | 1 | 1 | 1 | 1 | 1 |
| MG 1655 | E. coli K-12 | 1 | 0 | 0 | 0 | 0 | 0 |
| A | O157:H7 | 10 | 10 | 10 | 10 | 10 | 10 |
| O157:NM | 3 | 3 | 3 | 3 | 3 | 3 | |
| B | O26:H11 | 3 | 0 | 3 | 3 | 3 | 3 |
| O103:H2 | 3 | 0 | 3 | 3 | 3 | 3 | |
| O111:NM | 3 | 3 | 3 | 3 | 3 | 3 | |
| O121:H19 | 3 | 3 | 3 | 3 | 3 | 3 | |
| O145:NM | 3 | 3 | 3 | 3 | 3 | 3 | |
| C | O5:NM | 2 | 2 | 2 | 2 | 2 | 2 |
| O91:H21 | 4 | 4 | 0 | 0 | 0 | 0 | |
| O104:H21 | 1 | 1 | 0 | 0 | 0 | 0 | |
| O113:H21 | 4 | 4 | 0 | 0 | 0 | 0 | |
| O121:NM | 2 | 2 | 2 | 2 | 2 | 2 | |
| O165:H25 | 1 | 0 | 1 | 1 | 1 | 1 | |
| D | O7:H4 | 1 | 1 | 0 | 0 | 0 | 0 |
| O69:H11 | 1 | 0 | 1 | 0 | 0 | 1 | |
| O103:H25 | 2 | 2 | 2 | 2 | 2 | 2 | |
| O113:H4 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O117:H7 | 2 | 0 | 0 | 0 | 0 | 0 | |
| O119:H25 | 1 | 0 | 1 | 0 | 0 | 1 | |
| O132:NM | 1 | 0 | 0 | 0 | 0 | 0 | |
| O146:H21 | 1 | 0 | 0 | 0 | 0 | 1 | |
| O171:H2 | 1 | 1 | 0 | 0 | 0 | 0 | |
| O172:NM | 1 | 0 | 1 | 1 | 1 | 1 | |
| O174:H8 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O-rough:H2 | 1 | 0 | 1 | 1 | 1 | 1 | |
| E | O6:H34 | 1 | 0 | 0 | 0 | 0 | 0 |
| O8:H19 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O39:H49 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O46:H38 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O76:H7 | 1 | 0 | 1 | 0 | 0 | 1 | |
| O84:NM | 1 | 1 | 1 | 0 | 0 | 1 | |
| O88:H25 | 1 | 1 | 0 | 0 | 0 | 0 | |
| O98:H25 | 1 | 1 | 1 | 0 | 0 | 1 | |
| O113:NM | 1 | 0 | 0 | 0 | 0 | 0 | |
| O136:H12 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O136:NM | 1 | 0 | 0 | 0 | 0 | 0 | |
| O153:H31 | 1 | 0 | 0 | 0 | 0 | 0 | |
| O156:NM | 1 | 1 | 0 | 0 | 0 | 0 | |
| O163:NM | 1 | 0 | 0 | 0 | 0 | 0 | |
FIG. 3.
Distribution of complete, incomplete, and absent OI-122 in VTEC seropathotypes.
As with eae, in cases where a serotype was represented by more than one strain, all strains belonging to the same serotype had identical patterns of distribution of OI-122 genes (Table 4).
Frequency distribution of eae in seropathotypes, and relationship between eae and COI-122.
As with COI-122, the difference in the frequency of eae between seropathotypes A and B (which are associated with epidemic disease) and seropathotypes C, D, and E (which are not associated with epidemic disease) was highly significant (P < 0.0001; OR, undefined). The difference in the frequency of eae between seropathotypes A, B, and C (which are all associated with HUS) and seropathotypes D and E (which are not) was also highly significant (P < 0.0001; OR = 6.6; 95% CI, 2.2 to 19.2).
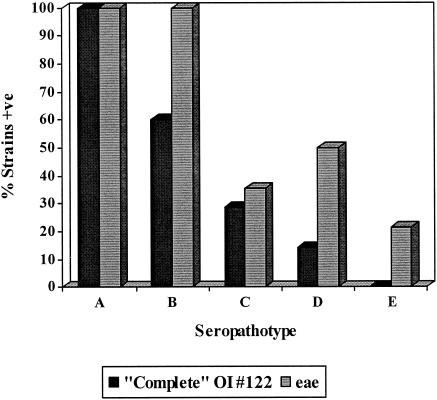
The overall frequency of COI-122 was 28 of 70 (40%), compared to a frequency of 43 of 70 (61.4%) for the eae gene. All COI-122-positive strains were eae positive. On the other hand, 15 eae-positive strains were COI-122 negative. Of 10 eae-positive strains in seropathotypes D and E, only 2 contained COI-122. The seropathotype distributions of eae and COI-122 are shown in Fig. 4.
FIG. 4.
Distributions of COI-122 and eae in VTEC seropathotypes.
Diagnostic application of COI-122 and eae in the detection of strains belonging to seropathotypes A, B, and C.
For the collection of 70 strains in this study, the use of eae to detect pathotypes A, B, and C (i.e., pathotypes that are associated with HUS) has a sensitivity of 79%, a specificity of 64%, and positive and negative predictive values of 61 and 39%, respectively. In contrast, the use of COI-122 to detect seropathotypes A, B, and C has a sensitivity of only 62% but a specificity of 93% and positive and negative predictive values of 40 and 60%, respectively.
DISCUSSION
The concept of a seropathotype classification, based on the reported occurrence of specific serotypes in human disease and on their association with HUS and with outbreaks, was used as an approach for better understanding the scientific basis for the apparent differences in virulence between groups of VTEC serotypes, with the recognition that this approach may have limitations. Disease incidence and severity are likely to be functions of a complex interplay between host, pathogen, and ecological factors and may not necessarily be explained on the basis of pathogen attributes alone. Furthermore, incidence may vary depending on the frequency with which specific diagnostic tests are used. Nevertheless, sufficient information has been published on the subject of VTEC infections to lend credence to the principle of the proposed seropathotype classification, though the boundaries between seropathotypes may be fluid. For example, seropathotypes A and B are both associated with outbreaks and HUS. The main difference between these two seropathotypes is that serotypes O157:H7 and possibly O157:NM are more commonly reported than are serotypes in seropathotype B. However, this distinction may not be valid in Australia, where serotype O111:NM was reported to be more common than O157:H7 (15).
Taking these limitations into account, our underlying hypothesis in this study is that differences in virulence between seropathotypes are related to the presence or absence of specific PAIs. This is based on increasing evidence that differences in virulence between groups of strains within species may be related to the presence of specific PAIs (26). The insertion of OI-122 into a tRNA locus is consistent with its being a PAI (20). Thirteen of its 26 ORFs are genes associated with mobile genetic elements, and the remaining 13 comprise the 4 virulence genes tested in this study and 9 genes of unknown function. Detection of flanking sequences was not considered a useful approach for identifying OI-122, because both upstream and downstream terminal sequences in OI-122 consist of genes associated with mobile genetic elements. The latter may be present in multiple copies, and their presence may not necessarily predict the presence of specific virulence genes which are essential components of a PAI. We therefore screened strains for OI-122 by using the only four genes (other than those associated with mobile genetic elements) that had a putative function based on homology. Thus, the underlying assumption in this study is that the presence of all four putative virulence genes is indicative of the presence of COI-122.
Based on this assumption, our results show that the presence of COI-122 is strongly correlated with VTEC seropathotypes (A and B) that are associated with epidemic disease and with seropathotypes (A, B, and C) associated with HUS. The study also confirms the close correlations between eae (a stable marker of the LEE PAI) (27) and seropathotypes that are associated with epidemic disease (A and B) and between eae and those associated with HUS (A, B, and C). The association between OI-122 and LEE-positive organisms (including EHEC and enteropathogenic E. coli) has been reported recently by Morabito et al. (44). It is noteworthy that within serotypes that were represented by more than one strain, all strains showed identical patterns for OI-122 (either complete, incomplete, or absent) and for eae. This supports our approach of postulating virulence differences between seropathotypes based on the presence or absence of specific PAIs rather than on the basis of individual virulence genes. In contrast to OI-122 and eae, VT genes and plasmid-encoded putative virulence genes (hlyA, espP, and katP) showed variability among individual strains belonging to the same serotype in several instances, indicating that these genes are not suitable for exploring virulence differences between seropathotypes.
Seropathotype A is characterized by the presence of COI-122 and eae. Strains of 3 of 5 serotypes (9 of 15 strains) in seropathotype B, 2 of 6 serotypes (4 of 14 strains) in seropathotype C, and 1 of 12 serotypes (2 of 14 strains) in seropathotype D were also positive for COI-122 and eae. This could mean that such strains have the same virulence potential as seropathotype A strains or that there are other, hitherto unknown factors that make seropathotype A strains more virulent than strains of the other seropathotypes. As shown in Fig. 4, there was a progressive increase in the frequency of strains with incomplete OI-122s from seropathotype A to E. The nature of incomplete OI-122 was different for different seropathotypes and serotypes. One-third of the 27 strains with incomplete OI-122s were lacking only one of the four OI-122 genes tested, whereas two-thirds were negative for two or three of the four genes. Most of the strains (6 of 9) lacking only one gene were in seropathotype B, whereas all 18 strains that lacked two or three genes were distributed in seropathotypes C, D, and E. The significance of these findings remains to be established.
A practical benefit of understanding the scientific basis for the difference in virulence between seropathotypes is that it can aid in the identification of suitable DNA targets for selective detection of strains (in seropathotypes A, B, and C) that pose a significant risk of severe and/or epidemic disease in humans. In the collection of 70 strains employed in this study, eae had a sensitivity and specificity of 79 and 64%, respectively, for detection of seropathotype A, B, and C strains, in contrast to a sensitivity and specificity of 62 and 93%, respectively, for COI-122. Clearly, neither test is completely reliable for field use. The eae gene is a stable marker for the LEE PAI. In contrast, detection of COI-122 requires testing for four genes, which limits its practical use, but this limitation may be overcome by using a multiplex PCR assay. The EDL 933 genome (50) has at least another seven putative PAIs whose pathogenic significance and value in distinguishing seropathotypes are unknown. Investigation of the seropathotype distribution of these seven putative PAIs may identify tests more practical than either eae or COI-122 for selectively detecting seropathotypes A, B, and C in a reliable manner.
The LEE PAI is associated with the characteristic AE lesions in several pathogens. These include EHEC, EPEC, Hafnia alvei, rabbit diarrheagenic E. coli (RDEC), the murine pathogen Citrobacter rodentium, and EPEC-like pathogens of cattle and dogs (40, 45). Although the sizes of LEEs in these organisms may be different, the core regions are highly conserved and consist of 41 genes that comprise the structural, effector, and regulatory components of a type III secretion system (23, 24) that mediates the AE lesion (40, 45). The core regions of LEE are devoid of elements associated with mobile genetic elements (27). In contrast, the core region of OI-122 contains several genes associated with mobile genetic elements (Fig. 1) (50), which could make this PAI less stable than LEE. The fact that OI-122 was incomplete in 27 (38.6%) strains is probably a reflection of instability resulting from the presence of mobile genetic elements. On the other hand, in serotypes that contain more than one strain, the pattern of genes in the incomplete structures appears to be conserved, suggesting that the latter became stabilized at some point in their evolutionary history.
While the core region of LEE from different AE bacteria is highly conserved, the flanking regions are divergent among various AE bacteria, e.g., EPEC strain E2348/69 (serotype O127:H6), EHEC strain EDL 933 (serotype O157:H7), and the RDEC strain RDEC-1 (serotype O15:NM) (49, 62, 71). In EPEC strain E2348/69, the right end of the LEE core region contains a transposon before its insertion into the selC locus. The right end of LEE in EDL 933 contains 13 ORFs belonging to a putative P4 family prophage before joining selC. Partial sequencing of the right end of the LEE core region of RDEC-1, on the other hand, revealed an ORF which is homologous to the efa1/lifA gene of EDL 933 OI-122 (71). Tauschek et al. (62) found that the LEE of an RDEC-1-like strain (83/89) of serotype O15:NM is much larger (59,540 bp) than the LEE regions of EDL 933 and E2348/69, which are less than 44,000 bp. Complete sequencing of the right end revealed that the 83/89 LEE is inserted into pheU tRNA and contains a ca. 15-kb region which includes two putative virulence genes of EDL 933 OI-122, namely, efa1/lifA (ORFs Z4332 and Z4333) and sen (Z4326) (62). Such an arrangement has been referred to as a mosaic PAI by Morabito and colleagues (44), who found evidence of a LEE-OI-122 mosaic PAI in eight different serogroups of EPEC and non-O157 EHEC, including serogroups O26 and O103. A mosaic PAI (containing LEE adjoined to an incomplete OI-122) would thus be a plausible explanation for the incomplete OI-122 observed in some seropathotype B strains (of serotypes O26:H11 and O103:H2) which, like the mosaic PAI in the RDEC-like strains, were found to be positive for efa1/lifA (ORFs Z4332 and Z4333) and sen (Z4326) but negative for pagC (Z4321). The close clonal relationship of RDEC-1 to EHEC serotype O26:H11 (http://www.shigatox.net/stec/) further supports this hypothesis.
To study their evolutionary relationship, Whittam and colleagues have studied the clonal relationship of VTEC strains that have been characterized by multilocus enzyme electrophoresis or by multilocus sequence typing (67, 68; STEC Reference Center [http://www.shigatox.net/stec/index.html]). Four major clonal groups have been identified: EHEC 1, EHEC 2, STEC 1, and STEC 2 (67; STEC Reference Center). Dendrograms exhibiting these clonal groups may be viewed at the STEC Reference Center website. The correlation between Whittam's clonal groups and some of the serotypes and seropathotypes in this study is of interest. EHEC 1, comprising serotypes O157:H7 and O157:NM, corresponds to seropathotype A. EHEC 2 contains serotype O26:H11, which is classified as seropathotype B. However, other serotypes in seropathotype B, such as O103:H2 and O145:NM, are outside the EHEC 2 clonal group, as is serotype O121:H19 (59). The STEC 1 clone (also known as the H21 clone) contains serotypes O91:H21 and O113:H21, which are major components of seropathotype C. However, STEC 1 also contains serotype O146:H21, one strain of which was included in the present study and classified as seropathotype D. Strains of serotype O91:H21 and O113:H21 contain only one of the OI-122 virulence genes (Z4321; pagC-like), whereas the O146:H21 strain contains none of the OI-122 putative virulence genes. Another H21 strain, of serotype O104:H21, is, like the O91:H21 and O113:H21 strains, classified as seropathotype C, and, like the latter, contains only the pagC-like gene Z4321. But, unlike serotype O91:H21 and O113:H21 strains, the O104:H21 strain is not in the H21 STEC 1 clonal group. It is, however, clonally related to a strain of serotype O132:NM. A strain of this serotype in our study is classified in seropathotype D and contains none of the OI-122 genes tested. Whittam's STEC 2 clone contains a serotype O103:H2 strain that belongs to seropathotype B. The tendency toward a linkage between serotypes, seropathotypes, clonal groups, and PAIs is consistent with Whittam's model, which envisages the clonal evolution of STEC pathogens through the acquisition of novel horizontally acquired genetic elements (67).
VTs and LEE provide the only proven virulence strategies for VTEC infection. VTs act systemically to produce cytopathology in capillary endothelial cells in the kidneys, bowel, and other organs and tissues, causing HUS and hemorrhagic colitis (30). LEE orchestrates the activities of a type III secretion system to produce AE cytopathology in the colonic mucosa (45). Several other putative virulence factors, both plasmid encoded and chromosomally encoded, have been described, mostly in E. coli O157:H7, but their role in pathogenesis remains unclear. E. coli O157: H7 and other EHEC isolates contain a large plasmid carrying several candidate virulence genes. In strain EDL 933, the 92-kb F-like plasmid pO157 contains 100 ORFs, 20 of which encode putative virulence factors (10, 29, 38). These include EHEC hemolysin (encoded by the hlyCABD operon), catalase-peroxidase (katP), a serine protease (espP), a 13-gene cluster (etpC to etpO) related to the type II secretion pathway, and an ORF encoding a large (3,169-amino-acid) predicted product that shares homology with large clostridial toxins (LCT). The large plasmid is present in several other VTEC serotypes, but its composition is very heterogeneous (4, 7). Plasmids from strains causing HUS have lacked one or more of hly, katP, and espP, suggesting that the latter are not, by themselves, critical for the development of disease (4, 7). The present study showed that within serotypes, different strains had different hly, katP, and espP contents, indicating that the latter are not suitable for distinguishing seropathotypes. Other putative virulence genes include efa1, a chromosomal gene (corresponding to Z4332 and Z4333 in the present study) reported in a serotype O111:NM strain that has been associated with adherence to Chinese hamster ovary cells (47), and iha (60), a putative chromosomally encoded adhesin in E. coli O157:H7 that is similar to the iron-regulated gene A (irgA) of Vibrio cholerae. Like many other enteric pathogens, E. coli O157: H7 is able to utilize heme and/or hemoglobin as iron sources (35). ChuA, an outer membrane protein, is thought to be a specific receptor for heme uptake in this organism (43, 65) and also to be a marker for human-pathogenic strains (35).
The publication of the genome sequence of E. coli O157:H7 has revealed, in addition to LEE, at least eight genomic islands, including OI-122, that contain candidate virulence genes (21, 50). Two islands, corresponding to EDL 933 OI-154 and OI-43, have been studied further. OI-43 is a large genomic island of ∼87.5 kb which is inserted at the serW tRNA locus and contains 106 ORFs, including prophage elements, urease and tellurite resistance operons, and the iha adhesin gene. Taylor et al. (63) investigated 15 E. coli O157:H7 strains for the presence of the tellurite resistance operon, consisting of the terABCDEF genes. Six strains carried a single copy of the tellurite resistance operon, five carried two copies, and in four strains the tellurite resistance operon was not detected. The possible significance of variability in the tellurite resistance operon to the virulence of E. coli O157:H7 is unknown. The distribution of this operon in different VTEC serotypes has not been reported. Doughty et al. (14) investigated OI-154, a fimbrial operon (lpfABCD) with homology to the long polar fimbria of Salmonella. A homologue of this operon, identified in a VTEC serotype O113:H21 strain, was found in 26 of 28 LEE-negative VTEC strains and in 8 of 11 non-O157:H7 LEE-positive VTEC strains. An lpfA mutant showed decreased adherence to epithelial cells, suggesting that Lpf may function as an adhesin.
The results of our study show an association between OI-122 and VTEC seropathotypes linked to epidemic and/or serious disease. OI-122 is thus an additional candidate virulence element whose role in disease remains to be established by further study. OI-122, or its component genes, may be a marker for these seropathotypes, or it may be involved in pathogenesis. OI-122 has four genes that exhibit homology to virulence genes and nine genes of unknown function, some of which may also be virulence genes. One of the four candidate virulence genes, Z4321, shows 45.71% identity (in residues 7 to 177 of 177) to the PagC membrane protein of Salmonella serovar Typhimurium. PagC is thought to be involved in promoting the survival of Salmonella in macrophages (42, 51). Z4326 shows 38.2% identity (in residues 5 to 543 of 549) to residues 22 to 559 (of 565) of S. flexneri enterotoxin 2 (ShET2), encoded by the plasmid-mediated sen gene (46). Z4332 shows 99.3% identity (in residues 1 to 433 of 433) to residues 1 to 433 (of 3,223) of Efa1 (EHEC factor for adherence), described for a VTEC strain of serotype O111:NM (47), and to an EPEC lymphocytotoxin (33). Z4333 shows 100% identity (in residues 1 to 275 of 275) to residues 437 to 711 (of 3,223) of Efa1. Efa1 is thought to be involved in bovine bowel colonization, because efa1 mutants exhibit markedly reduced association with the bovine intestinal epithelium (57). Further studies in various experimental systems are needed in order to understand the functions of all the putative virulence genes of OI-122 and to determine if and how OI-122 can provide a selective virulence advantage to VTEC.
Acknowledgments
Songhai Shen is a Health Canada Post-Doctoral Fellow.
We thank T. Honda, T. Whittam, H. Smith, F. Jamieson, S. Aleksic, L. Beutin, J. Preiksaitis, P. Van Caeseele, G. Horsman, and P. Desmarchelier for generously providing strains used in this study.
REFERENCES
- 1.Ahmed, S., and M. Donaghy. 1998. An outbreak of Escherichia coli O157:H7 in Central Scotland, p. 59-65. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 2.Banatvala, N., P. M. Griffin, K. D. Greene, T. J. Barrett, W. F. Bibb, J. H. Green, and J. G. Wells. 2001. The United States National Prospective Hemolytic Uremic Syndrome Study: microbiologic, serologic, clinical, and epidemiologic findings. J. Infect. Dis. 183:1063-1070. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., S. Zimmermann, and K. Gleier. 1998. Human infections with Shiga toxin-producing Escherichia coli other than serogroup O157 in Germany. Emerg. Infect. Dis. 4:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielaszewska, M., H. Schmidt, A. Liesegang, R. Prager, W. Rabsch, H. Tschape, A. Cizek, J. Janda, K. Blahova, and H. Karch. 2000. Cattle can be a reservoir of sorbitol-fermenting, Shiga toxin-producing Escherichia coli O157:H− strains and a source of human diseases. J. Clin. Microbiol. 38:3470-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 8.Brunder, W., H. Schmidt, and H. Karch. 1996. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 142:3305-3315. [DOI] [PubMed] [Google Scholar]
- 9.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7, cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 10.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calderwood, S., D. W. K. Acheson, G. T. Keusch, T. J. Barrett, P. M. Griffin, N. A. Strockbine, B. Swaminathan, J. B. Kaper, M. M. Levine, B. S. Kaplan, H. Karch, A. D. O'Brien, T. G. Obrig, Y. Takeda, P. I. Tarr, and I. K. Wachsmuth. 1996. Proposed new nomenclature for SLT (VT) family. ASM News 62:118-119. [Google Scholar]
- 12.Carter, A. O., A. A. Borczyk, J. A. K. Carlson, B. Harvey, J. C. Hockin, M. A. Karmali, C. Krishnan, D. A. Korn, and H. Lior. 1987. A severe outbreak of Escherichia coli O157:H7-associated hemorrhagic colitis in a nursing home. N. Engl. J. Med. 317:1496-1500. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1993. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—Western United States, 1992-1993. JAMA 269:2194-2196. [PubMed] [Google Scholar]
- 14.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70:6761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, E. J., R. M. Robins-Browne, E. V. O'Loughlin, V. Bennett-Wood, J. Bourke, P. Henning, G. G. Hogg, J. Knight, H. Powell, and D. Redmond. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing, W. H. 1986. Edwards and Ewing's identification of Enterobacteriaceae. Elsevier Science Publishing Company, New York, N.Y.
- 17.Fleiss, J. L. 1981. Statistical methods for rates and proportions, p. 24-26. John Wiley and Sons, New York, N.Y.
- 18.Gerber, A., H. Karch, F. Allerberger, H. M. Verweyen, and L. B. Zimmerhackl. 2002. Clinical course and the role of Shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997-2000, in Germany and Austria: a prospective study. J. Infect. Dis. 186:493-500. [DOI] [PubMed] [Google Scholar]
- 19.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 20.Hacker, J., and J. Kaper. 2001. The concept of pathogenicity islands, p. 1-11. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 21.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 22.Health Canada, Population and Public Health Branch. 2000. Waterborne outbreak of gastroenteritis associated with a contaminated municipal water supply, Walkerton, Ontario, May-June 2000. Can. Communicable Dis. Rep. 26:170-173. [PubMed] [Google Scholar]
- 23.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis, K. G., and J. B. Kaper. 1996. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun. 64:4826-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, R., R. C. Clarke, J. B. Wilson, S. C. Read, K. Rahn, S. A. Renwick, K. A. Sandhu, D. Alves, M. A. Karmali, H. Lior, S. A. McEwen, J. S. Spika, and C. L. Gyles. 1996. Growing concerns and recent outbreaks involving non-O157:H7 serotypes of verotoxigenic Escherichia coli. J. Food Prot. 59:1112-1122. [DOI] [PubMed] [Google Scholar]
- 26.Kaper, J. B., and J. Hacker. 1999. Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 27.Kaper, J. B., J. L. Mellies, and J. Nataro. 1999. Pathogenicity islands and other mobile genetic elements of diarrheagenic Escherichia coli, p. 33-58. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 28.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 29.Karch, H., H. Schmidt, and W. Brunder. 1998. Plasmid-encoded determinants of Escherichia coli O157:H7, p. 183-194. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 30.Karmali, M. A. 1989. Infection by Verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between hemolytic uremic syndrome and infection by Verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 32.Karmali, M. A., M. Petric, B. T. Steele, and C. Lim. 1983. Sporadic cases of hemolytic uremic syndrome associated with fecal cytotoxin and cytotoxin-producing Escherichia coli. Lancet i:619-620. [DOI] [PubMed] [Google Scholar]
- 33.Klapproth, J. M., I. C. Scaletsky, B. P. McNamara, L. C. Lai, C. Malstrom, S. P. James, and M. S. Donnenberg. 2000. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 68:2148-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konowalchuk, J., J. I. Speirs, and S. Stavric. 1977. Vero response to a cytotoxin of Escherichia coli. Infect. Immun. 18:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law, D., and J. Kelly. 1995. Use of heme and hemoglobin by Escherichia coli O157 and other Shiga-like-toxin-producing E. coli serogroups. Infect. Immun. 63:700-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lion, T., and O. A. Haas. 1990. Nonradioactive labeling of probe with digoxigenin by polymerase chain reaction. Anal. Biochem. 188:335-337. [DOI] [PubMed] [Google Scholar]
- 37.Lopez, E. L., M. M. Contrini, and M. F. de Rosa. 1998. Epidemiology of Shiga toxin-producing Escherichia coli in South America, p. 30-37. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 38.Makino, K., K. Ishii, T. Yasunaga, M. Hattori, K. Yokoyama, C. H. Yutsudo, Y. Kubota, Y. Yamaichi, T. Iida, K. Yamamoto, T. Honda, C. G. Han, E. Ohtsubo, M. Kasamatsu, T. Hayashi, S. Kuhara, and H. Shinagawa. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy, T. A., N. L. Barrett, J. L. Hadler, B. Salsbury, R. T. Howard, D. W. Dingman, C. D. Brinkman, W. F. Bibb, and M. L. Cartter. 2001. Hemolytic-uremic syndrome and Escherichia coli O121 at a lake in Connecticut, 1999. Pediatrics 108:E59. [DOI] [PubMed] [Google Scholar]
- 40.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michino, H., K. Araki, S. Minami, T. Nakayama, Y. Ejima, K. Hiroe, H. Tanaka, N. Fujita, S. Usami, M. Yonekawa, K. Sadamoto, S. Takaya, and N. Sakai. 1998. Recent outbreaks of infection caused by Escherichia coli O157:H7 in Japan, p. 73-81. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 42.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mills, M., and S. M. Payne. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 177:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morabito, S., R. Tozzoli, E. Oswald, and A. Caprioli. 2003. A mosaic pathogenicity island made up of the locus of enterocyte effacement and a pathogenicity island of Escherichia coli O157:H7 is frequently present in attaching and effacing E. coli. Infect. Immun. 71:3343-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nataro, J. P., J. Seriwatana, A. Fasano, D. R. Maneval, L. D. Guers, F. Noriega, F. Dubovsky, M. M. Levine, and J. G. Morris. 1995. Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect. Immun. 63:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholls, L., T. H. Grant, and R. Robins-Brown. 2001. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 48.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. Kirkpatrick, G. Posfal, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 51.Pulkkinen, W. S., and S. I. Miller. 1991. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J. Bacteriol. 173:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Read, S. C., R. C. Clarke, A. Martin, S. A. De Grandis, J. Hii, S. McEwen, and C. L. Gyles. 1992. Polymerase chain reaction for detection of verocytotoxigenic Escherichia coli isolated from animal and food sources. Mol. Cell. Probes 6:153-161. [DOI] [PubMed] [Google Scholar]
- 53.Read, S. C., C. L. Gyles, R. C. Clarke, H. Lior, and S. McEwen. 1990. Prevalence of verocytotoxigenic Escherichia coli in ground beef, pork, and chicken in southwestern Ontario. Epidemiol. Infect. 105:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 57.Stevens, M. P., P. M. van Diemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tarr, C. L., T. M. Large, C. L. Moeller, D. W. Lacher, P. I. Tarr, D. W. Acheson, and T. S. Whittam. 2002. Molecular characterization of a serotype O121:H19 clone, a distinct Shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70:6853-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tarr, P. I., S. S. Bilge, J. C. Vary, S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tarr, P. I., and M. A. Neill. 1996. The problem of non-O157:H7 shiga-toxin (verocytotoxin)-producing Escherichia coli. J. Infect. Dis. 174:1136-1139. [DOI] [PubMed] [Google Scholar]
- 62.Tauschek, M., R. A. Strugnell, and R. Robins-Brown. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 63.Taylor, D. E., M. Rooker, M. Keelan, L. K. Ng, I. Martin, N. T. Perna, N. T. Burland, and F. R. Blattner. 2002. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184:4690-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 66.Tozzi, A. E., A. Caprioli, F. Minelli, A. Gianviti, L. De Petris, A. Edefonti, G. Montini, A. Ferretti, T. De Palo, M. Gaido, and G. Rizzoni. 2003. Shiga toxin-producing Escherichia coli infections associated with hemolytic uremic syndrome, Italy, 1988-2000. Emerg. Infect. Dis. 9:106-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whittam, T. S. 1998. Evolution of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains, p. 195-209. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 68.Whittam, T. S., I. K. Wachsmuth, and R. A. Wilson. 1988. Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J. Infect. Dis. 157:1124-1133. [DOI] [PubMed] [Google Scholar]
- 69.Wilson, J. B., R. C. Clarke, S. Renwick, K. Rahn, R. P. Johnson, M. A. Karmali, H. Lior, D. Alves, C. Gyles, K. S. Sandhu, S. A. McEwen, and J. Spika. 1996. Verocytotoxigenic Escherichia coli infection in dairy farm families. J. Infect. Dis. 174:1021-1027. [DOI] [PubMed] [Google Scholar]
- 70.World Health Organization. 1999. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC), p. 1-30. Report of a WHO Scientific Working Group Meeting, Berlin, Germany, 22-23 June, 1998. World Health Organization, Geneva, Switzerland.
- 71.Zhu, C., T. S. Agin, S. Elliot, L. A. Johnson, T. Thate, J. B. Kaper, and E. C. Boedeker. 2001. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect. Immun. 69:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]