Abstract
Classically, detection of Entamoeba histolytica is performed by microscopic examination for characteristic cysts and/or trophozoites in fecal preparations. Differentiation of E. histolytica cysts and those of nonpathogenic amoebic species is made on the basis of the appearance and the size of the cysts. However, by classical means objective tools for confirmation and quality control do not exist. Therefore, a reverse line blot hybridization assay was developed to detect a variety of Entamoeba species and genetic variants known to infect humans. The assay was performed after amplification with general Entamoeba-specific primers. The assay could identify four genetic variants of Entamoeba polecki-like cysts as well as E. histolytica, Entamoeba dispar, Entamoeba hartmanni, Entamoeba moshkovskii and Entamoeba coli and even mixed infections in a range of controls and fecal samples. This technique can be used as an additional standard for diagnosis, epidemiology, and quality control for amoebic infections.
The main purpose of detection and differentiation of Entamoeba species in stool samples is the detection of the causative agent of amoebic dysentery, Entamoeba histolytica. It has been estimated that 40 million to 50 million people develop clinical amoebiasis each year, resulting in up to 100,000 deaths (15). Classically, diagnosis of an intestinal infection with E. histolytica is made by microscopic examination of feces, in which one must recognize and differentiate the cysts or trophozoites of E. histolytica from those of morphologically different nonpathogenic species. Although cysts and trophozoites of Entamoeba species that comply with all the textbook morphological characteristics can be found, in a majority of cases their appearances are tremendously more diverse. Therefore, the identification of these cysts and trophozoites requires a lot of skill and patience by the microscopist. In recent years, these difficulties in detection and differentiation of E. histolytica from morphologically different nonpathogenic species have become more compound, with the challenge being to differentiate E. histolytica from the morphologically identical species Entamoeba dispar. After decades of dispute, starting with the observations of Emile Brumpt (4), the biochemical, immunological, and genetic differences between E. histolytica and E. dispar, previously known as pathogenic and nonpathogenic E. histolytica, respectively, have proved to be sufficient to formally set them apart as two separate species (6, 7). Several targets for specific DNA amplification protocols for the differentiation of E. histolytica and E. dispar have been described and have been used with DNA samples extracted from amoebic abscess pus, fecal cultures, and stools (1-3, 8, 10). During the last 7 years, our laboratory in Leiden, The Netherlands, has received many stool samples for the species-specific diagnosis of E. histolytica and E. dispar infections (13). DNA was isolated from all stool samples by using spin columns, and a PCR-solution hybridization enzyme-linked assay was performed to identify and differentiate E. histolytica and E. dispar. Although most of the samples showed an E. histolytica- or E. dispar-specific PCR product, in some cases no specific product was found in either of these PCRs. In these cases microscopy revealed Entamoeba cysts that were classified as Entamoeba coli, Entamoeba hartmanni, or Entamoeba polecki-like (12); other protozoan cysts were also found. A method that used direct sequencing after DNA amplification with general Entamoeba-specific primers to confirm the morphological findings for non-E. histolytica, non-E. dispar uninucleated Entamoeba cysts was described (12). This method could be used only for the detection of infections with a single species, as mixtures of sequences were found in samples with multiple infections; moreover, the method is time-consuming and laborious. Here we describe a reverse line blot method for the detection and identification of Entamoeba species, even in mixed infections, after DNA amplification with general Entamoeba-specific primers and hybridization of the product obtained with probes specific for Entamoeba, uninucleated Entamoeba, and other Entamoeba species with the genetic variants known to infect humans.
MATERIALS AND METHODS
Controls and samples.
Control samples (Table 1) were obtained from culture (E. histolytica and E. dispar), cloned small-subunit (SSU) rRNA genes (Entamoeba moshkovskii, E. polecki, and Entamoeba chattoni), or human fecal samples (E. hartmanni, E. coli [HU-1; CDC type], E. coli (IH; 96/135 type), E. polecki-like variant 2, and E. polecki-like variant 3). The products of all control samples obtained by PCR with Entamoeba-specific primers were sequenced as described before (12). The sequences were compared with the sequences in GenBank and sequences published elsewhere (12).
TABLE 1.
Names and origins of control DNA samples and GenBank accession number or reference for target sequence with a complete match with the sequence of the PCR product after amplification with Entamoeba-specific primers Entam1 and Entam2
| Organism | Origin | GenBank accession no. or reference for sequence with a match |
|---|---|---|
| E. histolytica | HM1 axenic culture | X64142 |
| E. dispar | Polyxenic culture from human stool sample | Z49256 |
| E. hartmanni | Human stool sample | AF149907 |
| E. moshkovskii | SSU rRNA clone of E. moshkovskii Laredo strain | AF149906 |
| E. coli (HU-1; CDC type) | Human stool sample | AF149915 |
| E. coli (IH; 96/135 type) | Human stool sample | AF149914 |
| E. polecki | SSU rRNA clone of E. polecki NIH:1293:1 | AF149913 |
| E. polecki-like variant 2 | Human stool sample | 12 |
| E. polecki-like variant 3 | Human stool sample | 12 |
| E. chattoni | SSU rRNA clone of E. chattoni NIH:0191:1 | AF149912 |
Twenty human fecal samples were obtained from rural villages in northern Ghana. High prevalences of E. histolytica/E. dispar, E. coli, or E. hartmanni were found in these villages by light microscopy of iodine-stained wet mount preparations of the formalin-ether concentrate (14). Furthermore, we used nine human fecal samples that were sent to our laboratory for molecular differentiation of presumed E. histolytica/E. dispar cysts but in which no amplification product was found with E. histolytica- or E. dispar-specific primers. In these cases microscopy revealed uninucleated Entamoeba cysts in which the appearances of the nuclei, the inclusion bodies, and the chromatid bodies suggested that these were unlikely to be immature cysts of E. histolytica or E. dispar. Nine human fecal samples with E. histolytica/E. dispar cysts with which amplification products were obtained with E. histolytica-specific primers were also tested.
DNA isolation.
For DNA isolation, 200 μl of fecal suspension (≈0.5 g/ml of phosphate-buffered saline containing 2% polyvinylpolypyrrolidone [Sigma]) was heated for 10 min at 100°C. After sodium dodecyl sulfate (SDS)-proteinase K treatment (2 h at 55°C), DNA was isolated with QIAamp Tissue Kit spin columns (Qiagen, Hilden, Germany) (11).
PCR amplification.
General Entamoeba-specific primers were designed from the SSU rRNA gene sequences of E. polecki, E. chattoni, E. moshkovskii, E. dispar, E. histolytica, E. hartmanni, and E. coli (GenBank accession nos. AF149913, AF149912, AF149906, Z49256, X64142, AF49906, and AF149915, respectively). Forward primer Entam1 (biotin-5′-GTT GAT CCT GCC AGT ATT ATA TG-3′) and reverse primer Entam2 (biotin-5′-CAC TAT TGG AGC TGG AAT TAC-3′), which are specific for conserved regions, were chosen so that the DNA of all Entamoeba species would be amplified. Amplification reactions were performed in a volume of 40 μl containing PCR buffer (1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, HotStarTaq Master Mix [Qiagen]), 25 pmol of each primer, and 2 μl of the DNA sample. Amplification consisted of 15 min at 95°C and 38 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C, with a final step of 2 min at 72°C.
Reverse line blot hybridization assay.
A general Entamoeba-specific probe was designed from a conserved region of the SSU rRNA gene sequences of E. polecki, E. chattoni, E. moshkovskii, E. dispar, E. histolytica, E. hartmanni, and E. coli so that DNA amplified from any of the Entamoeba species would be detected. Furthermore, one probe was designed to detect DNA of uninucleated cyst-producing Entamoeba by using the SSU rRNA gene sequences of the E. polecki-like variants. Additionally, 14 species- and/or variant-specific probes were designed by using the respective sequences of the SSU rRNA genes of those species and variants (Table 2). Covalent binding of the specific probes to the membrane and hybridization with the amplification products were performed as described by others (9), with some modifications. Briefly, 50 to 750 pmol of the 5′ amino-linked oligonucleotide probes (Table 2) were covalently coupled to an activated Biodyne C membrane with a miniblotter (Immunetics, Cambridge, Mass.). After the oligonucleotide probes were bound to the membrane, the membrane was incubated for 10 min in 100 mM NaOH solution and then washed in 2× SSPE (360 mM NaCl, 20 mM Na2HPO4, 2 mM EDTA) containing 0.1% SDS at 60°C. The membrane was again placed in the miniblotter with the slots at right angles to the oligonucleotide lines. Twenty microliters of the PCR product was diluted in 150 μl of 2× SSPE-0.1% SDS, denatured for 10 min at 95°C, and immediately cooled on ice. The diluted and denatured PCR products were hybridized with the probes on the membrane for 1 h at 45°C. The membrane was washed with 2× SSPE-0.5% SDS for 2 min at room temperature, with preheated 2× SSPE-0.5% SDS at 50°C for 15 min, and twice with 2× SSPE-0.5% SDS for 2 min each time at room temperature before incubation for 15 min at room temperature with streptavidin-peroxidase (Roche) diluted 1:10,000 in 2× SSPE-0.5% SDS. The membrane was again washed twice with 2× SSPE-0.5% SDS for 5 min each time and was washed once with 2× SSPE for 5 min before incubation for 2 min with enhanced chemiluminescence detection liquid (Amerhsam International, Den Bosch, The Netherlands). Thereafter, hybridization was visualized by exposing the membrane to X-ray film (Fuji Photo Film Co. Ltd., Tokyo, Japan). The membrane with the probes could be used again at least five times after removal of the hybridized PCR products. First, the membrane was incubated twice for 30 min each time in 1% SDS solution at 80°C. Then, after 15 min of incubation in 20 mM EDTA solution at room temperature, the membrane was sealed and stored at 4°C.
TABLE 2.
Sequences of the Entamoeba species- and variant-specific oligonucleotides and GenBank accession number or reference for target sequence on the basis of which the probe has been designed
| Oligonucleotide no. and name | Oligonucleotide sequence | GenBank accession no. or reference for target sequence |
|---|---|---|
| 1. Entamoeba2 | TTTMVARATGGCTACCACTTCTA | All numbers below |
| 2. E. histolytica | ATGGCCAATTCATTCAATGA | X64142 |
| 3. E. dispar | TACAAAGTGGCCAATTTATGTAAGTA | Z49256 |
| 4. E. hartmanni (2) | GTGAAGAGAAAGGATATCCAAAGT | AF149907 |
| 5. E. moshkovskii | AGTCGGCCACTCTCTTCAC | AF149906 |
| 6. E. coli | CGGTTTTCACCCCTTGTC | AF149915 |
| 7. E. coli2a | CGCTATCCTCGTCTTTTGGC | AF149915 |
| 8. E. coli3 | TACCACTTTTTTTTGAATGAG | AF149915 |
| 9. E. coli IH | CGGGTAACGCCTTCAGTC | AF149914 |
| 10. E. coli IH2a | CGCTTTCCCTCGCTTTACGT | AF149914 |
| 11. E. coli IH3 | TACCACTTCTTTGTGAATAAG | AF149914 |
| 12. Uninucleate | GAATAGCTTTTTGAGAAGAAGGTTAAA | 12 |
| 13. Uninucleate Ep | AATAGAATCGATATTTATATTGATTCAAATG | AF149913 |
| 14. Uninucleate 2 | TTGGTCTATTCGATCAATTCAATT | 12 |
| 15. Uninucleate 3 | GGATTTGTTTAATAACAGATTCAATTG | 12 |
| 16. Uninucleate Ec | GGATTTGTTTTATAACAAGTTCAATTG | AF149912 |
RESULTS
Control samples.
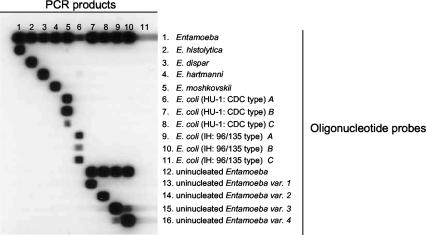
The expected amplicon of approximately 550 bp was produced from all samples (data not shown). Sequence analysis of the PCR products of all control samples obtained with the Entamoeba-specific primers (data not shown) revealed a complete match with the corresponding GenBank sequences (Table 1). Figure 1 shows the reactivities of the control samples for Entamoeba, uninucleated Entamoeba, and Entamoeba species with the genetic variants. There was no cross hybridization between the various species, between the genetic variants of E. coli, or between the genetic variants of the uninucleated Entamoeba variants.
FIG. 1.
Reverse line blot hybridization assay for the detection and identification of Entamoeba species and genetic variants. The oligonucleotide probes were coupled to the membrane in a horizontal direction (the numbers on the right refer to the numbers for the oligonucleotide names in Table 2), and the PCR samples were applied in the vertical direction. Lane 1, E. histolytica; lane 2, E. dispar; lane 3, E. hartmanni; lane 4. E. moshkovskii; lane 5, E. coli (HU-1; CDC type); lane 6, E. coli (IH; 96/135 type); lane 7, E. polecki (E. polecki like variant 1); lane 8, E. polecki-like variant 2; lane 9, E. polecki-like variant 3; lane 10, E. chattoni (E. polecki-like variant 4); lane 11, negative control.
Fecal samples.
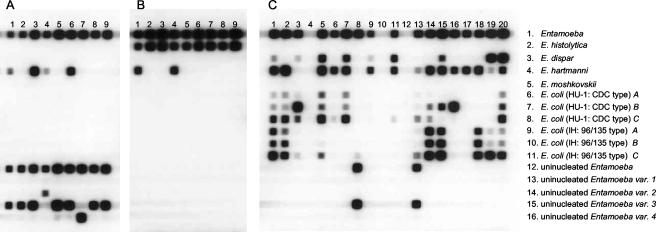
All nine samples with uninucleated Entamoeba cysts produced the expected amplicon of approximately 550 bp (data not shown). Figure 2A shows the reactivities of these PCR products for the Entamoeba species and the genetic variants. All of the samples reacted with the Entamoeba-specific probe, the uninucleated Entamoeba-specific probe, and one of the uninucleated Entamoeba variant-specific probes. Of these, one sample reacted with the E. chattoni-specific probe (uninucleated variant 4), one sample reacted with the variant 2-specific probe, and seven samples reacted with the variant 3-specific probe. Furthermore, four samples reacted with the E. hartmanni-specific probe.
FIG. 2.
Reverse line blot hybridization assay for the detection and identification of Entamoeba species and genetic variants. (A) Fecal samples in which uninucleated Entamoeba cysts were found by microscopy; (B) fecal samples in which E. histolytica or E. dispar cysts were found by microscopy and that produced an amplicon after PCR with E. histolytica-specific primers; (C) fecal samples from rural villages in northern Ghana. The oligonucleotide probes were coupled to the membrane in horizontal direction (the numbers on the right refer to the numbers for the oligonucleotide names in Table 2), and the PCR samples were applied in the vertical direction.
All nine human fecal samples with E. histolytica/E. dispar cysts for which amplification with the E. histolytica-specific primers was found showed the expected amplicon after PCR (data not shown). Figure 2B shows the reactivities of these PCR products for Entamoeba species and the genetic variants. All of the DNA samples reacted with the Entamoeba-specific probe and the E. histolytica-specific probe. Furthermore, two samples reacted with the E. hartmanni-specific probe.
Seventeen of 20 fecal samples from Ghana produced the expected amplicon (data not shown). Figure 2C shows the reactivities of the PCR products from these samples for the Entamoeba species and the genetic variants. Three samples without the visible amplicon on the agarose gel did not react with any of the probes. Seventeen samples which had produced the expected amplicon hybridized with the Entamoeba-specific probe and a variety of the other genus- and variant-specific probes (Table 3).
TABLE 3.
Results of reverse line blot analysis of PCR products obtained from 20 human fecal samples from rural villages in northern Ghana with Entamoeba species- and variant-specific oligonucleotides
| Combination of species found | No. of samplesa |
|---|---|
| E. dispar, E. hartmanni, and E. coli | ......7 |
| E. dispar and E. hartmanni | ......1 |
| E. dispar and E. coli | ......1 |
| E. hartmanni, E. coli, and E. polecki variant 2 | ......1 |
| E. hartmanni and E. coli | ......4 |
| E. hartmanni | ......2 |
| E. polecki variant 2 | ......1 |
| None | ......3 |
Number of samples that hybridized with the Entamoeba-specific probe or other genus- and variant-specific probes.
DISCUSSION
Classically, detection and identification of Entamoeba species is based on the morphological characteristics of cysts and trophozoites found by microscopic examination of stool preparations. The outcome of this detection and identification depends greatly on the skill and expertise of the microscopist. For decades, confirmation of microscopy results consisted of reexamination of the sample by a more experienced microscopist. Objective techniques for confirmation of microscopy results were not available. Moreover, morphologically identical species or genetic variants could not be distinguished by microscopy alone. Recently, the use of direct DNA sequencing after amplification with general Entamoeba-specific primers for the identification of uninucleated Entamoeba species has been described (12). However, this method could be used only for the detection of infections with single species and is time-consuming and laborious. Therefore, we have developed a reverse line blot hybridization assay to detect and identify Entamoeba species after DNA amplification with general Entamoeba-specific primers and hybridization of the products with probes specific for all Entamoeba species and genetic variants known to infect humans.
The assay described here can detect and identify Entamoeba species known to infect humans on the basis of the detection of differences in the DNA sequences of the SSU rRNA gene by a reverse line blot hybridization assay after amplification with general Entamoeba-specific primers. Furthermore, there is no cross hybridization between E. histolytica, E. dispar, E. hartmanni, E. moshkovskii, two genetic variants of E. coli, and four genetic variants of uninucleated Entamoeba (including E. polecki sensu lato and E. chattoni sensu lato).
The presence of E. polecki-like Entamoeba species (genetic variants of uninucleated Entamoeba) could be confirmed in nine samples in which no amplification was found with E. histolytica- or E. dispar-specific primers and in which only uninucleated Entamoeba cysts were found by microscopy.
Although human infections with uninucleated Entamoeba are regarded as rare zoonotic infections, 2 of 20 samples from humans in rural villages in northern Ghana revealed the presence of E. polecki-like variant 3. Until now, four genetic variants of uninucleated cysts producing Entamoeba are known to infect humans (12). The source of these uninucleated Entamoeba genetic variants is unknown. In order to determine the source, the Entamoeba reverse line blot assay could be used to detect and identify Entamoeba in samples from animals. Other genetic variants may exist, and therefore, a general uninucleated Entamoeba probe was designed for the distinction of these amoebas from the multinucleated cyst-producing Entamoeba.
The variety of hybridization of the PCR products obtained from samples from rural villages in northern Ghana with the E. coli-specific oligonucleotide probes shows that there is a large intraspecific variation in E. coli, which has been shown before by Clark and Diamond (5). With the knowledge of this genetic variation in E. coli, the possibility of the existence of E. coli strains that do not react with one of the E. coli-specific probes used in this study cannot be excluded. However, hybridization with the general Entamoeba-specific probe in such cases indicates the need for further sequence analysis to reveal new genetic variants. In the future, a general octanucleated Entamoeba-specific probe could be designed to detect all genetic variants of E. coli.
In all nine samples with E. histolytica/E. dispar cysts in which amplification with E. histolytica-specific primers was found, the presence of E. histolytica and coinfections with E. hartmanni in two samples could be confirmed. This demonstrates that the Entamoeba reverse line blot assay can also detect E. histolytica in human fecal samples.
An Entamoeba reverse line blot hybridization assay which can detect a variety of Entamoeba species and genetic variants known to infect humans in human stool samples after amplification with general Entamoeba-specific primers is presented. This assay can serve as a truly objective tool for the confirmation of microscopy results and can give insight into the epidemiology of Entamoeba species and genetic variance in Entamoeba.
In the future, other PCRs and specific probes for the detection of other protozoa, e.g., Endolimax nana and Iodamoeba butschlii, could be added to increase the range of parasites whose presence can be confirmed by this technique.
Acknowledgments
We acknowledge Jos Drabbels for his help in setting up the reverse line blot technique. We thank Juventus Siem, who provided the Ghanaian fecal samples and to Graham Clark for kindly providing the SSU clones as control DNA samples. We thank K. Templeton for critical reading of the manuscript.
REFERENCES
- 1.Acuna-Soto, R., J. Samuelson, P. De Girolami, L. Zarate, F. Millan-Velasco, G. Schoolnick, and D. Wirth. 1993. Application of the polymerase chain reaction to the epidemiology of pathogenic and nonpathogenic Entamoeba histolytica. Am. J. Trop. Med. Hyg. 48:58-70. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, A., D. C. Warhurst, F. Guhl, and I. A. Frame. 1995. Polymerase chain reaction-solution hybridization enzyme-linked immunoassay (PCR-SHELA) for the differential diagnosis of pathogenic and non-pathogenic Entamoeba histolytica. Trans. R. Soc. Trop. Med. Hyg. 89:187-188. [DOI] [PubMed] [Google Scholar]
- 3.Britten, D., S. M. Wilson, R. McNerney, A. H. Moody, P. L. Chiodini, and J. P. Ackers. 1997. An improved colorimetric PCR-based method for detection and differentiation of Entamoeba histolytica and Entamoeba dispar in feces. J. Clin. Microbiol. 35:1108-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumpt, E. 1925. Etude sommaire de l' “Entamoeba dispar” n. sp., amibe a kystes quadrinuclees, parasite de l'homme. Bull. Acad. Med. (Paris) 94:942-952. [Google Scholar]
- 5.Clark, C. G., and L. S. Diamond. 1997. Intraspecific variation and phylogenetic relationships in the genus Entamoeba as revealed by riboprinting. J. Eukaryot. Microbiol. 44:142-154. [DOI] [PubMed] [Google Scholar]
- 6.Diamond, L. S., and C. G. Clark. 1993. A redescription of Entamoeba histolytica Schaudinn, 1903 (emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J. Eukaryot. Microbiol. 40:340-344. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Ruiz, A., and S. G. Wright. 1998. Disparate amoebae. Lancet 351:1672-1673. [DOI] [PubMed] [Google Scholar]
- 8.Rivera, W. L., H. Tachibana, and H. Kanbara. 1998. Application of the polymerase chain reaction (PCR) in the epidemiology of Entamoeba histolytica and Entamoeba dispar infections. Tokai J. Exp. Clin. Med. 23:413-415. [PubMed] [Google Scholar]
- 9.Schouls, L. M., D. P. Van, I., S. G. Rijpkema, and C. S. Schot. 1999. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J. Clin. Microbiol. 37:2215-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verweij, J. J., J. Blotkamp, E. A. Brienen, A. Aguirre, and A. M. Polderman. 2000. Differentiation of Entamoeba histolytica and Entamoeba dispar cysts using polymerase chain reaction on DNA isolated from faeces with spin columns. Eur. J. Clin. Microbiol. Infect. Dis. 19:358-361. [DOI] [PubMed] [Google Scholar]
- 11.Verweij, J. J., D. S. S. Pit, L. Van Lieshout, S. M. Baeta, G. D. Dery, R. B. Gasser, and A. M. Polderman. 2001. Determining the prevalence of Oesophagostomum bifurcum and Necator americanus infections using specific PCR amplification of DNA from faecal samples. Trop. Med. Int. Health 6:726-731. [DOI] [PubMed] [Google Scholar]
- 12.Verweij, J. J., A. M. Polderman, and C. G. Clark. 2001. Genetic variation among human isolates of uninucleated cyst-producing Entamoeba species. J. Clin. Microbiol. 39:1644-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verweij, J. J., L. Vanlieshout, C. Blotkamp, E. A. T. Brienen, S. Vanduivenvoorden, M. Vanesbroeck, and A. M. Polderman. 2000. Differentiation of Entamoeba histolytica and Entamoeba dispar using PCR-SHELA and comparison of antibody response. Arch. Med. Res. 31:S44-S46. [DOI] [PubMed] [Google Scholar]
- 14.Verweij, J. J., F. Oostvogel, E. A. T. Brienen, A. Nang-Neifubah, J. Ziem, and A. M. Polderman. Prevalence of Entamoeba histolytica and Entamoeba dispar in northern Ghana. Trop. Med. Int. Health, in press. [DOI] [PubMed]
- 15.Walsh, A. L. 1988. Prevalence in Entamoeba histolytica infection, p. 93-105. In J. I. Ravdin (ed.), Amebiasis: human infection by Entamoeba histolytica. John Wiley & Sons, Inc., New York, N.Y.