Abstract
Q fever is a potentially severe disease which can occur in large outbreaks of acute infections and is a possible bioterrorism agent. In order to lessen the delay in diagnosing acute Q fever, we compared LightCycler Nested PCR (LCN-PCR), a rapid nested PCR assay that uses serum sampled early during the disease as a specimen and the LightCycler as a thermal cycler, to serology by indirect immunofluorescence. We used the 20-copy htpAB-associated element as the DNA target. The detection sensitivity of this method was one Coxiella burnetii DNA copy. We applied this method to the first serum samples taken from 100 patients diagnosed in our laboratory as having acute Q fever on the basis of clinical manifestations and serology and to 80 controls. The LCN-PCR had a specificity of 100%. The sensitivity was 26% when no antibodies were detected but only 5% with seropositive patients (P < 10−2). The technique was most efficient in the first 2 weeks following the onset of symptoms (P = 0.02), when its sensitivity was 24% compared with 14% for serology. With combined use of LCN-PCR and serology within the first 2 weeks, the sensitivity was significantly increased over that with serology alone (P < 10−2). Thus, we propose a strategy for improving the early diagnosis of acute Q fever where LCN-PCR should be performed together with serology in the first 2 weeks of the disease but should be reserved for seronegative patients in the next 2 weeks and not used later than 4 weeks following onset, when serology is highly sensitive.
Q fever is a ubiquitous zoonosis caused by Coxiella burnetii, a short and pleomorphic, strictly intracellular bacillus (20). The reservoir of this bacterium is extensive but only partially known and includes mammals, birds, and arthropods, mainly ticks (20). C. burnetii is desiccation resistant and is shed in urine, feces, milk, and especially in birth products of mammals (11). The most commonly identified sources of human infection are farm animals, such as cattle, goats, and sheep, but pets, including cats and dogs (10), have also been demonstrated as potential sources of urban outbreaks. The organism is highly infectious: only one organism is required to produce infection under experimental conditions (13). C. burnetii is currently considered a potential warfare agent and is classified as a category B biological agent by the Center for Diseases Control and Prevention. In humans, infection results mainly from inhalation of contaminated aerosols from amniotic fluid, placenta, or contaminated wool, but the disease may also be acquired by the digestive route (3). At greatest risk are persons in contact with farm animals, as well as laboratory personnel who work with infected animals (9). In Europe, acute Q fever cases are more frequently reported in spring and early summer. The disease may occur at all ages but is more frequent in men than in women. The clinical manifestations of Q fever are varied and nonspecific and may be acute, most often presenting as pneumonia or hepatitis, or chronic, most often presenting as endocarditis. Inapparent and subclinical infections are common. Acute Q fever is usually benign (14), but patients with cardiovascular abnormalities are at risk of chronic infection.
Usually, the microbiological diagnosis of Q fever relies upon serology, the most commonly used method being the indirect immunofluorescence assay (IFA). However, although highly reliable, this technique provides only indirect evidence of infection, and antibodies are absent in the early phases of the disease. PCR assays, especially those targeting the htpAB-associated repetitive element, which exists in 20 copies in the genome of C. burnetii (GenBank accession number AE016828), have previously been demonstrated to be very sensitive (21). However, molecular detection in serum, which is the specimen most easily obtained from patients and which may be conserved frozen for long periods, lacks sensitivity (5). Recently, in order to increase our detection sensitivity, we have developed for Bartonella endocarditis an efficient and rapid nested PCR assay using serum as the template and the LightCycler (Roche Diagnostics, Basel, Switzerland) as the thermal cycler, named the LightCycler nested PCR assay (LCN-PCR) (22). In this assay, the two primer pairs had different hybridization temperatures, reaction tubes were not opened during the whole amplification process, and we used one of the LightCycler's advantages, i.e., the rapidity, since both amplification and reamplification were performed within 90 min.
The aim of the present study was to compare the efficiency of LCN-PCR targeting the htpAB-associated repetitive element and that of serology by IFA for early diagnosis of acute Q fever.
MATERIALS AND METHODS
Patients.
We selected, from our serum collection, the earliest serum sample taken from each of 100 patients diagnosed as having acute Q fever between January 2002 and January 2003, for whom at least 200 μl of serum remained. Among these, 69 had contracted Q fever during an outbreak that occurred from July to October 2002 in the Chamonix valley in the French Alps (H. Tissot-Dupont and D. Raoult, unpublished data). This outbreak, probably linked to sheep flock migration, involved 101 patients. The other 31 patients included in our study were infected in various areas in France, independently from each other. When several serum specimens were available for a patient, we used the earliest serum sample with respect to the onset of symptoms. For each patient, we recorded the sex, age, time interval between onset of symptoms and serum sampling, and presence of fever, chills, headache, myalgias, nausea, atypical pneumonia, and transaminase levels of >40 IU. In order to estimate the specificity of our PCR assay, we also applied it to 40 patients with pneumonia caused by other microorganisms. Ten of the patients with pneumonia were infected with Chlamydia pneumoniae, 10 were infected with Legionella pneumophila, and 20 were infected with Mycoplasma pneumoniae. We also tested 40 blood donors with negative Q fever serology. Informed consent was obtained from all patients
Case definition.
Acute C. burnetii infection was diagnosed on the basis of the association of a fever of >39°C with at least two other symptoms (chills, headache, myalgias, atypical pneumonia, and/or elevated hepatic transaminase levels) and with exhibition of a phase II immunoglobulin G (IgG) titer of ≥1:200 and an IgM titer of ≥1:50 when only one serum specimen was available or a seroconversion when two sera sampled two or more weeks apart were available (6, 17). Patients were considered seronegative when titers of IgM were <1:25 and titers of IgG were <1:50.
Serology.
Serology by microimmunofluorescence (MIF) was carried out as previously reported (19).
Molecular methods. (i) DNA extraction
Total genomic DNA was extracted from serum samples using the QIAamp blood kit (Qiagen, Hilden, Germany) as described by the manufacturer. Two hundred microliters of serum was used. Fifty microliters of elution buffer was used to resuspend the DNA. Genomic DNAs were stored at 4°C until their use as templates in PCR assays.
(ii) LCN-PCR.
DNA samples were handled carefully to avoid the risk of cross-contamination. DNA extraction, mix preparation, and PCR were performed in different rooms to prevent PCR carryover contamination. No positive control was used to prevent lateral contamination (i.e., contamination caused by PCR products amplified in other tubes in the same assay, as previously observed [22]). DNA extracted from serum specimens from blood donors was used every seven specimens as a negative control. Each 20-μl reaction mixture was made of 4 μl of DNA master SYBR Green, 4.8 μl of 3 mM MgCl2, 1 μl of each of the four primers, IS111F1 (5′-TACTGGGTGTTGATATTGC-3′), IS111R1 (5′-CCGTTTCATCCGCGGTG-3′), IS111F2 (5′-GTAAAGTGATCTACACGA-3′), and IS111R2 (5′-TTAACAGCGCTTGAACGT-3′) at 0.5 μM, 5.2 μl of sterile distilled water, and 2 μl of DNA. The IS111F1 and IS111R1 primers, which were designed to amplify a 485-bp fragment of the htpAB-associated repetitive element (GenBank accession number M80806), were used for the first amplification, and reamplification was performed using the IS111F2 and IS111R2 primers, which amplify a 260-bp fragment. Following an initial denaturation step at 95°C for 8 min, our rapid nested PCR program was made of 35 cycles of denaturation at 95°C for 15 s (temperature transition, 20°C/s), annealing at 52°C for 5 s, and extension at 72°C for 18 s followed by 35 cycles of denaturation at 95°C for 15 s (temperature transition, 20°C/s), annealing at 48°C for 5 s, and extension at 72°C for 18 s. The amplification was completed by holding for 10 min at 68°C to allow complete extension of the PCR products. Since the LightCycler was not able to infer the results from the nested PCR, in particular the melting curve, amplicons from the second amplification were separated by electrophoresis on 1% agarose gels and then purified by using a QIAquick PCR purification kit (Qiagen) as described by the manufacturer.
In order to estimate the sensitivity of our PCR assay but to avoid contamination of the assay performed on serum by amplicons from a previous amplification, we applied it a posteriori, after the patients' specimens had been tested, to 10-fold dilutions in serum from a seronegative blood donor of a suspension of 106 C. burnetii strain Nine Mile (ATCC VR615) bacteria. C. burnetii was purified using sonication and a sucrose gradient as previously described (7) and quantified by counting the number of bacteria present in 10 μl of Gimenez dye-stained 10-fold dilutions of a culture of C. burnetii in a 150-cm2 tissue culture flask under a microscope at magnification ×1,000. The extraction method, primers, and conditions were those described above.
(iii) Sequencing of PCR products and sequence analysis.
PCR products were sequenced in both directions using the d-Rhodamine Terminator Cycle sequencing ready reaction kit (Perkin-Elmer, Coignieres, France) as described by the manufacturer. Sequencing products were resolved using an ABI 3100 automated sequencer (Perkin-Elmer). Sequence analysis was performed with the software package ABI Prism DNA sequencing analysis software, version 3.0 (Perkin-Elmer), and multisequence alignment was made with CLUSTAL W software, version 1.81 (18).
Statistical analysis.
We evaluated the influence of the clinical symptoms and the presence of antibodies on PCR positivity using the χ2 test. To estimate the influence of the age of patients and of the time interval between onset of symptoms and serum sampling on the sensitivity of the LCN-PCR, we used Student's t test. We also studied the influence of each variable by using univariate logistic regression analysis. In order to identify variables independently associated with PCR results, we conducted multivariate logistic regression analysis. The STATA software (version 7.0; Stata Corporation, College Station, Tex.) was used for analysis.
RESULTS
Patients' characteristics.
Sixty-eight of the 100 patients were male. Their mean age (± standard deviation) was 48 ± 18 years (range, 12 to 90 years; median, 47 years). All patients were febrile, 48 reported chills, 55 suffered from headache, and 58 suffered from myalgias. Nine patients were diagnosed as having atypical pneumonia on the chest X-ray, and 55 exhibited elevated transaminase levels.
Serology results.
The mean time interval between onset of symptoms and serum sampling (± standard deviation) was 17.9 ± 18.4 days (range, 2 to 71 days; median, 10.5 days). Among the 100 patients, 39 exhibited positive titers of antibody to C. burnetii in the tested serum sample. The sensitivity of IgM detection by serology with respect to the time interval between onset of symptoms and serum sampling is detailed in Table 1. Titers of IgM ranged from 0 to 1:800, whereas titers of IgG ranged from 0 to 1:3,200. For all patients, titers of IgG antibody to phase I were lower than those to phase II. The 61 seronegative patients had been diagnosed as having Q fever on the basis of a seroconversion observed using a serum sampled later.
TABLE 1.
Sensitivities of IgM detection by serology and LCN-PCR with respect to the time interval between serum sampling and onset of symptoms
| Result | % Sensitivitya for time interval (wk) of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Titer of IgM, ≥ 1:50 | 7 (3/43) | 26 (6/23) | 50 (4/8) | 100 (6/6) | 100 (5/5) | 100 (3/3) | 100 (3/3) | 100 (2/2) | 100 (3/3) | 100 (2/2) | 100 (2/2) |
| Positive LCN-PCR | 19 (8/43) | 35 (8/23) | 0 (0/8) | 0 (0/6) | 20 (1/5) | 0 (0/3) | 0 (0/3) | 0 (0/2) | 33 (1/3) | 0 (0/2) | 0 (0/2) |
Numbers of positive sera/total number of tested sera are indicated in parentheses.
LCN-PCR assay.
The overall sensitivity of LCN-PCR was 18% (18 of 100). The sensitivity of the test varied according to the time interval between onset of symptoms and serum sampling (Table 1). However, the sensitivity of the test was 26% among seronegative specimens (16 of 61) but only 5% among seropositive samples (2 of 39) (Table 2). Sequences obtained from all PCR products were identical to that of C. burnetii. LCN-PCR exhibited a specificity of 100%, since all 80 negative controls were PCR negative.
TABLE 2.
Influence of the clinical presentation, presence of antibodies, and age of serum on the LCN-PCR results
| Variable | Value for positive LCN-PCR group (n = 18) | Value for negative LCN-PCR group (n = 82) | Univariate analysis
|
Multivariate analysis, P value | |
|---|---|---|---|---|---|
| P value | Odds ratio (range) | ||||
| Sex ratio (M/F)a | 1.6 | 1.7 | 0.8 | 0.9 (0.3-2.6) | 0.7 |
| Mean age ± standard deviation (yr) | 46.1 ± 12.5 | 47.9 ± 18.9 | 0.8 | 0.8 | |
| % of patients with: | |||||
| Chills | 55.5 | 46.3 | 0.5 | 1.4 (0.5-4.0) | 0.8 |
| Headache | 61.1 | 53.6 | 0.6 | 1.3 (0.5-3.8) | 0.9 |
| Myalgias | 50.0 | 59.7 | 0.4 | 0.7 (0.2-1.9) | 0.5 |
| Pneumonia | 16.7 | 7.3 | 0.2 | 2.5 (0.5-11.2) | 0.3 |
| Liver involvement | 50.0 | 56.1 | 0.6 | 0.8 (0.3-2.1) | 0.6 |
| Presence of antibodies to C. burnetii | 11.1 | 45.1 | <10−2 | 0.2 (0.06-0.8) | 0.1 |
| Mean time interval between onset and serum sampling ± SD (days) | 12.2 ± 14.2 | 19.2 ± 19.0 | 0.1 | 0.8 | |
| % of patients for which: | |||||
| Sera were sampled ≤7 days after onset of symptoms | 44.4 | 42.7 | 0.9 | 1.1 (0.3-3.3) | 0.9 |
| Sera were sampled ≤10 days after onset of symptoms | 55.5 | 48.7 | 0.6 | 1.3 (0.4-4.1) | 0.8 |
| Sera were sampled ≤14 days after onset of symptoms | 88.9 | 60.9 | 0.02 | 5.1 (1.02-34.6) | 0.9 |
M/F, male/female.
When estimating the detection threshold of this assay, we were able to obtain positive PCR products from the 10−5 dilution of our C. burnetii suspension (equivalent of 1 DNA copy of the bacterium).
Combination of both tests.
When the results of either LCN-PCR or serology were positive, the sensitivity was 38% (25 of 66) for the first 2 weeks, 71% (10 of 14) for the next 2 weeks, and 100% (20 of 20) for sera sampled later than 4 weeks.
Statistical analysis.
Since fever was part of the inclusion criteria and thus was present in all studied patients, it was not included in the statistical analysis. The sex, age, clinical presentation, and time interval between onset of symptoms and serum sampling had no influence on PCR results (Table 2). When comparing patients with respect to the time interval from onset of symptoms to serum sampling and the presence of IgM antibodies (Table 3), significantly fewer sera sampled <14 days since onset exhibited antibodies than did those sampled between 15 and 28 days (9 of 66 versus 10 of 14; P < 10−2), and significantly fewer of those sampled between 15 and 28 days since onset exhibited antibodies than did those sampled later than 28 days (10 of 14 versus 20 of 20; P = 0.03). Using univariate logistic regression analysis, we compared Q fever patients with respect to their PCR result and the time interval between onset of symptoms and serum sampling or presence of IgM antibodies. We observed that the absence of antibodies and a sampling interval of ≤14 days following the onset of symptoms were statistically associated with positive PCR results (P = 0.03 and 0.02, respectively) (Table 2). When estimating the role of IgM in LCN-PCR positivity regardless of the time interval, we obtained a P value of <10−2 (16 of 18 versus 23 of 82). However, with multivariate logistic regression analysis, neither the presence of antibodies nor the time interval from sampling was independently associated with the PCR result (Table 2). The combination of LCN-PCR and serology in the first 2 weeks of infection significantly increased the diagnostic sensitivity (25 of 66 for LCN-PCR plus serology versus 9 of 66 for serology alone; P < 10−2).
TABLE 3.
Distribution of serum specimens according to the presence of IgM to phase II C. burnetii, LCN-PCR result, and time interval from onset of symptoms
| Category or characteristic | No. of specimens
|
|||
|---|---|---|---|---|
| Days 1 to 14 | Days 15 to 28 | Later than 28 days | Total | |
| Tested sera | 66 | 14 | 20 | 100 |
| Positive LCN-PCR and titer of IgM to phase II C. burnetii of ≥1:50 | 0 | 0 | 2 | 2 |
| Positive LCN-PCR and titer of IgM to phase II C. burnetii of <1:50 | 16 | 0 | 0 | 16 |
| Subtotal positive LCN-PCR | 16 | 0 | 2 | 18 |
| Negative LCN-PCR and titer of IgM to phase II C. burnetii of ≥1:50 | 9 | 10 | 18 | 37 |
| Negative LCN-PCR and titer of IgM to phase II C. burnetii of <1:50 | 41 | 4 | 0 | 45 |
| Subtotal negative LCN-PCR | 50 | 14 | 18 | 82 |
DISCUSSION
In the present study, we describe the successful adaptation of LCN-PCR to the diagnosis of acute Q fever and compare this technique to IFA, the reference serological method for this disease.
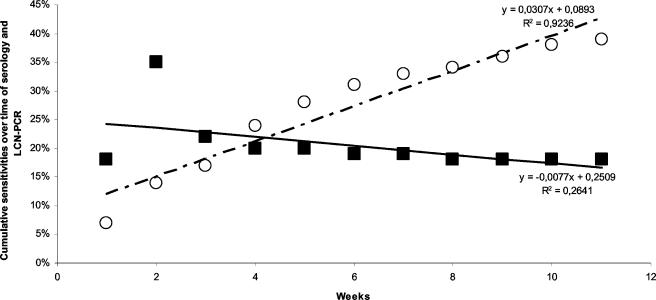
Among the 100 patients with definite acute Q fever included in our series, we could detect C. burnetii in the serum of 18. Serum is one of the easiest human samples to obtain and, when sampled early in the evolution of a systemic disease, is likely to contain DNA copies of systemic pathogens (16). Therefore, it is important to be able to use this sample for direct diagnosis. However, the presence of inhibitors in serum, although in smaller quantity than in whole blood (1), and the usually small amount of bacterial DNA present in serum limit the efficiency of PCR for this assay. In contrast with the usual PCR, nested PCR approaches with serum have been effective for the diagnosis of plague (15) and rickettsial diseases (16). However, nested PCR assays are subject to lateral contamination during the amplification. None of the serum specimens from 40 blood donors and 40 patients with pneumonia caused by other pathogens which we used as negative controls was positive, thus demonstrating the high specificity of LCN-PCR for the detection of C. burnetii. The htpAB-associated element, present in 20 highly conserved copies throughout the C. burnetii genome, appeared to be a valuable DNA target for PCR assays. By targeting this multicopy DNA fragment, previously successfully used for biopsy specimens, we were able to detect as little as one DNA copy of C. burnetii in serum. However, when compared with LCN-PCR results obtained for Bartonella endocarditis, where the test had a sensitivity of 58.1% (22), the sensitivity of our assay (18%) may appear low. We may explain this difference by the fact that Bartonella endocarditis is an intravascular infection with persistent bacteremia, and thus, the amount of circulating bacteria is likely to be higher than in acute Q fever, in which bacteria are rapidly eliminated from the bloodstream and patients most often spontaneously recover (12). We observed that the sensitivity of the test was higher when applied prior to seroconversion (26%) and much lower among seropositive samples (5%). In acute Q fever, phase II antibodies appear between weeks 2 and 3 after the onset of symptoms (19), which is verified in our series, where serology clearly lacks sensitivity in the first 2 weeks of infection. When comparing the sensitivities of LCN-PCR and serology with respect to the time interval between onset of symptoms and serum sampling (Fig. 1), LCN-PCR was mostly useful in the first 2 weeks of the disease (P = 0.02), and serology was effective after the second week (P < 10−2) and optimal after the fourth week (P = 0.03). In addition, LCN-PCR positivity was significantly associated with the absence of antibodies (P = 0.03). Whether antibodies themselves play a direct role is unclear, since the multivariate analysis showed that the presence of antibodies was not independently linked to negative PCR results (Table 2) but was influenced by the time interval between onset of symptoms and serum sampling (P < 10−2).
FIG. 1.
Cumulative sensitivities of LCN-PCR and serology. Each white circle represents the ratio of sera exhibiting titers of positive IgM antibodies to phase II C. burnetii of >1:50 among all samples tested until a given week. Black squares represent the ratio of positive LCN-PCR sera among all samples tested until a given week. The bold line represents the trend line for LCN-PCR positivity, whereas the dashed line represents the trend line for sera exhibiting titers of IgM antibodies to phase II C. burnetii of >1:50. Curve equations and determination coefficients (R2) are indicated.
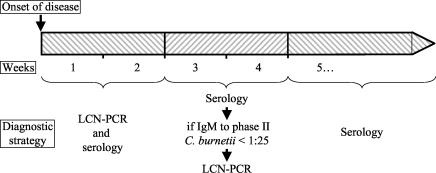
In the initial 2-week period after onset of disease, the combination of our assay and serology significantly increased the sensitivity of the diagnosis of acute Q fever. In the following 2 weeks, significantly more sera contained IgM antibodies to C. burnetii than in the first 2 weeks, but 29% of specimens were seronegative, whereas all samples taken after the fourth week were seropositive. Thus, we propose a diagnostic strategy to improve the early diagnosis of acute Q fever in patients with clinical manifestations suggestive of acute Q fever (Fig. 2): for serum specimens collected in the first 2 weeks of infection, both LCN-PCR and serology should be performed; from weeks 3 to 4 after onset, serology should be performed first and LCN-PCR should be reserved for seronegative specimens; for serum specimens collected later than 4 weeks after the onset of symptoms, serology but not LCN-PCR should be used as diagnostic test.
FIG. 2.
Diagnostic strategy for the early diagnosis of acute Q fever.
Two of the LCN-PCR-positive serum specimens had been taken during the fifth and ninth weeks, respectively, following the onset of symptoms. Using real-time PCR, Harris et al. (8) were able to detect C. burnetii in the bone marrow of patients years after acute infection, suggesting that this latent infection may be a step toward chronic infection. Clinically, our two patients recovered from acute infection, but a serological follow-up will be performed in the coming months.
The need for sensitive and rapid diagnostic tools for acute Q fever has recently been emphasized, both because C. burnetii may be responsible for severe infections (2, 4) and because it is currently regarded as a potential agent of bioterrorism. The htpAB-associated element-based LCN-PCR, using a rapid one-step procedure in the LightCycler thermocycler which prevents amplicon carryover, can be carried out with serum and may help lessen the delay in diagnosing acute Q fever. Since C. burnetii multiplies within mononuclear phagocytes, our technique also will need to be evaluated on buffy coat specimens. We propose that LCN-PCR should be applied to subjects suspected to have Q fever, especially in the case of outbreaks, including terrorist attacks. We recommend that it should be applied in priority to specimens sampled in the first 2 weeks of acute Q fever together with serology and as a secondary tool after serology for samples collected between weeks 3 and 4.
Acknowledgments
We thank Gilbert Greub for statistical support, Hervé Tissot-Dupont for providing patients' clinical information, Thi-Phong Huyhn for her technical help, and Patrick J. Kelly for reviewing the manuscript.
REFERENCES
- 1.Abu Al-Soud, W., and P. Radstrom. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernit, E., J. Pouget, F. Janbon, H. Dutronc, P. Martinez, P. Brouqui, and D. Raoult. 2002. Neurological involvement in acute Q fever—a report of 29 cases and review of the literature. Arch. Intern. Med. 162:693-700. [DOI] [PubMed] [Google Scholar]
- 3.Fishbein, D. B., and D. Raoult. 1992. A cluster of Coxiella burnetii infections associated with exposure to vaccinated goats and their unpasteurized dairy products. Am. J. Trop. Med. Hyg. 47:35-40. [DOI] [PubMed] [Google Scholar]
- 4.Fournier, P. E., J. Etienne, J. R. Harle, and D. Raoult. 2000. Myocarditis, a rare but severe manifestation of Q fever: report of 8 cases and review of the literature. Clin. Infect. Dis. 32:1440-1447. [DOI] [PubMed] [Google Scholar]
- 5.Fournier, P. E., H. Lelievre, S. J. Eykyn, J. L. Mainardi, T. J. Marrie, F. Bruneel, C. Roure, J. Nash, D. Clave, E. James, C. Benoit-Lemercier, L. Deforges, H. Tissot-Dupont, and D. Raoult. 2001. Epidemiologic and clinical characteristics of Bartonella quintana and Bartonella henselae endocarditis: a study of 48 patients. Medicine (Baltimore) 80:245-251. [DOI] [PubMed] [Google Scholar]
- 6.Fournier, P. E., T. J. Marrie, and D. Raoult. 1998. Diagnosis of Q fever. J. Clin. Microbiol. 36:1823-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fournier, P. E., and D. Raoult. 1999. Predominant immunoglobulin A response to phase II antigen of Coxiella burnetii in acute Q fever. Clin. Diagn. Lab. Immunol. 6:173-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris, R. J., P. A. Storm, A. Lloyd, M. Arens, and B. P. Marmion. 2000. Long-term persistence of Coxiella burnetii in the host after primary Q fever. Epidemiol. Infect. 124:543-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatchette, T. F., R. C. Hudson, W. M. Scheld, N. A. Campbell, J. E. Hatchette, S. Ratnam, D. Raoult, C. Donovan, and T. J. Marrie. 2001. Goat-associated Q fever: a new disease in Newfoundland. Emerg. Infect. Dis. 7:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins, D., and T. J. Marrie. 1990. Seroepidemiology of Q fever among cats in New Brunswick and Prince Edward Island. Ann. N. Y. Acad. Sci. 590:271-274. [DOI] [PubMed] [Google Scholar]
- 11.Marrie, T. J., H. Durant, J. C. Williams, E. Mintz, and D. M. Waag. 1988. Exposure to parturient cats: a risk factor for acquisition of Q fever in maritime Canada. J. Infect. Dis. 158:101-108. [DOI] [PubMed] [Google Scholar]
- 12.Maurin, M., and D. Raoult. 1999. Q fever. Clin. Microbiol. Rev. 12:518-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ormsbee, R. A., M. G. Peacock, R. Gerloff, G. Tallent, and D. Wike. 1978. Limits of rickettsial infectivity. Infect. Immun. 19:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raoult, D. 1990. Host factors in the severity of Q fever. Ann. N. Y. Acad. Sci. 590:33-38. [DOI] [PubMed] [Google Scholar]
- 15.Raoult, D., G. Aboudharam, E. Crubezy, G. Larrouy, B. Ludes, and M. Drancourt. 2000. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of Medieval Black Death. Proc. Natl. Acad. Sci. USA 97:12800-12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raoult, D., P. E. Fournier, F. Fenollar, M. Jensenius, T. Prioe, J. J. de Pina, G. Caruso, N. Jones, H. Laferl, J. E. Rosenblatt, and T. J. Marrie. 2001. Rickettsia africae, a tick-borne pathogen in travelers to sub-Saharan Africa. N. Engl. J. Med. 344:1504-1510. [DOI] [PubMed] [Google Scholar]
- 17.Raoult, D., H. Tissot-Dupont, C. Foucault, J. Gouvernet, P. E. Fournier, E. Bernit, A. Stein, M. Nesri, J. R. Harle, and P. J. Weiller. 2000. Q fever 1985-1998—clinical and epidemiologic features of 1,383 infections. Medicine 79:109-123. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tissot-Dupont, H., X. Thirion, and D. Raoult. 1994. Q fever serology: cutoff determination for microimmunofluorescence. Clin. Diagn. Lab. Immunol. 1:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss, E., and J. W. Moulder. 1984. Order I Rickettsiales, Gieszczkiewicz 1939, p. 687-703. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 21.Willems, H., D. Thiele, R. Frolich-Ritter, and H. Krauss. 1994. Detection of Coxiella burnetii in cow's milk using the polymerase chain reaction. J. Vet. Med. B 41:580-587. [DOI] [PubMed] [Google Scholar]
- 22.Zeaiter, Z., P. E. Fournier, G. Greub, and D. Raoult. 2003. Diagnosis of Bartonella endocarditis by a real-time nested PCR assay using serum. J. Clin. Microbiol. 41:919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]