Abstract
Serotype 1 pneumococci are a major cause of serious disease and have been associated with outbreaks but are rarely carried. The high attack rate and lack of coverage of this serotype by the heptavalent conjugate vaccine prompted the characterization of a geographically diverse collection of 166 serotype 1 isolates from recent cases of invasive disease. The isolates were resolved by multilocus sequence typing into 16 clones, which clustered into three major lineages with very different geographic distributions. Lineage A isolates were exclusively from Europe and North America, lineage B isolates were predominantly from Africa and Israel, and lineage C isolates were mainly from Chile. There was no clear association between the presence of individual clones within a country and the prevalence of serotype 1 disease.
Serotype 1 has remained one of the most common pneumococcal serotypes causing disease since its description and association with serious pneumonia in 1913 (5), although its rank order among serotypes causing invasive disease differs among countries and has changed over time within countries (13). It is one of the few pneumococcal serotypes associated with outbreaks of disease in small or closed communities (3, 4, 11, 12, 18, 20, 22, 26), and increases in serotype 1 disease have been associated with nationwide increases in invasive disease (14, 16). Three recent reports have indicated that serotype 1 pneumococci are the most common cause of complicated pneumonia and pulmonary empyema in children (2, 7, 28), and serotype 1 has been associated with rare disease syndromes such as peritonitis and salpingitis in adolescent girls (19, 25, 30).
Streptococcus pneumoniae is commonly carried in the nasopharynges of healthy children and adults; however, a characteristic epidemiological feature of serotype 1 is that it is very rarely found among healthy carriers, even in locations where serotype 1 frequently causes invasive disease (11, 14, 18, 21, 27). The fact that serotype 1 is among the most common causes of invasive disease in many parts of the world, although it is rarely carried by either adults or children, and the ability of serotype 1 to cause outbreaks suggest that this serotype has an unusually high attack rate.
The ability to differentiate pneumococcal capsular types has allowed investigators to explore the serotype-specific epidemiology of pneumococcal disease in different countries. These studies have informed the development of the conjugate vaccines, as the vaccines cover those serotypes most commonly recovered in cases of invasive disease (13). However, the currently licensed pediatric heptavalent vaccine does not provide protection against serotype 1 disease, although higher-valency vaccines will do so. The prevalence of serious disease caused by a serotype that appears to have a high attack rate and that is not covered by the currently licensed conjugate polysaccharide vaccine is of some concern, particularly because the use of conjugate vaccines may influence the extent to which individual serotypes are carried and transmitted within a community. Therefore, we explored the clonal diversity of invasive serotype 1 pneumococci and attempted to determine whether specific clones are associated with the high prevalence of serotype 1 disease in certain parts of the world.
MATERIALS AND METHODS
The study collection was acquired from colleagues in 16 locations who were asked to contribute their 10 most recent invasive serotype 1 S. pneumoniae isolates to this study. We also requested data on the rank order of serotype 1 pneumococci causing invasive disease in their local regions during the time the isolates they submitted to this study were recovered. All pneumococcal isolates were identified in their respective microbiology laboratories by standard microbiological techniques. Isolates were sent to the University of Oxford, where the serotype was confirmed by the Quellung reaction using a serotype 1 antiserum (Statens Serum Institut, Copenhagen, Denmark), genomic DNA was extracted using the DNeasy tissue kit (Qiagen, West Sussex, England), and molecular typing was performed. A reference strain of serotype 1 S. pneumoniae (NCTC 7465), deposited in the National Collection of Type Cultures, London, United Kingdom, in 1948, was also included in the study collection.
The pneumococcal isolates were characterized by using multilocus sequence typing (MLST), a precise method of genetic characterization which involves PCR amplification and sequencing of internal fragments of seven housekeeping genes, aroE, gdh, gki, recP, spi, xpt, and ddl, to identify the allele at each locus (8). The different alleles (sequences) are assigned numbers, and the allele numbers at the seven loci indicate the allelic profile, or sequence type (ST). A clone (ST) is defined as a group of S. pneumoniae isolates that have the identical MLST allelic profile. All of the allele and ST designations are given according to the MLST website (http://www.mlst.net).
RESULTS
Each collaborator provided 7 to 14 unique patient isolates of serotype 1 S. pneumoniae, resulting in a study collection of 166 isolates from 14 different countries (Table 1). The pneumococci were recovered from patients with invasive disease and were isolated from normally sterile body sites, primarily blood (82.5%) and cerebrospinal fluid (15.1%). The isolates were collected from patients who were 11 days to 85 years old. The proportion of isolates in each of three age groups was as follows: ≤5 years, 27.1%; 6 to 64 years, 56.0%; ≥65 years, 13.9%. For 3.0% of isolates, the ages of the patients were unknown.
TABLE 1.
Description of a global collection of invasive serotype 1 S. pneumoniae isolates
| Country | No. of isolates | No. (%) of isolates of the following specimen typea
|
No. (%) of isolates in the following age group:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Blood | CSF | BF | ≤5 yr | 6-64 yr | ≥65 yr | Unknown | ||
| Canada (Quebec province) | 10 | 10 | 8 | 2 | ||||
| Canada (Toronto) | 10 | 8 | 2 | 6 | 4 | |||
| Chile | 11 | 9 | 2 | 6 | 3 | 2 | ||
| Denmark | 9 | 9 | 1 | 5 | 3 | |||
| England | 10 | 8 | 2 | 9 | 1 | |||
| France | 10 | 10 | 1 | 9 | ||||
| Germany | 10 | 9 | 1 | 6 | 4 | |||
| Israel | 12 | 12 | 6 | 6 | ||||
| Kenya | 12 | 9 | 3 | 5 | 7 | |||
| Norway | 10 | 10 | 6 | 4 | ||||
| Poland | 7 | 7 | 1 | 2 | 2 | 2 | ||
| South Africa | 10 | 5 | 5 | 9 | 1 | |||
| Spain | 14 | 13 | 1 | 3 | 8 | 3 | ||
| The Netherlands | 13 | 9 | 4 | 2 | 8 | 3 | ||
| United States | 10 | 10 | 2 | 7 | 1 | |||
| United States (Navajo Indians) | 8 | 6 | 2 | 3 | 4 | 1 | ||
| Total | 166 | 137 (82.5) | 25 (15.1) | 4 (2.4) | 45 (27.1) | 93 (56.0) | 23 (13.9) | 5 (3.0) |
CSF, cerebrospinal fluid; BF, other normally sterile body fluid.
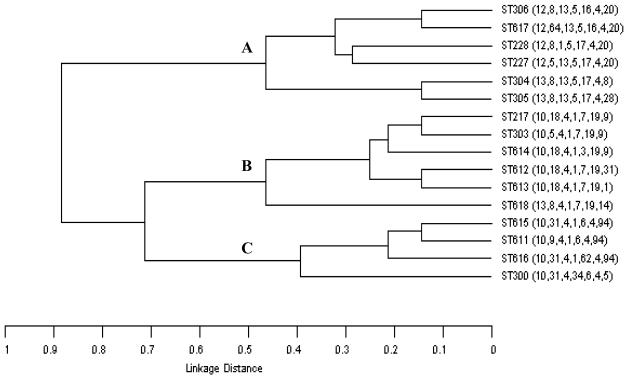
Sixteen different STs (clones) were identified among the entire collection of S. pneumoniae isolates, but six clones, STs 306, 227, 217, 228, 615, and 304, made up nearly 90% (n = 147) of the collection (Table 2). The STs clustered into three major lineages (A, B, and C) (Fig. 1), defined as a group of STs that shared four or more alleles. Lineages B and C also appeared to be related, since most of the STs in these two lineages shared alleles at three of the seven loci (Table 3). Interrogation of the pneumococcal MLST database identified only an additional four STs of serotype 1, and these also clustered within the same three major lineages. The same groupings could be obtained by using BURST (available at http://www.mlst.net), and ST217 was assigned by BURST as the ancestral genotype from which the other STs of lineage B arose. Robust predictions for the ancestors of lineages A and C could not be obtained.
TABLE 2.
ST distribution of invasive serotype 1 S. pneumoniae isolates
| Country of origin | No. of isolates of the following ST:
|
Total no. of isolates | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST306 | ST617 | ST228 | ST227 | ST304 | ST305 | ST217 | ST303 | ST614 | ST612 | ST613 | ST618 | ST615 | ST611 | ST616 | ST300 | ||
| Denmark | 6 | 1 | 1 | 1 | 9 | ||||||||||||
| The Netherlands | 3 | 3 | 1 | 3 | 2 | 1 | 13 | ||||||||||
| Norway | 8 | 1 | 1 | 10 | |||||||||||||
| France | 3 | 2 | 3 | 2 | 10 | ||||||||||||
| Spain | 6 | 7 | 1 | 14 | |||||||||||||
| Germany | 8 | 2 | 10 | ||||||||||||||
| Poland | 5 | 2 | 7 | ||||||||||||||
| England | 9 | 1 | 10 | ||||||||||||||
| Canada (Toronto) | 10 | 10 | |||||||||||||||
| Canada (Quebec) | 1 | 3 | 5 | 1 | 10 | ||||||||||||
| United States | 1 | 6 | 1 | 1 | 1 | 10 | |||||||||||
| United States (Navajo Indians) | 8 | 8 | |||||||||||||||
| Kenya | 8 | 3 | 1 | 12 | |||||||||||||
| South Africa | 5 | 3 | 2 | 10 | |||||||||||||
| Israel | 11 | 1 | 12 | ||||||||||||||
| Chile | 11 | 11 | |||||||||||||||
| Total | 41 | 1 | 15 | 40 | 10 | 3 | 29 | 1 | 3 | 4 | 1 | 1 | 12 | 2 | 2 | 1 | 166 |
FIG. 1.
Dendrogram describing the relatedness of each of the 16 clones. A linkage distance of 0.14 or 0.29 indicates identical alleles at 6 of 7 or 5 of 7 loci, respectively. Lineage A includes ST306, 617, 228, 227, 304, and 305; lineage B includes ST217 (ancestral clone), 303, 614, 612, 613, and 618; and lineage C includes ST615, 611, 616, and 300. ST615 includes the serotype 1 reference strain from 1948.
TABLE 3.
STsa characterized among a global collection of 166 serotype 1 S. pneumoniae isolates
| Genetic lineage | ST | No. of isolates | Yr of isolation | Allelic profile
|
Location(s) where ST was isolated (no. of isolates) | MLST comments | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | ||||||
| A | 306 | 41 | 1995-2001 | 12 | 8 | 13 | 5 | 16 | 4 | 20 | Europe (39), Canada (1), United States (1) | Major invasive serotype 1 clone in Sweden in the late 1990s |
| 617 | 1 | 2001 | 12 | 64 | 13 | 5 | 16 | 4 | 20 | Europe | Newly recognized ST | |
| 228 | 15 | 1996-2001 | 12 | 8 | 1 | 5 | 17 | 4 | 20 | Europe (12), Canada (3) | Previously noted in Denmark and Spain | |
| 227 | 40 | 1995-2001 | 12 | 5 | 13 | 5 | 17 | 4 | 20 | England (9), Canada (15), United States (14), Europe (2) | Previously noted in the United Kingdom and Denmark | |
| 304 | 10 | 1996-2001 | 13 | 8 | 13 | 5 | 17 | 4 | 8 | Europe (8), Canada (1), United States (1) | Previously noted in Sweden | |
| 305 | 3 | 2000 | 13 | 8 | 13 | 5 | 17 | 4 | 28 | Europe | Previously noted in Sweden | |
| B | 217 | 29 | 1990-2001 | 10 | 18 | 4 | 1 | 7 | 19 | 9 | Africa (13), Israel (11), Europe (5) | Previously noted in Denmark and Sweden |
| 303 | 1 | 2001 | 10 | 5 | 4 | 1 | 7 | 19 | 9 | United States | Single-locus variant of ST217; previously noted in Sweden | |
| 614 | 3 | 2001 | 10 | 18 | 4 | 1 | 3 | 19 | 9 | Africa | Single-locus variant of ST217 | |
| 612 | 4 | 1998-2001 | 10 | 18 | 4 | 1 | 7 | 19 | 31 | Africa (3), Israel (1) | Single-locus variant of ST217 | |
| 613 | 1 | 2001 | 10 | 18 | 4 | 1 | 7 | 19 | 1 | Africa | Single-locus variant of ST217 | |
| 618 | 1 | Unknown | 13 | 8 | 4 | 1 | 7 | 19 | 14 | Europe | Newly recognized ST | |
| C | 615 | 12 | 2000-2001 | 10 | 31 | 4 | 1 | 6 | 4 | 94 | Chile (11), United States (1) | ST of the 1948 serotype 1 reference strain |
| 611 | 2 | 2001 | 10 | 9 | 4 | 1 | 6 | 4 | 94 | Africa | Single-locus variant of ST615 | |
| 616 | 2 | 1995-1996 | 10 | 31 | 4 | 1 | 62 | 4 | 94 | Europe | Single-locus variant of ST615 | |
| 300 | 1 | 2001 | 10 | 31 | 4 | 34 | 6 | 4 | 5 | England | Double-locus variant of ST615; previously noted in Sweden | |
A total of 16 STs were found.
There was a striking geographic structure within the lineages. All 110 isolates within lineage A were recovered from Europe, the United States, or Canada, whereas 82.1% (32 of 39) of the isolates of lineage B were from Africa or Israel. There were relatively few isolates in lineage C, and most of these were members of ST615, which with one exception was recovered exclusively from South America (Table 3).
Lineage A included two major clones, ST227 and ST306, whose distribution also showed a marked geographic structure (Table 3). Of the isolates from England and North America, 79.2% were members of clone ST227, including all eight isolates from the American Navajo Indians and three isolates from the Canadian Inuit, the aboriginal inhabitants of the North American Arctic. All of the isolates submitted from Toronto belonged to ST227. Conversely, 95.1% of the ST306 isolates were from continental Europe (Table 2). Besides ST306, two other major clones within lineage A, ST228 and ST304, were recovered predominantly from continental Europe. Twelve ST228 isolates were from continental Europe, and the remaining three were from Quebec, a province with close links to France. Similarly, of the 10 ST304 isolates, 8 were from continental Europe and 1 was from the province of Quebec. Together, these three clones (STs 306, 228, and 304) comprised 80.8% of all continental European isolates.
Within lineage B, one major clone, ST217, predominated in Kenya, South Africa, and Israel; 70.6% of the 34 African and Israeli isolates belonged to ST217. Three closely related STs, identical to ST217 at six of the seven loci, described a further eight isolates, all of which were recovered in Africa or Israel. There were relatively few isolates within lineage C, the majority of which were from Chile and were assigned to clone ST615. The NCTC type strain of serotype 1 was also found to be a member of this clone.
The rank order of serotype 1 among serotypes causing invasive disease was also obtained for 14 of the 16 contributors (1, 6, 10, 15, 24, 26, 29; B. Beall, D. Caugant, A. Fenoll, H. Konradsen, A. McGreer, V. Prado, and A. von Grottberg, personal communication). Serotype 1 was ranked among the most common invasive pneumococcal serotypes in Denmark, Israel, Kenya, Norway, and the Inuit in Nunavik, Quebec province (1st), England (2nd), South Africa (2nd [blood] and 4th [cerebrospinal fluid]), Germany (3rd), Chile and Spain (5th), American Navajo Indian adults (among the first 6), and France (7th) but ranked 15th among invasive pneumococci in Toronto, Canada, and 17th among isolates from the United States. The rank order among isolates from Poland and The Netherlands was unknown.
DISCUSSION
For nearly a century, serotype 1 has consistently ranked among the most prevalent pneumococcal serotypes causing severe disease in both children and adults worldwide. However, in contrast to most other invasive serotypes, serotype 1 is very rarely recovered from healthy carriers. The disparity between the extent of disease and the extent of carriage suggests that serotype 1 pneumococci have a high attack rate; acquisition is rare, but if the organism is acquired, there is a much greater chance of progression to invasive disease than with most other serotypes. It could be argued that the inability to observe serotype 1 in carriage is an artifact of cross-sectional carriage studies (e.g., it could be acquired frequently but very rapidly cleared from the nasopharynx), but a high attack rate is supported by the observation that serotype 1 is one of the very few pneumococcal serotypes that have been associated with outbreaks of disease.
Only two previous studies have used molecular methods to explore clonal diversity in serotype 1 pneumococci. The first was a study of pediatric isolates from Israel in 1995 to 1999, where serotypes 1 and 5 were the most common pediatric invasive serotypes during the 10 years prior to the study. Sixty-six invasive isolates and 13 carriage isolates (representing 0.3% of the 4,600 carried pneumococci isolated) were analyzed by pulsed-field gel electrophoresis (PFGE), and all were considered to belong to a single clonal type (21). The second molecular study was from Sweden (14) and was undertaken in response to a significant nationwide increase in invasive pneumococcal disease from 1989 to 1997. All serotype 1 isolates recovered in Sweden in 1987 (n = 3), 1992 (n = 6), and 1997 (n = 51) were characterized by PFGE, and 20 were subsequently selected for MLST characterization. Sixty-seven percent of the type 1 isolates from 1997 belonged to ST306, and this clone was not detected in either 1987 or 1992; therefore, a 10-fold nationwide increase in serotype 1 invasive disease was attributed to the appearance and increase in prevalence of clone ST306. Swabbing the nasopharynges of children in day care centers in Stockholm during the time when serotype 1 disease was increased resulted in no recovery of serotype 1 isolates among 246 pneumococcal strains carried.
The very low carriage of serotype 1 pneumococci may lead to more distinctive geographic differences in the clones that circulate than are found for other pneumococci. The movement of people between countries or continents frequently transfers clones of the commonly carried serotypes, but this effect would be expected to be very much less for serotype 1, as it is so rarely carried. Indeed, there was a striking geographic structure among the major serotype 1 lineages, with marked differences in the geographic distribution of individual clones. This structure was most apparent in lineage A, where the clones recovered from continental Europe were different from those recovered from North America and England.
The very low prevalence of serotype 1 carriage is also expected to have other consequences. Selective pressures for the emergence of antibiotic resistance, exerted mainly on the nasopharyngeal flora of children, should be low in serotype 1 compared to that in the commonly carried serotypes. As expected, low levels of antibiotic resistance have been reported among serotype 1 isolates in several countries where this serotype is commonly recovered in cases of disease, including those where the overall prevalence of antimicrobial resistance among pneumococcal isolates is high (2, 14, 17, 21, 23, 24). Resistance was low among all of the six major clones described in this study and at least three of the minor clones (14; N. Porat, R. Reinert, and A. Wasas, personal communication; http://www.mlst.net).
Serotype 1 clones may be more stable than those of other serotypes, as diversification of the genome of pneumococcal clones occurs mainly by the accumulation of small recombinational imports from other pneumococci (9), which requires concomitant colonization by isolates with distinct genotypes. This would be expected to be rare for serotype 1; however, a larger data set would be required to determine whether recombination is a less frequent mode of evolutionary change among serotype 1 isolates than among other pneumococci.
We had hoped to be able to identify an association between the prevalence of serotype 1 invasive disease within a country and the predominance of individual serotype 1 clones. Serotype 1 ranked among the most common serotypes causing invasive disease in the countries from which this collection of pneumococci was obtained, with the exception of North America, but including American Navajo Indians. ST306 and ST217 are prevalent clones in continental Europe, Africa, and Israel, and all of the countries in which either of these two clones is found rank serotype 1 among the most invasive serotypes. However, ST227 is the most common clone in England, the United States, and Canada, yet while serotype 1 ranks 2nd in England, it ranks only 15 to 17th in North America. Thus, among this diverse collection of serotype 1 isolates, it was difficult to conclude that any one clone is more likely than any other to be responsible for the prevalence of serotype 1 invasive disease.
Acknowledgments
This work was supported by grants from the Wellcome Trust (to B.G.S.).
We are very grateful to the many individuals who contributed to the provision of S. pneumoniae isolates for this study: in Africa, Anne von Gottberg, Keith Klugman, Lesley McGee, Jack Obiero, Anthony Scott, and Avril Wasas; in Canada, Louise Jette, Donald Low, Allison McGeer, Jean-Francois Proulx, and Barbara Willey; in Chile, Aurora Maldonado, Valeria Prado, and Mabel Seoane; in Denmark, Helle Bossen Konradsen and Anne Marie Jorgensen; in England, Derrick Crook, David Griffiths, and Kyle Knox; in France, Edouard Bingen; in Germany, Claudia Cremer and Ralf Reinert; in Israel, Ron Dagan and Nurith Porat; in The Netherlands, Peter Hermans, Marcel Sluijter, and Lodewijk Spanjaard; in Norway, Dominique Caugant and E. Arne Hoiby; in Poland, Waleria Hryniewicz, and Ewa Sadowy; in Spain, Asuncion Fenoll; and in the United States, Bernard Beall, T. Kuusisto, and Kate O'Brien.
REFERENCES
- 1.Benin, A. L., K. L. O'Brien, J. P. Watt, R. Reid, E. R. Zell, S. Katz, C. Donaldson, A. Parkinson, A. Schuchat, M. Santosham, and C. G. Witney. 2003. Effectiveness of the 23-valent polysaccharide vaccine against invasive pneumococcal disease in Navajo adults. J. Infect. Dis. 188:81-89. [DOI] [PubMed] [Google Scholar]
- 2.Byington, C. L., L. Y. Spencer, T. A. Johnson, A. T. Pavia, D. Allen, E. O. Mason, S. Kaplan, K. C. Carroll, J. A. Daly, J. C. Christenson, and M. H. Samore. 2002. An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations. Clin. Infect. Dis. 34:434-440. [DOI] [PubMed] [Google Scholar]
- 3.Dagan, R., S. Gradstein, I. Belmaker, N. Porat, Y. Siton, G. Weber, J. Janco, and P. Yagupsky. 2000. An outbreak of Streptococcus pneumoniae serotype 1 in a closed community in southern Israel. Clin. Infect. Dis. 30:319-321. [DOI] [PubMed] [Google Scholar]
- 4.DeMaria, A., K. Browne, S. L. Berk, E. J. Sherwood, and W. R. McCabe. 1980. An outbreak of type 1 pneumococcal pneumonia in a men's shelter. JAMA 244:1446-1449. [PubMed] [Google Scholar]
- 5.Dochez, A. R., and L. J. Gillespie. 1913. A biologic classification of pneumococci by means of immunity reactions. JAMA 61:727-730. [Google Scholar]
- 6.Doit, C., C. Loukil, P. Geslin, and E. Bingen. 2002. Phenotypic and genetic diversity of invasive pneumococcal isolates recovered from French children. J. Clin. Microbiol. 40:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eltringham, G., A. Kearns, R. Freeman, J. Clark, D. Spencer, K. Eastham, J. Harwood, and J. Leeming. 2003. Culture-negative childhood empyema is usually due to penicillin-sensitive Streptococcus pneumoniae capsular serotype 1. J. Clin. Microbiol. 41:521-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 9.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, D., N. Givon-Lavi, N. Bilenko, and R. Dagan. 2001. A decade (1989-1998) of pediatric invasive pneumococcal disease in 2 populations residing in 1 geographic location: implications for vaccine choice. Clin. Infect. Dis. 33:421-427. [DOI] [PubMed] [Google Scholar]
- 11.Gilman, B. B., and G. W. Anderson. 1938. A community outbreak of type 1 pneumococcus infection. Am. J. Hyg. 28:345-358. [Google Scholar]
- 12.Gratten, M., F. Morey, J. Dixon, K. Manning, P. Torzillo, R. Matters, J. Erlich, J. Hanna, V. Asche, and I. Riley. 1993. An outbreak of serotype 1 Streptococcus pneumoniae infection in central Australia. Med. J. Aust. 158:340-342. [DOI] [PubMed] [Google Scholar]
- 13.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part 1. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 14.Henriques Normark., B., M. Kalin, A. Ortqvist, T. Akerlund, B. Olsson Liljequist, J. Hedlund, S. B. Svenson, J. Zhou, B. G. Spratt, S. Normark, and G. Kallenius. 2001. Dynamics of penicillin-susceptible clones in invasive pneumococcal disease. J. Infect. Dis. 184:861-869. [DOI] [PubMed] [Google Scholar]
- 15.Jette, L. P., G. Delage, L. Ringuette, R. Allard, P. De Wals, F. Lamothe, V. Loo, and the Pneumococcus Study Group. 2001. Surveillance of invasive Streptococcus pneumoniae infection in the province of Quebec, Canada, from 1996 to 1998: serotype distribution, antimicrobial susceptibility, and clinical characteristics. J. Clin. Microbiol. 39:733-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konradsen, H. B., and M. S. Kaltoft. 2002. Invasive pneumococcal infections in Denmark from 1995 to 1999: epidemiology, serotypes and resistance. Clin. Diagn. Lab. Immunol. 9:358-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linares, J., R. Pallares, T. Alonso, J. L. Perez, J. Ayats, F. Gudiol, P. F. Viladrich, and R. Martin. 1992. Trends in antimicrobial resistance of clinical isolates of Streptococcus pneumoniae in Bellvitge Hospital, Barcelona, Spain (1979-1990). Clin. Infect. Dis. 15:99-105. [DOI] [PubMed] [Google Scholar]
- 18.Mackenzie, G. M. 1940. Epidemiology of an outbreak of pneumococcal pneumonia in a rural community. Trans. Assoc. Am. Physicians 55:199-208. [Google Scholar]
- 19.McFarlane, A. C., L. K. Hamra, E. Reiss-Levy, and D. Hansman. 1979. Pneumococcal peritonitis in adolescent girls. Med. J. Aust. 1:100-101. [DOI] [PubMed] [Google Scholar]
- 20.Mercat, A., J. Nguyen, and B. Dautzenberg. 1991. An outbreak of pneumococcal pneumonia in two men's shelters. Chest 99:147-151. [DOI] [PubMed] [Google Scholar]
- 21.Porat, N., R. Trefler, and R. Dagan. 2001. Persistence of two invasive Streptococcus pneumoniae clones of serotypes 1 and 5 in comparison to that of multiple clones of serotypes 6B and 23F among children in southern Israel. J. Clin. Microbiol. 39:1827-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proulx, J. F., S. Dery, L. P. Jette, J. Ismael, M. Libman, and P. De Wals. 2002. Pneumonia epidemic caused by a virulent strain of Streptococcus pneumoniae serotype 1 in Nunavik, Quebec. Can. Commun. Dis. Rep. 28:129-131. [PubMed] [Google Scholar]
- 23.Reinert, R. R., A. Al-Lahham, M. Lemperle, C. Tenholte, C. Briefs, S. Haupts, H. H. Gerards, and R. Lutticken. 2002. Emergence of macrolide and penicillin resistance among invasive pneumococcal isolates in Germany. J. Antimicrob. Chemother. 49:61-68. [DOI] [PubMed] [Google Scholar]
- 24.Scott, J. A., A. J. Hall, A. Hannington, R. Edwards, S. Mwarumba, B. Lowe, D. Griffiths, D. Crook, and K. Marsh. 1998. Serotype distribution and prevalence of resistance to benzylpenicillin in three representative populations of Streptococcus pneumoniae isolates from the coast of Kenya. Clin. Infect. Dis. 27:1442-1450. [DOI] [PubMed] [Google Scholar]
- 25.Sirotnak, A. P., S. C. Eppes, and J. D. Klein. 1996. Tuboovarian abscess and peritonitis caused by Streptococcus pneumoniae serotype 1 in young girls. Clin. Infect. Dis. 22:993-996. [DOI] [PubMed] [Google Scholar]
- 26.Sleeman, K., K. Knox, R. George, E. Miller, P. Waight, D. Griffiths, A. Efstratiou, K. Broughton, R. T. Mayon-White, E. R. Moxon, and D. W. Crook, on behalf of the Public Health Laboratory Service and the Oxford Pneumococcal Surveillance Group. 2001. Invasive pneumococcal disease in England and Wales: vaccination implications. J. Infect. Dis. 183:239-246. [DOI] [PubMed] [Google Scholar]
- 27.Smillie, W. G., G. H. Warnock, and H. J. White. 1938. A study of type 1 pneumococcus epidemic at the state hospital at Worcester, Massachusetts. Am. J. Pub. Health 28:293-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan, T. Q., E. O. Mason, Jr., E. R. Wald, W. J. Barson, G. E. Schutze, J. S. Bradley, L. B. Givner, R. Yogev, K. S. Kim, and S. L. Kaplan. 2002. Clinical characteristics of children with complicated pneumonia caused by Streptococcus pneumoniae. Pediatrics 110:1-6. [DOI] [PubMed] [Google Scholar]
- 29.von Kries, R., M. Hermann, A. Hachmeister, A. Siedler, H. J. Schmitt, A. Al-Lahham, and R. R. Reinert. 2002. Prediction of the potential benefit of different pneumococcal conjugate vaccines on invasive pneumococcal disease in German children. Pediatr. Infect. Dis. J. 21:1-7. [DOI] [PubMed] [Google Scholar]
- 30.Westh, H., L. Skibsted, and B. Korner. 1990. Streptococcus pneumoniae infections of the female genital tract and in the newborn child. Rev. Infect. Dis. 12:416-422. [DOI] [PubMed] [Google Scholar]