Abstract
Phylogenetic analysis of the sequences of the 5′ noncoding regions (5′NCR) of 149 samples from hepatitis C virus (HCV) RNA-positive chronic carriers representing northern, southern, eastern, and western India showed that type 3 and type 1 are the predominant genotypes circulating in India, with an overall prevalence of 53.69 and 38.25%, respectively. Type 4 viruses (6.04%) were seen only in southern India. Sequence analysis of the core region of 51 of the above isolates enabled us to classify them further into subtypes as 1b (number of isolates [n] = 10), 1a (n = 6), 3a (n = 9), 3g (n = 14), 3f (n = 1), and 4d (n = 3). Three new subtypes were identified for the first time and designated as 3i (n = 5), 3j (n = 2), and 6l (n = 1). Sequencing the 5′NCR could differentiate HCV types, whereas classification at the level of subtype was possible with sequence analysis of the core region.
Hepatitis C virus (HCV) is a major causative agent of chronic hepatitis that can progress to liver cirrhosis and hepatocellular carcinoma. HCV has a substantial nucleotide sequence diversity, with 68 to 79% overall sequence similarity among strains. Regions encoding the putative envelope proteins (E1 and E2/NS1) are the most variable (6, 15, 37), whereas the 5′ noncoding region (5′NCR) is the most conserved (9, 14). Six major genotypes and more than 100 different subtypes of the virus have been identified worldwide (3, 5, 7, 19, 28, 30). The relative prevalences of these genotypes vary among different geographic regions. Subtypes 1a, 1b, 2a, 2c, and 3a account for more than 90% of the HCV infections in North and South America, Europe, Russia, China, Japan, Australia, and New Zealand (19). Most infections in Egypt, North Africa, Central Africa, and the Middle East are of genotype 4. Type 5 has been described in South Africa (1, 10, 11). Genotype 6 isolates are primarily found in Southeast Asia. In the Indian subcontinent, subtype 1b seems to be the most prevalent type, but many isolates related to genotype 3 have been described from northern and southern India, Bangladesh, Pakistan, and Nepal (2, 5, 7, 23, 25-27, 34, 36). Studies of HCV variants have been made in the neighboring countries of Thailand (21, 22), Vietnam (35), Indonesia (16), and Burma. Type 1 (a, b, c), type 2, type 3 (a, b, c, d, e, f, g), and type 6 variants are prevalent in these areas. Subtype 1b is the major subtype in China, while type 2 is also prevalent in some areas (8).
Many methods of genotyping are based on amplified 5′NCR sequences because of the high sensitivity of PCR assays in this conserved region (30). Phylogenetic analysis of sequence information obtained from the 5′NCR correlates fairly well with that obtained from the core and the NS3, NS4, and NS5 regions of the viral genome in the determination of major genotypes (13). However, insufficient sequence variation in the 5′NCR limits its usefulness for determining differences among various subtypes (4, 5). The identification and characterization of HCV types and subtypes have major implications for HCV diagnostics as well as for the development of a vaccine(s).
The aim of this cross-sectional study was to identify the genotypes circulating in India and to determine the suitability of the HCV genomic region for routine determination of HCV genotypes.
Serum samples.
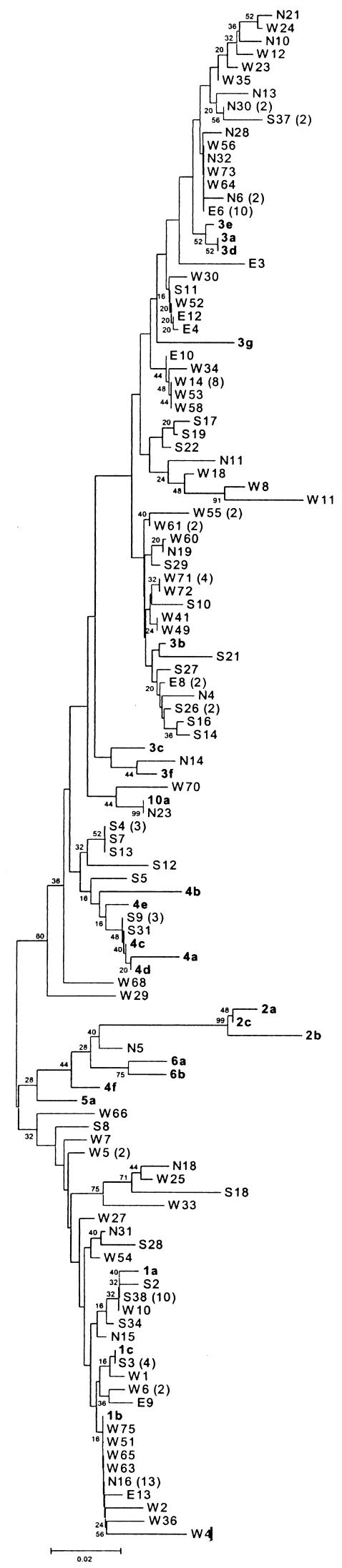
Serum samples (stored in aliquots at −70°C) from chronic HCV carriers showing HCV RNA positivity and representing four different areas of India were included in the study. The north was represented by Delhi and Lucknow (number of isolates [n] = 30); the south was represented by Bangalore, Coimbatore, and Cochin (n = 36); the east was represented by Calcutta (n = 12); and the west was represented by Pune, Mumbai, Ahmedabad, and Baroda (n = 71). The isolates from north, south, west, or east India were designated as N, S, W, or E, respectively, to identify the origin of the samples (Fig. 1 and 2 ).
FIG.1.
Phylogenetic analysis of 5′NCR (nt −241 to −60; 182 nt) sequences of 149 HCV isolates. The isolates obtained from the areas of north, south, west, or east India are designated as N, S, W, or E, respectively, to identify the origins of the samples. Numbers in parentheses indicate the number of isolates branching similarly. Sequences for each major subtype were selected from the GenBank database for analysis. The accession numbers of the reference sequences (with subtypes) are as follows: AF011753 (1a), AJ132996 (1b), AY051292 (1c), AF238485 (2a), AF238486 (2b), L38330 (2c), AF046866 (3a), D49374 (3b), D16612 (3c), D16620 (3d), D16618 (3e), D16614 (3f), X91421 (3g), D49753 (10a), Y11604 (4a), M84845 (4b), M84862 (4c), M84832 (4d), M84828 (4e), M84829 (4f), Y13184 (5a), Y12083 (6a), and D84262(6b). All reference sequences are marked in bold.
FIG. 2.
Phylogenetic analysis of partial core gene (nt 1 to 360; 360 nt) sequences of 51 HCV isolates. The isolates obtained from the areas of north, south, west, or east India are designated as N, S, W, or E, respectively, to identify the origins of the samples. Isolates grouping in the newly identified subtypes are circled. All GenBank reference sequences are marked in bold. The accession numbers of reference sequences (with subtypes) are as follows: AF011753 (1a), AJ132996 (1b), AY051292 (1c), AF238485 (2a), AF238486 (2b), L38330 (2c),D49746 (2e), D49754 (2f), AB031663 (2k), AF046866 (3a), D49374 (3b), D16612 (3c), D16620 (3d), D16618 (3e), D16614 (3f), X91423 (3g), D49753 (10a), Y11604 (4a), U10235 (4b), U10238 (4c), U10192 (4d), U10237 (4e), U10240 (4f), Y13184 (5a), Y12083 (6a), D84262 (6b), D84263 (6d), D63822 (6e), D84265 (6h), and D84264 (6k).
RT-PCR, sequencing, and phylogenetic analysis.
For the detection of HCV RNA, nested reverse transcriptase (RT)-PCR was carried out with 5′NCR primers as described by Bukh et al. (6). Briefly, total RNA was extracted from 100 μl of serum with TRIzol reagent (GIBCO BRL, Life Technologies). Single-tube nested RT-PCR was carried out, and HCV RNA-positive PCR products (nucleotides [nt] −276 to −21; 256 bp) were purified in a column with a gel extraction kit (QIAGEN, Valencia, California) and used as templates for sequencing in the Big-Dye Terminator cycle sequencing ready reaction kit (Applied Biosystems). Samples were analyzed on an automated sequencer (ABI PRISM 310 genetic analyzer; Applied Biosystems). Products were sequenced from both strands to get consensus sequences. The sequence from nt −241 to −60 (182 nt) was taken for analysis. A total of 149 isolates were sequenced in the 5′NCR.
Fifty-one serum samples were also amplified and sequenced in the core region. These samples included isolates of different genotypes and nontypeable isolates as well as those branching differently from the prototype sequences based on the 5′NCR analysis. The core region (405 nt) was amplified with the following primers: outer sense primer (nt −54 to −34), CC1 (5′ACT GCC TGA TAG GGT GCT TGC 3′); outer antisense primer (nt 410 to 391), CC2 (5′ ATG TAC CCC ATG AGG TCG GC 3′); inner sense primer (nt −22 to −4), CC3 (5′ AGG TCT CGT AGA CCG TGC A 3′); and inner antisense primer (nt 383 to 364), CC4 (5′ CAC GTT AGG GTA TCG ATG AC 3′). The core region sequence from nt 1 to 360 was taken for analysis.
Indian isolates sequenced in the present study were aligned with the representative number of sequences for each major genotype and subtype selected from the GenBank database with the help of the Multalign program. Pairwise comparisons for percent nucleotide homology and evolutionary distance were made. The phylogenetic analysis of HCV isolates was performed with MEGA 2.1 software (18). Jukes-Cantor algorithms were utilized, and phylogenetic trees were constructed by the neighbor-joining method. The reliability of different phylogenetic groupings was evaluated by using the bootstrap resampling test from the MEGA program (1,000 bootstrap replications).
Genotyping based on 5′NCR analysis.
On the basis of phylogenetic analysis, the 149 Indian isolates were classified as follows: 38.25% (n = 57) were type 1, 53.69% (n = 80) were type 3, 6.04% (n = 9) were type 4, and 0.671% (n = 1) were type 6 (Fig. 1 and Table 1). Two isolates, W68 and W29, remained nontypeable. The percent nucleotide identity (PNI) was 98.19% ± 0.45% within Indian type 1 sequences, 98.12% ± 0.50% for type 3 sequences, and 98.75% ± 0.48% for type 4 sequences. The PNI between different genotypes was 94.67% ± 0.11% for type 1 and type 3, 94.80% ± 0.11% for type 1 and type 4, and 95.26% ± 0.12% for type 3 and type 4. (The overall nucleotide similarity among different Indian isolates was 94.55% ± 0.96%.) There was a stretch of hypervariable region from nt −166 to −86 in the 5′NCR which had motifs for different genotypes. Indian isolates showed 9.31% ± 0.21% nucleotide variability in this region. The comparatively conserved stretch from nt −241 to −165 showed only 3.30% ± 1.06% variation.
TABLE 1.
Prevailing HCV genotypes in India based on 5′NCR sequence analysis
| Type | Distribution (%) of samples by regiona
|
|||
|---|---|---|---|---|
| North | South | West | East | |
| 1 | 30 (9) | 38.88 (14) | 43.66 (31) | 25 (3) |
| 3 | 66.66 (20) | 36.11 (13) | 53.52 (38) | 75 (9) |
| 4 | 25 (9) | |||
| Nontypeable | 3.33 (1) | 2.82 (2) | ||
The numbers in parentheses indicate the sizes of the samples.
It was not possible to classify the type 1 and type 4 isolates further into different subtypes. In the case of the type 3 isolates, there was a clear clustering of isolates into subtypes 3b and 3g. However, the other subtypes, 3a, 3d, and 3e, clustered together. Therefore, all of the 149 isolates were classified only into major genotypes. Type 3 and type 1 seem to be the predominant genotypes circulating in different parts of India, with a prevalence of 53.69 and 38.25%, respectively. Type 3 and type 1 were present in almost equal proportions in the south (36.11 and 38.88%, respectively) and also in the west (53.52 and 43.66%, respectively). However, the north (66.66 and 30% for type 3 and type 1, respectively) and the east (75 and 25% for type 3 and type 1, respectively) showed a greater occurrence of cases of type 3 than of type 1. Variants of genotype 3 are prevalent in the neighboring countries to India's north and east, such as Bangladesh, Pakistan, Thailand, Nepal, Indonesia, and Vietnam (26, 30, 31, 33-35). The higher percentages of type 3 cases in the northern and eastern parts of India could probably be due to geographical proximity to neighboring countries and a long history of travel, trade, and communication among the people of these areas. More or less equal proportions of type 1 and type 3 isolates in the western and southern parts of India indicate that type 3 variants are spreading all over India and most probably will exceed the occurrence of type 1 in the future.
Analysis of the sequences in the core region.
The isolates could be classified into subtypes (with the number of isolates identified by area) as type 1a (6 isolates: north, 1; south, 2; and west, 3), type 1b (10 isolates: north, 1; south, 3; and west, 6), type 3a (9 isolates: north, 3; south, 1; east, 2; and west, 3), type 3g (14 isolates: north, 2; south, 3; east, 1; and west, 8), type 3f (1 isolate from the north), 3i (5 isolates from the west), 3j (2 isolates: north, 1; and south, 1), type 4d (3 isolates from the south), and 6l (1 isolate from the north) (Fig. 2). The PNI within subtypes was 97.54% ± 0.48% for subtype 1a, 94.51% ± 0.57% for subtype 1b, 96.13% ± 0.59% for subtype 3a, 96.04% ± 0.61% for subtype 3g, 98.20% ± 0.57% for subtype 3i, 95.32% ± 1.04% for subtype 3j, and 98.75% ± 0.48% for subtype 4d. The two isolates, W68 and W29, which remained nontypeable with 5′NCR analysis, were typed as 1b and 3g, respectively.
Isolate N5 clustered separately with type 6 isolates. However, it showed less than 88% homology with any of the reference sequences belonging to this type and was thus classified as a new subtype 6l. The patient circulating this virus was unavailable for follow-up and, hence, the origin of the virus could not be ascertained. Isolates S9 and N23 clustered with the reference sequence of isolate JK072, with a homology of 95.60% ± 1.03% and 97% ± 0.87%, respectively. JK072, an isolate from Jakarta, Indonesia, initially classified as type 10a (31), was subsequently grouped as a divergent subtype of genotype 3 (12, 29). This distant subtype 3 group was newly designated 3j in the present study. One isolate, S9, showed a different branching in the 5′NCR tree (type 4) than that indicated by the core tree (3j). The possibility of infection with multiple HCV genotypes or with a recombinant virus in this patient is being explored (17). Five isolates, W8, W14, W61, W53, and W58, clustered separately from the 3f and 3g clusters. The PNI within the group was 98.20%, and the PNI with the nearest neighbors was −92.80% for 3f and −92.33% for 3g, indicating a need to assign these five isolates to a separate subgroup. They were grouped into a new subtype, 3i. Isolate N14 showed a PNI of 95.36% with 3f and 94.37% with 3g. This isolate was classified as subtype 3f.
Type 4 viruses were seen only in the southern part of India (from only one city, Coimbatore; 9 of 25 isolates) and showed very high sequence homology among them (98.75% ± 0.48%), indicating a common source of infection. The isolates, S7, S31, and S12, clustered tightly with the sequence of 4d, with a homology of 96.60% ± 0.59%. It may be noted here that type 4 viruses are prevalent in Middle Eastern countries, where a large number of people from southern Indian travel and work. Transmission from such countries, where type 4 is endemic, was ruled out as none of the patients or their contacts reported such a history. In fact, the patients had not traveled to any other country or even outside the state. Thus, though these isolates seem to be local, their origin as well as subsequent spread needs to be carefully investigated. Recently, isolates belonging to subtype 1c have also been obtained from northern India (our unpublished observations).
A typing method based on the sequence of the core region could reliably identify subtypes as well as major genotypes since the sequence divergence was greater than the divergence of the 5′NCR sequence. Though the preferred regions for genotyping are E1, NS4, and NS5, which are more variable than the core, it is not always possible to amplify them. The main reason is a lack of conservation in the primer binding sites. We found that a common set of primers could be used for amplification of the core region for all the samples. All (51/51) HCV RNA-positive samples (based on 5′NCR analysis) were amplified with these primers. The partial core gene (nt 1 to 360) sequence analysis could differentiate all the reference sequences into their respective types and subtypes. The intersubtype homology ranged from 90 to 94% for types 1, 2, 4, and 5 in this region. However, this range was very high for the subtypes of 3 (85 to 92%). The GenBank sequences of isolates from Southeast Asia (grouped as 7a, 7b, 7c, 7d, 8a, 8b, 9a, 9b, 9c, and 11a) (31, 33, 35) also clustered clearly into type 6 as per their recent classification (12, 20, 24, 29, 32), while the group of isolates from Indonesia (Jakarta) typed earlier as 10a also clustered clearly as type 3 (as per their recent grouping).
Importantly, this study described three new subtypes for the first time, designated as 3i (n = 5), 3j (n = 2), and 6l (n = 1). The occurrence of many type 3 variants in India is not surprising because most of the variants other than subtype 3a and subtype 3b have been reported from neighboring countries. The possibility of identifying more variants cannot be ruled out in the present situation. A multicenter study representing larger numbers of isolates from every Indian state and community seems essential to generate countrywide data. Thus, we conclude that multiple HCV genotypes are prevalent in India and that 5′NCR sequence analysis is satisfactory for the routine and quick genotyping of isolates; however, sequencing of the core region is more suitable for further classification of the virus into subtypes.
Nucleotide sequence accession numbers.
The 5′NCR and core region sequences have been deposited in the GenBank database under accession numbers AY190377 to AY190507 and AY190328 to AY190376, respectively.
Acknowledgments
The authors thank Fulford India Pvt. Ltd. for the management of some of the samples and B. Kundu for technical assistance.
REFERENCES
- 1.Abdulkarim, A. S., N. N. Zein, J. J. Germer, C. P. Kolbert, L. Kabbani, K. L. Krajnik, A. Hola, M. N. Agha, M. Tourogman, and D. H. Persing. 1998. Hepatitis C virus genotypes and hepatitis G virus in hemodialysis patients from Syria: identification of two novel hepatitis C virus subtypes. Am. J. Trop. Med. Hyg. 59:571-576. [DOI] [PubMed] [Google Scholar]
- 2.Amarapurkar, D., M. Dhorda, A. Kirpalani, A. Amarapurkar, and S. Kankonkar. 2001. Prevalence of hepatitis C genotypes in Indian patients and their clinical significance. J. Assoc. Physicians India 49:983-985. [PubMed] [Google Scholar]
- 3.Bukh, J., S. U. Emerson, and R. H. Purcell. 1997. Genetic heterogeneity of hepatitis C virus and related viruses, p. 167-175. In M. Rizzetto, R. H. Purcell, J. L. Gerin et al. (ed.), Viral hepatitis and liver disease. Minerva Medica, Turin, Italy.
- 4.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 5.Bukh, J., R. H. Purcell, and R. H. Miller. 1993. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc. Natl. Acad. Sci. USA 90:8234-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukh, J., R. H. Purcell, and R. H. Miller. 1992. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc. Natl. Acad. Sci. USA 89:87-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, Q., X. Zhang, L. Tian, M. Yuan, G. Jin, and Z. Lu. 2002. Variant analysis and immunogenicity prediction of envelope gene of HCV strains from China. J. Med. Virol. 67:490-500. [DOI] [PubMed] [Google Scholar]
- 9.Cha, T. A., J. Kolberg, B. Irvine, M. Stempien, E. Beall, M. Yano, Q. L. Choo, M. Houghton, G. Kuo, J. H. Han, and M. S. Urdea. 1991. Use of a signature nucleotide sequence of hepatitis C virus for detection of viral RNA in human serum and plasma. J. Clin. Microbiol. 29:2528-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamberlain, R. W., N. Adams, A. A. Saeed, P. Simmonds, and R. M. Elliot. 1997. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J. Gen. Virol. 78:1341-1347. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain, R. W., N. J. Adams, L. A. Taylor, P. Simmonds, and R. M. Elliott. 1997. The complete coding sequence of hepatitis C virus genotype 5a, the predominant genotype in South Africa. Biochem. Biophys. Res. Commun. 236:44-49. [DOI] [PubMed] [Google Scholar]
- 12.de Lamballerie X., R. N. Charrel, H. Attoui, and P. De Micco. 1997. Classification of hepatitis C virus variants in six major types based on analysis of the envelope 1 and nonstructural 5B genome regions and complete polyprotein sequences. J. Gen. Virol. 78:45-51. [DOI] [PubMed] [Google Scholar]
- 13.Germer, J. J., and N. N. Zein. 2001. Advances in the molecular diagnosis of hepatitis C and their clinical implications. Mayo Clin. Proc. 76:911-920. [DOI] [PubMed] [Google Scholar]
- 14.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irvine, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q. L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly (A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 88:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, and K. Shimotohno. 1991. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem. Biophys. Res. Commun. 175:220-228. [DOI] [PubMed] [Google Scholar]
- 16.Hotta, H., R. Handajani, M. I. Lusida, W. Soemarto, H. Doi, H. Miyajima, and M. Homma. 1994. Subtype analysis of hepatitis C virus in Indonesia on the basis of NS5b region sequences. J. Clin. Microbiol. 32:3049-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Maertens, G., and L. Stuyver. 1997. Genotypes and genetic variation of hepatitis, p. 183-233. In T. J. Harrison and A. Zuckerman (ed.), The molecular medicine of viral hepatitis. John Wiley and Sons, Chichester, England.
- 20.Mizokami, M., T. Gojobori, K. Ohba, K. Ikeo, X. M. Ge, T. Ohno, E. Orito, and J. Y. Lau. 1996. Hepatitis C virus types 7, 8 and 9 should be classified as type 6 subtypes. J. Hepatol. 24:622-624. [DOI] [PubMed] [Google Scholar]
- 21.Mori, S., N. Kato, A. Yagyu, T. Tanaka, Y. Ikeda, B. Petchclai, P. Chiewsilp, T. Kurimura, and K. Shimotohno. 1992. A new type of hepatitis C virus in patients in Thailand. Biochem. Biophys. Res. Commun. 183:334-342. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, H., H. Tokita, M. Sakamoto, M. Horikita, M. Kojima, H. Iizuka, and S. Mishiro. 1993. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J. Gen. Virol. 74:2385-2390. [DOI] [PubMed] [Google Scholar]
- 23.Panigrahi, A. K., J. Roca, S. K. Acharya, S. Jameel, and S. K. Panda. 1996. Genotype determination of hepatitis C virus from northern India: identification of a new subtype. J. Med. Virol. 48:191-198. [DOI] [PubMed] [Google Scholar]
- 24.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, I. T. Shin, P. Simmonds, D. Smith, L. Stuyver, A. Weiner, et al. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 25.Sawant, P, P. M. Rathi, and A. Upadhyaya. 1999. Hepatitis B and hepatitis C genotypes in cirrhosis in Western India: results of a pilot study. J. Assoc. Physicians India 47:580-583. [PubMed] [Google Scholar]
- 26.Shah, H. A., W. Jafri, I. Malik, L. Prescott, and P. Simmonds. 1997. Hepatitis C virus (HCV) genotypes and chronic liver disease in Pakistan. J. Gastroenterol. Hepatol. 12:758-761. [DOI] [PubMed] [Google Scholar]
- 27.Shrestha, S. M., F. Tsuda, H. Okamoto, H. Tokita, M. Horikita, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis B virus subtypes and hepatitis C virus genotypes in patients with chronic liver disease in Nepal. Hepatology 19:805-809. [PubMed] [Google Scholar]
- 28.Simmonds, P., A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, D. S. Chen, et al. 1994. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19:1321-1324. [PubMed] [Google Scholar]
- 29.Simmonds, P., J. Mellor, T. Sakuldamrongpanich, C. Nuchaprayoon, S. Tanprasert, E. C. Holmes, and D. B. Smith. 1996. Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J. Gen. Virol. 77:3013-3024. [DOI] [PubMed] [Google Scholar]
- 30.Simmonds, P. 1995. Variability of hepatitis C virus. Hepatology 21:570-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokita, H., H. Okamoto, H. Iizuka, J. Kishimoto, F. Tsuda, L. A. Lesmana, Y. Miyakawa, and M. Mayumi. 1996. Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J. Gen. Virol. 77:293-301. [DOI] [PubMed] [Google Scholar]
- 32.Tokita, H., H. Okamoto, H. Iizuka, J. Kishimoto, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1998. The entire nucleotide sequences of three hepatitis C virus isolates in genetic groups 7-9 and comparison with those in the other eight genetic groups. J. Gen. Virol. 79:1847-1857. [DOI] [PubMed] [Google Scholar]
- 33.Tokita, H., H. Okamoto, P. Luengrojanakul, K. Vareesangthip, T. Chainuvati, H. Iizuka, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d) and ninth (9b, 9c) major genetic groups. J. Gen. Virol. 76:2329-2335. [DOI] [PubMed] [Google Scholar]
- 34.Tokita, H., S. M. Shrestha, H. Okamoto, M. Sakamoto, M. Horikita, H. Iizuka, S. Shrestha, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis C virus variants from Nepal with novel genotypes and their classification into the third major group. J. Gen. Virol. 75:931-936. [DOI] [PubMed] [Google Scholar]
- 35.Tokita, H., H. Okamoto, F. Tsuda, P. Song, S. Nakata, T. Chosa, H. Iizuka, S. Mishiro, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc. Natl. Acad. Sci. USA 91:11022-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valliammai, T., S. P. Thyagarajan, A. J. Zuckerman, and T. J. Harrison. 1995. Diversity of genotypes of hepatitis C virus in southern India. J. Gen. Virol. 76:711-716. [DOI] [PubMed] [Google Scholar]
- 37.Weiner, A. J., M. J. Brauer, J. Rosenblatt, K. H. Richman, J. Tung, K. Crawford, F. Bonino, G. Saracco, Q. L. Choo, M. Houghton, et al. 1991. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology 180:842-848. [DOI] [PubMed] [Google Scholar]