Abstract
The Chemagic Viral DNA/RNA kit was evaluated for extraction of cytomegalovirus (CMV), hepatitis B virus (HBV), and hepatitis G virus (HGV) by using the QIAamp DNA Blood Mini kit and the QIAamp Viral RNA Mini kit as reference protocols. The extraction efficiencies of the different kits for CMV DNA and HBV DNA were not distinguishable, but the extraction efficiency for HGV RNA was better with the Chemagen protocol. All clinical specimens tested HBV DNA- or HGV RNA-positive after QIAGEN protocols for extraction were confirmed by using the Chemagen protocol. The Chemagen kit failed to confirm one of 75 CMV DNA-positive specimens. Thus, a new competitive extraction method using a technology with a high potential for automation is available.
Combining sensitive detection and quantification of pathogen-encoded nucleic acids with short processing times, nucleic-acid amplification protocols are becoming a dominating technique in the diagnostic laboratory. Prerequisite to benefiting from the advantages of this technique are efficient protocols for extraction of nucleic acids. Many existing protocols are labor intensive, time consuming, or restricted to certain specimens or types of nucleic acids. The optimal protocol should offer high sensitivity for extraction of DNA and RNA from a broad range of specimens combined with low time consumption, reduced hands-on time, low price, and the potential for automation. Many procedures available so far use a precipitation step to gain pure nucleic acids from the extracts. Other protocols take advantage of the nucleic acid-binding potential of matrix material supplied in a column (1). A recent development is the coverage of magnetic beads with nucleic acid-binding matrices, which promises a high potential for automation (5).
In this study, we investigated the suitability of the recently released Chemagic Viral DNA/RNA kit (Chemagen, Baesweiler, Germany), which is available worldwide, to isolate viral DNA (for CMV and HBV) and viral RNA (for HGV) from human specimens for sensitive detection of viral genomes by HBV-specific, CMV-specific, and HGV-specific PCR.
One hundred ninety-two specimens tested DNA- or RNA-positive for the respective pathogen (for HBV, 100 serum samples; for CMV, 8 urine samples, 52 serum samples, and 15 plasma samples; for HGV, 17 serum samples) by virus-specific quantitative PCR (HBV [Cobas Amplicor; Roche Diagnostics, Basel, Switzerland] and CMV) (6) or qualitative PCR (HGV) in our routine diagnostic laboratory following extraction with the QIAamp DNA Blood Mini kit or the QIAamp Viral RNA Mini kit (QIAGEN, Hilden, Germany). Extraction capacity was evaluated for HBV with a dilution series of the EUROHEP standard (4 × 105 to 1 × 101 copies per ml of serum), for CMV with a dilution series of the standard plasmid pCR-gpB diluted in serum (2 × 105 to 2 × 102 copies per ml of serum), and for HGV with a dilution series of viral RNA from serum samples from a highly viremic patient (dilutions of 1−6 to 10−6). The serum for the dilution series tested CMV-, HBV-, and HGV-negative by virus-specific PCR.
Total DNA and total RNA were extracted by using magnetic bead technology with the Chemagic Viral DNA/RNA kit according to the manufacturer's recommendations. Briefly, samples (200-μl) were incubated with protease and lysis buffer. Magnetic beads were added to the mixture. After incubation, nucleic acids bound to the beads were separated with a magnetic separator. After two washing steps, the nucleic acids were eluted in a volume of 50 μl of elution buffer and the magnetic beads were separated from the solution. The approximate processing time for 24 samples is 100 min. As reference protocols, the QIAamp DNA Blood Mini kit was used for extraction of DNA and the QIAamp Viral RNA Mini kit was used for extraction of RNA from 200-μl samples as recommended by the manufacturer. The extracted nucleic acids were resuspended in a volume of 50 μl of aqua bidest. The approximate processing time for 24 samples is 100 min.
A fluorescence resonance energy transfer (FRET)-based CMV DNA amplification protocol with a sensitivity of 200 genome equivalents per ml of sample was used for detection of CMV genomes in 5 μl of extracted nucleic acids (6). Such a real-time approach allows detection of amplicons by FRET with a virus-specific primer pair labeled with fluorescent dyes. It can define the cycle number of the entry into the exponential phase of PCR and use it for quantification (2, 4). HBV detection was performed with an aliquot of 10 μl of extracted nucleic acid by nested PCR on a Geneamp 9700 (Perkin-Elmer) as described elsewhere (3). The detection limit is 40 genome equivalents per ml of serum. HGV detection was performed by reverse transcription (RT)-PCR on a Geneamp 9700 with the Titan OneTube RT-PCR system (Roche Biochemicals, Mannheim, Germany) as recommended by the manufacturer. Four microliters of HGV-specific primers from the hepatitis G virus-primer and capture probe set (Roche Biochemicals) and 10 μl of extracted RNA were added to the mix. The experimental PCR protocol was as follows: an initial 50 min at 30°C and 2 min at 68°C for RT, followed by 40 cycles of 45 s of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of extension at 68°C. The detection limit is 400 genome equivalents per ml of serum.
For evaluation of the analytical sensitivity of the Chemagic Viral DNA/RNA kit, nucleic acids were extracted from serum dilution series containing the CMV-, HBV-, and HGV-positive controls and subsequently amplified with virus-specific protocols. The identical samples were extracted with the QIAamp DNA Blood Mini kit (for CMV and HBV samples) and the QIAamp Viral RNA Mini kit (for HGV samples) as reference protocols. Each extraction was done at least in triplicate; all results were in agreement.
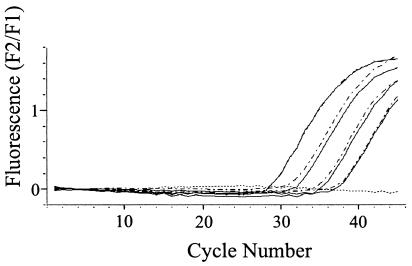
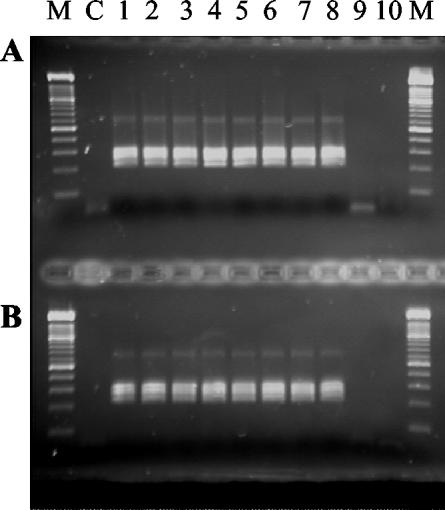
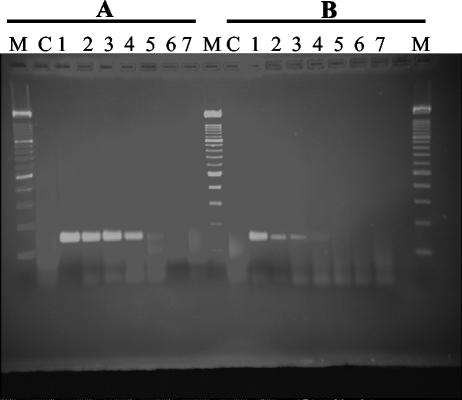
Extracts of all samples of the CMV-positive control dilution series obtained with the Chemagen and QIAGEN kits resulted in the amplification of PCR products, indicating the successful extraction of 2 × 102 to 2 × 105 copies per ml of serum (Fig. 1). Quantification results following extraction with both protocols differed less than 1.4-fold (0.5 cycles) for each serial dilution. Amplification reactions using extracts of the HBV-positive control resulted in detectable PCR products for 4 × 101 to 2 × 105 genome equivalents per ml of serum independently of the extraction protocol (Fig. 2). Thus, no difference in extraction efficiency could be observed between the two protocols for the extraction of HBV DNA or CMV DNA on the basis of identical sample volumes. Extracts of the dilution series of viral RNA from serum samples from a highly viremic HGV genome-positive patient obtained with the Chemagic DNA/RNA kit resulted in the amplification of PCR products, including a dilution of 10−4 (Fig. 3). The extraction efficiency of the QIAamp Viral RNA Mini kit was 10-fold lower.
FIG. 1.
Amplification of a serial dilution of the standard plasmid pCR-gpB by real-time PCR with CMV-specific primers (amplicon, 254 bp) on the LightCycler instrument (software, version 3.5; analysis method, fit points with manual noise band adjustment, hybridization probe format) following extraction with the Chemagic DNA/RNA kit and the QIAamp DNA Blood Mini kit. pCR-gpB concentrations (from left to right) were 2 × 105, 2 × 104, 2 × 103, and 2 × 102 copies per ml of serum. Continuous line, Chemagen extracts; broken line, QIAGEN extracts. Cycle number, cycle number of the amplification reaction; F2/F1, quotient of fluorescence channel F2 (hybridization probe) and fluorescence channel F1 (fluorescein).
FIG. 2.
Amplification of a serial dilution of the EUROHEP standard with HBV-specific primers in a nested PCR following extraction with the Chemagic DNA/RNA kit and the QIAamp DNA Blood Mini kit. (A) Chemagen extracts. (B) QIAGEN extracts. Lane M shows a 100-bp ladder, and lane C shows results for the negative control. Lanes 1 through 10 show results for different viral loads expressed in numbers of copies per ml: 1, 4 × 105; 2, 4 × 104; 3, 4 × 103; 4, 4 × 102; 5, 2 × 102; 6, 1 × 102; 7, 8 × 101; 8, 4 × 101; 9, 2 × 101; 10, 1 × 101.
FIG. 3.
Amplification of a serial dilution of serum from a highly viremic HGV genome-positive patient with HGV-specific primers following extraction with the Chemagic DNA/RNA kit and the QIAamp DNA Blood Mini kit. (A) Chemagen extracts. (B) QIAGEN extracts. Lane M shows a 100-bp ladder, and lane C shows results for the negative control. Lanes 1 through 7 show results for different dilution rates: 1, undiluted; 2, 10−1, 3, 10−2; 4, 10−3; 5, 10−4; 6, 10−5; 7, 10−6.
The clinical sensitivity of the Chemagic DNA/RNA kit was investigated by extraction of 8 urine samples, 15 plasma samples, and 52 serum samples that had previously tested positive for CMV DNA, 100 serum samples that had previously tested positive for HBV DNA, and 17 serum samples that had previously tested positive for HGV RNA in our routine diagnostic laboratory after extraction with the QIAamp DNA Blood Mini kit or the QIAamp Viral RNA Mini kit. All samples were extracted in parallel with the Chemagen protocol and the reference protocols. Extracts were qualitatively analyzed by virus-specific amplification protocols to minimize the effects of variation in PCR performance on the evaluation.
A total of 75 specimens ranging from 5 × 101 to 1 × 107 copies of CMV per ml (Table 1) have been analyzed. All of them resulted in a positive PCR following extraction with the QIAamp DNA Blood Mini kit. Apart from one sample previously quantified to contain 7 × 102 copies of CMV per ml of serum, PCR products could be obtained in the remaining 74 specimens following extraction with the Chemagic DNA/RNA kit. The entire set of 100 HBV-positive sera ranging from 5 × 101 to 9 × 107 copies per ml of serum resulted in HBV-specific PCR products after amplification of extracts generated with both the Chemagic DNA/RNA kit and the QIAamp DNA Blood Mini kit (Table 2). Following extraction of 17 HGV genome-positive sera with the Chemagic DNA/RNA kit, HGV-specific PCR products could be generated from all specimens. Identical results were obtained after extraction of the identical specimens with the QIAamp Viral RNA Mini kit.
TABLE 1.
Qualitative PCR results of CMV genome-positive specimens following extraction with the Chemagic DNA/RNA kit and the QIAamp DNA Blood Mini kit
| Viral loada (genome equivalents/ml) | No. of samples | PCR positive following extraction with:
|
|
|---|---|---|---|
| Chemagen | QIAGEN | ||
| ≤2 × 102 | 19 | 19 | 19 |
| >2 × 102-≤5 × 102 | 12 | 12 | 12 |
| >5 × 102-≤1 × 103 | 14 | 13 | 14 |
| >1 × 103-≤5 × 103 | 10 | 10 | 10 |
| >5 × 103-≤1 × 104 | 5 | 5 | 5 |
| >1 × 104 | 15 | 15 | 15 |
| Total | 75 | 74 | 75 |
The samples were grouped by viral loads previously obtained by quantitative CMV PCR following extraction with the QIAamp DNA Blood Mini kit in our routine laboratory.
TABLE 2.
Qualitative PCR results of HBV genome-positive specimens following extraction with the Chemagic DNA/RNA kit and the QIAamp DNA Blood Mini kit
| Viral loada (genome equivalents/ml) | No. of samples | No. PCR positive following extraction with:
|
|
|---|---|---|---|
| Chemagen | QIAGEN | ||
| ≤5 × 102 | 23 | 23 | 23 |
| >5 × 102-≤1 × 103 | 13 | 13 | 13 |
| >1 × 103-≤5 × 103 | 14 | 14 | 14 |
| >5 × 103-≤5 × 104 | 17 | 17 | 17 |
| >5 × 104-≤5 × 105 | 14 | 14 | 14 |
| >5 × 105-≤5 × 106 | 7 | 7 | 7 |
| >5 × 106 | 12 | 12 | 12 |
| Total | 100 | 100 | 100 |
The samples were grouped by viral loads previously obtained by quantitative HBV PCR following extraction with the QIAamp DNA Blood Mini kit in our routine laboratory.
We performed an evaluation of the Chemagic DNA/RNA kit by comparing the analytical and clinical sensitivity of the protocol to that of the QIAamp DNA Blood Mini kit and the QIAamp Viral RNA Mini kit. Determination of the extraction efficiency for CMV DNA, HBV DNA, and HGV RNA was done by PCR assays of standard dilution series and of clinical specimens, including serum, plasma, and urine samples. However, due to the limited numbers of urine and plasma samples used in this study, a well-grounded evaluation of the kit for extraction of nucleic acids from our data can be done only for serum.
Analysis of the nucleic acid extracts obtained by the extraction systems in use resulted in 100% detection of positive samples by the extraction systems of both companies for HBV- and HGV genome-positive specimens. Analysis of the CMV genome-positive specimens detected CMV in 98.7% of the samples after extraction with the Chemagic DNA/RNA kit and in 100% of the samples after extraction with the QIAamp DNA Blood Mini kit. Evaluation of analytical sensitivity revealed identical results for CMV and HBV with both extraction systems. Extraction with the Chemagic DNA/RNA kit was 10-fold more sensitive for HGV-positive sera than with the QIAamp Viral RNA Mini kit. An advantage of both protocols is the absence of a precipitation step, which often causes problems with yield and purity.
Besides sensitivity of extraction, costs are a factor to be considered when selecting an appropriate extraction protocol for a laboratory. The costs per extraction are 1.95 Euro for the Chemagic DNA/RNA kit, 1.86 Euro for the QIAamp DNA Blood Mini kit, and 2.52 Euro for the QIAamp Viral RNA Mini kit. These costs make the Chemagen protocol a competitive candidate for laboratories extracting DNA and RNA. A further advantage of the product is that simultaneous extraction of both species of nucleic acids can be performed in the same time as the reference protocols. Specimens to be analyzed for both viral DNA and RNA have to be extracted only once. Therefore, hands-on time is minimized, and efficient use of specimens difficult to obtain, e.g., ascites, is guaranteed.
With the novel Chemagic DNA/RNA kit a very competitive new product is available. The analytical and clinical sensitivity are equal to those of the reference protocols. The simultaneous extraction of DNA and RNA and the underlying magnetic beads technology make it an interesting system for manual extraction of nucleic acids in a routine diagnostic laboratory and especially for automated extraction of nucleic acids. Very recently, an extraction robot using the Chemagic DNA/RNA kit in a 96-well format with high efficiency of extraction and minimization of cross contamination in preliminary experiments has been released.
REFERENCES
- 1.Fahle, G. A., and S. H. Fischer. 2000. Comparison of six commercial DNA extraction kits for recovery of cytomegalovirus DNA from spiked human specimens. J. Clin. Microbiol. 38:3860-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Häusler, M., S. Scheithauer, K. Ritter, and M. Kleines. 2003. Molecular diagnosis of Epstein-Barr virus. Expert Rev. Mol. Diagn. 3:89-100. [DOI] [PubMed] [Google Scholar]
- 3.Heermann, K. H., Y. Hagos, and R. Thomssen. 1994. Liquid-phase hybridization and capture of hepatitis B virus DNA with magnetic beads and fluorescence detection of PCR product. J. Virol. Methods 50:43-57. [DOI] [PubMed] [Google Scholar]
- 4.Kleines, M., K. Ritter, and L. Schaade. 2002. Advances in quantification of cytomegalovirus (HCMV)-loads by real time PCR approaches. Recent Res. Devel. Microbiol. 6:107-114. [Google Scholar]
- 5.Legler, T. J., M. Kohler, and K. H. Heermann. 1999. High-throughput extraction, amplification, and detection (HEAD) of HCV-RNA in individual blood donations. J. Clin. Virol. 13:95-103. [DOI] [PubMed] [Google Scholar]
- 6.Schaade, L., P. Kockelkorn, K. Ritter, and M. Kleines. 2000. Detection of cytomegalovirus DNA in human specimens by LightCycler PCR. J. Clin. Microbiol. 38:4006-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]