Abstract
Oxacillin-resistant Staphylococcus aureus (ORSA) is a virulent pathogen responsible for both health care-associated and community onset disease. We used SmaI-digested genomic DNA separated by pulsed-field gel electrophoresis (PFGE) to characterize 957 S. aureus isolates and establish a database of PFGE patterns. In addition to PFGE patterns of U.S. strains, the database contains patterns of representative epidemic-type strains from the United Kingdom, Canada, and Australia; previously described ORSA clonal-type isolates; 13 vancomycin-intermediate S. aureus (VISA) isolates, and two high-level vancomycin-resistant, vanA-positive strains (VRSA). Among the isolates from the United States, we identified eight lineages, designated as pulsed-field types (PFTs) USA100 through USA800, seven of which included both ORSA and oxacillin-susceptible S. aureus isolates. With the exception of the PFT pairs USA100 and USA800, and USA300 and USA500, each of the PFTs had a unique multilocus sequence type and spa type motif. The USA100 PFT, previously designated as the New York/Tokyo clone, was the most common PFT in the database, representing 44% of the ORSA isolates. USA100 isolates were typically multiresistant and included all but one of the U.S. VISA strains and both VRSA isolates. Multiresistant ORSA isolates from the USA200, -500, and -600 PFTs have PFGE patterns similar to those of previously described epidemic strains from Europe and Australia. The USA300 and -400 PFTs contained community isolates resistant only to β-lactam drugs and erythromycin. Noticeably absent from the U.S. database were isolates with the previously described Brazilian and EMRSA15 PFGE patterns. These data suggest that there are a limited number of ORSA genotypes present in the United States.
Oxacillin-resistant Staphylococcus aureus (ORSA), more commonly referred to as methicillin-resistant S. aureus (MRSA) (even though methicillin is rarely tested in U.S. laboratories), is a frequent cause of infections both in health care and community settings and is endemic in many U.S. hospitals (12, 26, 27, 32, 48, 56). While a variety of strain typing methods have been used over the years to track the spread of ORSA (64), most outbreaks of ORSA have been characterized by bacteriophage typing or pulsed-field gel electrophoresis (PFGE). Although public health institutions in several countries, such as Australia (68), Denmark (41, 53, 67), The Netherlands (41, 66, 67), Canada (59), and the United Kingdom (41), have tracked ORSA strain types over the years, this has not been true in the United States for the most part.
Recent reports of vancomycin-intermediate S. aureus (VISA) (vancomycin MICs, 8 to 16 μg/ml) (5, 22, 28, 33, 50, 60, 65), of vanA-positive ORSA showing high-level vancomycin resistance (8, 9, 11), and of ORSA causing severe disease and death in children (6, 27, 42) all argued that better tracking of ORSA strains nationwide was needed to monitor the spread of such organisms. Thus, the Centers for Disease Control and Prevention (CDC), in collaboration with state health departments, has undertaken the goal of assembling a national database of S. aureus PFGE profiles similar to the PulseNet program for Escherichia coli O157:H7 (62).
The overall goal of this study was to assemble a database of ORSA PFGE profiles and to identify major lineages of ORSA present in the United States. PFGE was chosen over other typing methods, such as multilocus sequence typing (MLST), staphylococcal protein A gene (spa) typing, and restriction fragment length polymorphism-based methods, because the infrastructure and expertise for PFGE typing already exist within the state health departments in the United States. However, a national PFGE-based typing system for S. aureus would have to be validated with MLST and spa typing data to maintain continuity with the nomenclature already established in the literature. This would make the national database information useful for global tracking of ORSA, an organism with a limited number of lineages that exists endemically in health care and non-health-care settings. Thus, we typed a large number of ORSA isolates and developed a PFGE nomenclature scheme that was consistent with MLST and spa data yet was also useful for microbiologists and epidemiologists who were specifically studying ORSA infections in the United States. Herein, we present a framework for describing the lineages of ORSA present in the United States and correlating SmaI PFGE profiles with the spa and MLST types already established in the literature (18, 19, 40, 46, 47, 58).
MATERIALS AND METHODS
Bacterial isolates.
A total of 957 S. aureus isolates were examined in the study; 722 were oxacillin resistant and 235 were oxacillin susceptible according to NCCLS criteria (43). Of the 957 isolates, 300 S. aureus isolates were chosen at random from the CDC S. aureus strain collection. These strains were from outbreaks in hospitals, food-borne disease, and community-acquired infections and were submitted to CDC for typing between 1995 and 2000. They were selected mainly for geographical and source diversity. An additional 381 ORSA isolates were from health care and community outbreak investigations conducted by CDC and state health departments from 2001 to 2003. These included two vanA-positive VRSA strains from Michigan (8) and Pennsylvania (9), eight U.S. VISA isolates for which the vancomycin MICs were 8.0 μg/ml, and 22 isolates for which the vancomycin MICs were 4 μg/ml (22, 60). A further 221 isolates were from various CDC-sponsored surveillance studies. Also included in the database were one VISA isolate from Japan (28), one from France (50), one from Hong Kong, one from Korea (33), and two from Scotland; 15 clonal-type isolates (graciously provided from the collection of H. de Lencastre, Rockefeller University, New York, N.Y.) (13); 15 epidemic MRSA isolates (EMRSA; isolates obtained from H. Aucken, Public Health Laboratory Service, Colindale, United Kingdom); four representative epidemic Canadian MRSA isolates (CMRSA) (59) (courtesy of B. Willey, Mt. Sinai Hospital, Toronto, Ontario, Canada); 12 isolates from community outbreaks in Australia (39, 44) (courtesy of J. Bell, Women and Children's Hospital, Adelaide, Australia), and three staphylococcal SCCmec-type control isolates (31) (courtesy of K. Hiramatsu, Juntendo University, Tokyo, Japan).
Antimicrobial susceptibility testing.
The antimicrobial susceptibility profiles of the isolates were determined by broth microdilution with cation-adjusted Mueller-Hinton broth (BD BioSciences, Sparks, Md.) as described in the NCCLS publication M7-A5 (43). The antimicrobial agents tested were chloramphenicol, clindamycin, erythromycin, gentamicin, levofloxacin, linezolid, oxacillin, penicillin, quinupristin-dalfopristin, rifampin, tetracycline, trimethoprim-sulfamethoxazole, and vancomycin. Quality control strains included S. aureus ATCC 29213, E. coli ATCC 25922, and Enterococcus faecalis ATCC 29212. Multiresistance was defined as resistance to three or more classes of antimicrobial agents. Inducible clindamycin resistance was determined with a disk diffusion test in which an erythromycin disk was placed 12 mm from the edge of a clindamycin disk on a blood agar plate after inoculating the plate with a suspension of the test organism that was adjusted to the turbidity of a 0.5 McFarland standard. The presence of a D-shaped zone of inhibition indicated the induction of methylase production by erythromycin (69). Inducible clindamycin resistance was presumed to be mediated by an erm gene. If no clindamycin induction occurred, the resistance mechanism was assumed to be efflux of the macrolide via msrA (35). High-level resistance to spectinomycin (500 μg/ml), which is associated with the majority of the ORSA strains that carry the transposon Tn554 (49, 55), was determined by disk diffusion with use of a 300-μg disk (made in-house). The absence of a measurable zone of inhibition after 24 h of incubation at 35°C was indicative of resistance. S. aureus strains HDE1 and HPV107, which are known to be susceptible and resistant to spectinomycin, respectively (55), were used as controls.
PFGE.
A single colony of the test isolate was inoculated into 5 ml of brain heart infusion broth and incubated with vigorous shaking at 35 to 37°C for 24 h. The concentrations of the cell suspensions were adjusted with saline either by using a MicroScan Turbidity Meter (Dade Behring, Inc., Deerfield, Ill.) to a turbidity reading of 1.1 to 1.3 or by using a spectrophotometer to an absorbance of 0.9 to 1.1 at 610 nm. Two hundred microliters of the adjusted cell suspension was centrifuged at 12,000 × g for 2 to 4 min, and the supernatant was aspirated. The pellet was resuspended in 300 μl of Tris-EDTA (TE) buffer (10 mM Tris HCl, 1 mM EDTA [pH 8]) and equilibrated in a 37°C water bath for 10 min. Three microliters of recombinant (no. L-0761; Sigma, St. Louis, Mo.) or 4 μl of conventional (no. L-7386; Sigma) lysostaphin stock solution (1 mg/ml in 20 mM sodium acetate [pH 4.5]) and 300 μl of 1.8% (wt/vol) SeaKem Gold agarose (FMC, Rockland, Maine) in TE buffer (equilibrated to 55°C) were added to the cell suspension, gently mixed, and dispensed into the wells of either a large-plug mold (volume of each well, ∼250 μl) or into the wells of a small disposable mold (∼100 μl each). The plugs were allowed to solidify at room temperature for 10 to 15 min or in the refrigerator (4°C) for 5 min. The plugs were removed and placed into a tube containing at least 3 ml of EC lysis buffer (6 mM Tris HCl, 1 M NaCl, 100 mM EDTA, 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% sodium lauroylsarcosine) and incubated at 37°C for at least 4 h. The EC lysis buffer was poured off, and 4 ml of TE buffer was added. The TE washings were repeated at least three more times, and the plugs were stored at 4°C.
SmaI restriction enzyme digestion and gel electrophoresis conditions.
A plug slice was cut to the desired comb size (2 by 5 mm for a 10-, 20-, or 30-tooth comb or 2 by 10 mm for a 10- by 10-tooth comb) and equilibrated in 1× restriction buffer for at least 30 min. After removal of the 1× restriction buffer, 3 μl of SmaI restriction enzyme (Promega no. R6125, 10 U/μl; Promega Corp., Madison, Wis.) in 200 μl of 1× restriction buffer was added to each tube, and the tubes were incubated at 25°C for 2 to 3 h. A SeaKem Gold 1% (wt/vol) agarose gel was prepared in 0.5× TBE from 10× Tris-borate-EDTA buffer (Gibco BRL no. 15581044; Life Technologies, Inc., Gaithersburg, Md.). The plug slices were loaded directly on the end of the comb tooth before placing the comb into the comb holder, and the equilibrated agarose was poured carefully into the gel casting platform. PFGE was performed using a contour-clamped homogeneous electric field apparatus, i.e., DR-II, DR-III, or CHEF Mapper (Bio-Rad, Hercules, Calif.). Running parameters were as follows: 200 V (6 V/cm); temperature, 14°C; initial switch, 5 s; final switch, 40 s; and time, 21 h. After the electrophoresis run was completed, the gel was stained in a 1.5 μg/ml ethidium bromide solution (AMRESCO X328, 10 mg/ml; Amresco, Inc., Solon, Ohio) for 20 min in a covered container and destained in fresh distilled water for 45 min.
Data analysis.
Gels were photographed and digitized with the FOTO/Analyst Archiver system (Fotodyne, Inc., Hartland, Wis.) and saved as a TIFF file for analysis with BioNumerics software (Applied Maths, Kortrijk, Belgium). The reference standard S. aureus NCTC 8325, which was included in the first, seventh, fourteenth, twentieth, and last lanes of each gel, was normalized to the global-standard S. aureus NCTC 8325. Percent similarities were identified on a dendrogram derived from the unweighted pair group method using arithmetic averages and based on Dice coefficients. Band position tolerance and optimization were set at 1.25 and 0.5%, respectively. A similarity coefficient of 80% was selected to define the pulsed-field type (PFT) clusters after reviewing the epidemiologic data associated with each of the clusters of isolates.
spa typing.
The typing of the polymorphic region of the protein A gene was performed according to previously described procedures (58).
SCCmec typing.
SCCmec typing was performed as described by Okuma et al. (45).
MLST.
MLST was performed on selected isolates as described by Enright et al. (18).
Nomenclature of ORSA clones.
The nomenclature chosen is similar to that described by Enright et al. (20), Feil et al. (21), and Hiramatsu et al. (29), except that the genetic backgrounds of the strains in this study were determined by SmaI PFGE and designated as PFTs. The PFTs were further described as either ORSA or oxacillin-susceptible S. aureus (OSSA), and ORSA PFTs were subdivided into SCCmec types I, II, III, and IV.
RESULTS
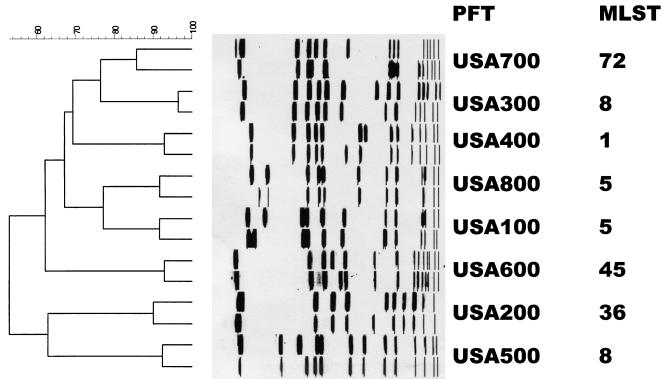
The SmaI macrorestriction fragment profiles of 957 S. aureus isolates were determined by PFGE. A dendrogram of percent similarity, calculated with Dice coefficients from the PFGE data using a cutoff of 80%, revealed eight major clusters of isolates, designated as PFTs USA100 through USA800 (Fig. 1). Of the 667 U.S. ORSA isolates, 622 (93%) clustered within these eight PFTs. In addition, 134 of the 235 OSSA isolates (57%) had PFGE patterns that fell within the same eight PFTs. The results of MLST, spa typing, SCCmec typing, antimicrobial resistance profiles, and other relevant properties of ORSA strains are summarized in Table 1. All but four PFTs (USA300 and -500, and -100 and -800) had a unique MLST sequence type and spa type motif. Five of the eight PFTs (USA100, -200, -500, -600, and -800) contained isolates that were predominantly obtained from health care-associated infections, while the isolates from two PFTs (USA300 and -400) were from community infections. USA700 isolates were obtained from patients in both community and health care settings.
FIG. 1.
Dendrogram of PFTs with type strain (most frequent pattern) and a variant strain. Also shown is the corresponding MLST for each PFT (18, 19, 20).
TABLE 1.
Characteristics of ORSA PFTsa
| Name of PFT | Total no. of U.S. ORSA strains in PFT (%) | MLST and allelic profileb | spa typec | Clone named |
|---|---|---|---|---|
| USA100 | 292 (44) | ST 5, 1- 4- 1- 4- 12- 1- 10 | TJMBMDMGMK | USA100 ORSA II |
| USA800 | 45 (6.7) | ST 5, 1- 4- 1- 4- 12- 1- 10 | TJMBMDMGMK | USA800 ORSA IVe |
| TJMBMDMGMK | ST 5 ORSA I | |||
| USA200 | 57 (8.5) | ST 36, 2- 2- 2- 2- 3- 3- 2 | WGKAKAOMQQQ | USA200 ORSA II |
| USA400 | 42 (6.3) | ST 1, 1- 1- 1- 1- 1- 1- 1 | UJJJFE | USA400 ORSA IVe |
| USA300 | 91 (13.6) | ST 8, 3- 3- 1- 1- 4- 4- 3 | YHGF MBQBLO | USA300 ORSA IVe |
| USA500 | 34 (5.1) | ST 8, 3- 3- 1- 1- 4- 4- 3 | YHGC MBQBLO | USA500 ORSA IV,e USA500 ORSA II |
| ST 8, 3- 3- 1- 1- 4- 4- 3 | YHGC MBQBLO | ST8 ORSA I, ST8 ORSA IV, ST8 ORSA III | ||
| ST 250, 3- 3- 1- 1- 4- 4- 16 | YH GF MBQBLO | ST250 ORSA I | ||
| ST 247, 3- 3- 1- 12- 4- 4- 16 | YHFGF MBQBLO | ST247 ORSA I | ||
| USA600 | 23 (3.4) | ST 45, 10- 14- 8- 6- 10- 3- 2 | A2AKEEMBKB | USA600 ORSA II |
| ST 45, 10- 14- 8- 6- 10- 3- 2 | A2AKEEMBKB | USA600 ORSA IVe | ||
| USA700 | 38 (5.7) | ST 72, 1- 4- 1- 8- 4- 4- 3 | UJGFMGGM | USA700 ORSA IVe |
| Unnamed | 14 (2) | Not tested | ZDMDMNKB | |
| Unnamed | 3 | ST 239, 2- 3- 1- 1- 4- 4- 3 | WGKAOMQ | ST239 ORSA III |
| ST 240, 2- 3- 1- 1- 2- 4- 3 | WGKAOMQ | ST240 ORSA III | ||
| Unnamed | 0 | ST 22, 7- 6- 1- 5- 8- 8- 6 | TJEJN12MN12MOM | ST 22 ORSA IV |
| Published name (references) | Antimicrobial resistance patternf | Published isolates (reference[s]) | Comments |
|---|---|---|---|
| New York/Japan (1, 51) | β-Lactams, ery, cli, lvx | 7 U.S. VISAs (22, 60, 65), 2 U.S. VRSAs (8, 9, 11), Japan VISA Mu50 (28), Korea VISA (33), CMRSA-2,g BK2464 (1, 13, 51), PA237 (52), JP48 (1) | Predominate U.S health care-associated PFT; endemic in many U.S. hospitals |
| Pediatric (24, 55) | β-Lactams | HDE1 (55), HDE288 (13, 55), COB94 (24) | |
| β-Lactams, ery, cli, gen | ERMSA3h | ||
| β-Lactams, ery, cli, lvx, gen | EMRSA16h | Second most common U.S. health care-associated PFT | |
| β-Lactams, ery | MW2 (6, 42) | Community onset; isolates from (i) Native Americans (25), (ii) children without risk factors (6, 27, 42), (iii) early Australian community onset (39, 44), and (iv) San Francisco urban poor (12) | |
| β-Lactams, ery | Community onset; isolates from (i) correctional facilities in Miss. (7), Ga., Tenn., Tex., and Calif. (10) and (ii) athletic teams in Pa. and Calif. (10) | ||
| β-Lactams, ery, cli, tet, lvx, gen, sxt | U.S. VISA (Ohio) | ||
| Archaic/Iberian (46, 47) | β-Lactams, ery, tet | EMRSA2, EMRSA6, EMRSA7, EMRSA12, EMRSA13, EMRSA14h | |
| Archaic/Iberian (46, 47, 57) | β-Lactams, ery, tet | EMRSA 8, PER34 (57), NCTC10442 (31) | SCCmec I identified (31) |
| Archaic/Iberian (46, 47, 57) | β-Lactams, ery, cli, tet, lvx, gen, sxt | EMRSA5,h E2125 (13), HPV 107 (13, 57) France VISA (50), Scotland 463 | |
| β-Lactams, ery, cli, lvx | CMRSA-1g | ||
| β-Lactams, ery | PLN49 (2) | Tenn. food outbreak (34) | |
| β-Lactams | Second genotype at Miss. correction facility outbreak (7); hospital-associated mec+, susceptible phenotypei | ||
| β-Lactams, ery | Alaska surveillance; Vt. wrestling team outbreak (37) | ||
| Brazilian/Hungarian (2, 46, 63) | β-Lactams, ery, cli, tet, chl, lvx, gen, sxt | EMRSA1, EMRSA11, EMRSA4,h HU25 (2), HSJ216, HUSA304 (13), 85/2082 (31), CMRSA-3,g Scotland 461 | SCCmec III identified (31) |
| Brazilian/Hungarian (2, 46, 63) | β-Lactams, ery, cli, tet, chl, lvx, gen, sxt | EMRSA9h | |
| β-Lactams | EMRSA15h (54) | Endemic to United Kingdom (54) |
PFT is based on SmaI PFGE with ≥80% similarity identified on an UPGMA-derived dendrogram using Dice coefficients.
Allelic profile is based on the DNA sequences of seven housekeeping genes: arcC, aroE, glpF, gmk, pta, tpi, and yqiL.
DNA sequence of polymorphic region of protein A gene. The spa type motif is shown in boldface.
The clone name consists of the following three parts: PFT or MLST sequence type, the oxacillin phenotype (all are ORSA in this table), and the SCCmec type (when available).
Isolates in this PFT subgroup were susceptible to spectinomycin.
β-Lactams (oxacillin and penicillin); chl, chloramphenicol; cli, clindamycin; ery, erythromycin; gen, gentamicin; lvx, levofloxacin; tet, tetracycline; sxt, trimethoprim-sulfamethoxazole.
CMRSA strains are epidemic type strains from Canada (59). Four strains are included in this study.
EMRSA strains are epidemic type strains from the United Kingdom (16, 54). Fifteen strains are included in this study.
J. Block, M. F. Orlando, L. K. McDougal, L. Jevitt, W. M. Dunne, S. Fitzsimmons, and J. Gerst, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. 2003, abstr. C-088.
Of isolates from health care-associated infections, USA100 was the largest and most diverse of the PFTs, containing 292 ORSA isolates from throughout the United States. For the seven housekeeping genes, these isolates shared a common MLST allelic profile (1- 4- 1- 4- 12- 1- 10), which is designated as ST 5, and a common spa motif (MDMGMK). USA100 isolates were usually spectinomycin resistant (consistent with SCCmec II) and multiresistant to commonly used therapeutic agents. This group included seven of eight U.S. VISA isolates and VISA isolates from Japan (Mu50) and Korea. USA100 also included the two U.S. VRSA isolates from Michigan and Pennsylvania. All isolates were resistant to erythromycin: 72% were constitutively clindamycin resistant, and 28% showed inducible clindamycin resistance. S. aureus isolates BK2464, PA237, and JA48, from the New York/Japan clone (1, 51, 52), belonged to this PFT.
USA800 isolates shared the same MLST sequence type and spa motif as the USA100 isolates. These isolates were primarily from community surveillance studies, were spectinomycin susceptible (suggesting the lack of Tn554 [consistent with SCCmec type I or IV]) and were generally resistant only to β-lactam drugs. Twenty-three percent were erythromycin resistant. When we tested the isolates, SCCmec typing showed that most of them were SCCmec type IV. S. aureus isolates HDE1, HDE288, and COB94 from the Pediatric clone (24, 55) belonged to this group. Other isolates, such as EMRSA3, which carried SCCmec type I and fell just outside the USA100 and -800 clusters, shared the same MLST and spa motif.
USA200, the second most common health care-associated PFT among U.S. isolates, contained ORSA isolates that were spectinomycin resistant (consistent with SCCmec II) and were multiresistant to therapeutic agents. All ORSA isolates were erythromycin resistant, with 98% showing constitutive resistance to clindamycin. The isolates had the same MLST profile, i.e., ST 36 (2- 2- 2- 2- 3- 3- 2), and spa type motif (WGKAKAOMQQQ) as isolates from the EMRSA16 epidemic clone.
Isolates from community onset infections belonging to PFT USA400 had the MLST ST 1 profile (1- 1- 1- 1- 1- 1- 1) and spa type motif (UJJJFE). These ORSA isolates were spectinomycin susceptible, carried SCCmec IV, and were not multiresistant. S. aureus MW2, an isolate from a rapid fatal infection in a child from Minnesota (6), was included in this group.
Although representative isolates from USA300 and USA500 had the same MLST allelic profile (ST 8, 3- 3- 1- 1- 4- 4- 3) and spa type motif (MBQBLO), these isolates clustered into separate, but contiguous, groups by SmaI PFGE. USA300 isolates carried SCCmec IV, were resistant to β-lactam drugs, were frequently resistant to erythromycin, and were predominantly from community onset skin infections. Eighty-five percent of the erythromycin-resistant isolates were susceptible to clindamycin and were not inducible with erythromycin (probably due to msrA). On the other hand, USA500 isolates were generally from health care-related infections. The majority of USA500 isolates were spectinomycin susceptible, indicating the absence of Tn554 (consistent with SCCmec type I or IV). Most of the isolates were resistant to clindamycin, erythromycin, gentamicin, levofloxacin, tetracycline, trimethoprim-sulfamethoxazole, and the β-lactams. The remaining U.S. VISA isolate and the three isolates for which the vancomycin MICs were 4 μg/ml had PFGE profiles belonging to this PFT. PFGE patterns of S. aureus isolates PER34, E2125, and HPV107 from the Archaic/Iberian clones (13, 14, 57); EMRSA isolates 2, 5, 6, 10, 12, 13, and 14; VISA isolates from Hong Kong and France; and the Iberian clonal-type isolate from Scotland clustered near the USA500 isolates. EMRSA isolates 2, 6, 12, 13, and 14 shared the same MLST profile (3- 3- 1- 1- 4- 4- 3) as the USA300 and USA500 isolates, which differed at a single locus from ST 250 (3- 3- 1-1- 4-4 -16) of isolate Per34. S. aureus E2125 and HPV107 (Archaic/Iberian clonal-type isolates) and the S. aureus EMRSA5 isolate have the MLST profile ST 247 (3- 3- 1- 12- 4- 4- 16), which differed at one locus from ST250 and two loci from ST 8. All of these isolates shared a similar spa type motif (MBQBLO).
USA600 contained 23 isolates. Representative strains were ST 45 (10- 14- 8- 6- 10- 3- 2) and spa type A2AKEEMBKB. All but four of the ORSA isolates in this PFT were spectinomycin resistant (SCCmec II) and multiresistant; the remaining four were spectinomycin susceptible. Isolates from USA700 shared a unique MLST type, i.e., ST 72 (1- 4- 1- 8- 4- 4- 3), and spa motif (UJGFMGGM) from community surveillance and a hospital-acquired outbreak.
There were two clusters within the remaining 45 U.S. ORSA isolates. The first cluster (14 isolates) included surveillance isolates, mainly from Alaska, and isolates from an outbreak of soft-skin infections among Vermont wrestlers (37). The second cluster included three isolates that clustered with S. aureus isolates HU25, HSJ216, and HUSA304 (Brazilian and Hungarian type strains [13, 63]); EMRSA isolates 1, 4, and 11; and MRSA and VISA isolates from Scotland. These isolates had the same MLST profile (ST 239) and spa type motif (WGKAOMQ). Twenty-eight isolates had either miscellaneous PFGE patterns or patterns that could not be identified within an 80% PFT cutoff.
Of the 235 U.S. OSSA isolates, 134 (57%) clustered within the eight PFTs. All of the PFTs, with the exception of USA500, contained OSSA. Twenty OSSA isolates were USA800, 1 isolate was USA100, 78 isolates were USA200, 9 isolates were USA300, 1 isolate was USA400, 22 isolates were USA600, and 3 isolates were USA700. Twenty-eight OSSA isolates clustered in a unique PFT. There were 10 OSSA clusters with 3 to 13 isolates, and 23 isolates with unique SmaI PFGE profiles.
DISCUSSION
Because of its high discriminatory power, PFGE is a valuable tool for investigating outbreaks of S. aureus infections, particularly in hospital settings (3, 4, 16, 27, 61, 64, 66). PFGE has also been used to discriminate among community- and health care-acquired ORSA strains (42). In previous work, de Lencastre and colleagues used PFGE to delineate lineages of ORSA that circulate in Europe, South America, and the United States, and they have given the lineages names, such as the Archaic clone, the Iberian clone, the Pediatric clone, and the New York/Tokyo clone (13). Similarly, Simor et al. have used PFGE to designate four lineages of ORSA in Canada, designated as CMRSA 1 through 4 (59). These and other studies have shown that PFGE can identify stable lineages of ORSA and can be used to track the spread of these lineages from continent to continent over extended periods of time.
In the past, the sharing of PFGE data among laboratories was difficult (15), and typing results for the same strains performed in different laboratories often lacked concordance (66). However, recent advances in gel analysis software programs allow the creation and storage of large databases of normalized fragment patterns in which similarity calculations and cluster analyses can be performed with relative ease. Normalization of the fragment patterns using established standards (such as S. aureus NCTC 8325) and the advent of new database sharing tools both serve to facilitate the exchange of PFGE strain typing data and epidemiologic information among reference laboratories, even in different countries. Thus, the traditional barriers to the sharing of PFGE patterns, even those run using different switching parameters, have to a large extent been overcome with powerful new software programs (62).
However, the appropriateness of using PFGE to study the long-term evolution of S. aureus lineages in general, and ORSA isolates in particular, remains a concern (19, 40). While MLST, which examines changes in the sequences of seven neutral loci on the S. aureus genome, may be more amenable to long-term population studies of ORSA, the higher discriminatory ability of PFGE profiles over that of MLST for S. aureus is advantageous. This holds true for epidemiologic studies of particular clusters of isolates, such as those isolates causing community onset disease (12, 27, 44) and for assessing the effectiveness of targeted-prevention programs. For example, USA300 and -500 isolates were assigned different PFTs because they separated into two distinct pulsed-field clusters and had different antimicrobial susceptibility patterns and epidemiology, even though they both shared ST 8 (Table 1). USA300 isolates are predominantly from community onset infections and are resistant only to β-lactam drugs and macrolides, while USA500 isolates differ from representatives of the Archaic and Iberian clones by only one or two locus variants (14, 20), tend to be from health care-associated infections, and are multidrug resistant. USA300 isolates have been identified in multiple community onset ORSA outbreaks associated with soft-skin infections in correctional facilities (7, 10), athletic teams (10), and nurseries (10). Isolates from prisons in Mississippi, Texas, Tennessee, and Georgia all shared this PFT, as did isolates from an outbreak of ORSA infections in football players. Thus, the epidemiology of USA300 isolates is quite distinct from the health care-associated USA500 isolates, even though they are indistinguishable by MLST typing. Similarly, although USA100 and -800 isolates shared a common MLST sequence type (ST 5) and spa motif (MDMGMK), the isolates carried different SCCmec structures and had different susceptibility profiles. USA100 isolates cluster with representatives of the multiresistant New York/Japan clone containing SSCmec II, while isolates from USA800 cluster with representatives of the Pediatric clone containing SSCmec IV. These critical epidemiologic differences are obscured by MLST. Thus, the PFGE typing system described here will likely be more useful than MLST, particularly for public health agencies, for targeted epidemiologic studies aimed at understanding the epidemiology of specific clonal groups, and for developing interventional studies to halt the transmission of the disease. Given the large data sets of PFGE profiles already established in the United States and around the world (15, 16, 41), the development of a surveillance system that can integrate these databases and make the information available to other reference centers is critical.
The other lineage associated with community onset disease was USA400. This PFT included ORSA isolates that caused severe, and in some cases fatal, disease in children from Minnesota and North Dakota, and was associated with skin disease in Native Americans from eastern Washington (state) (6, 25, 27, 42). Also in this group were isolates from Australian Aborigines (39, 44). Although not present in the U.S. database, isolates of this same MLST type (ST 1) were obtained in a community study in England from healthy carriers of S. aureus and those with health care-associated infections (21). This may be another instance in which MLST obscures epidemiologically relevant information regarding clusters of strains with different virulence characteristics. Many S. aureus strains responsible for primary skin infections and necrotizing pneumonia harbor the Panton-Valentine leukocidin determinant (17, 23, 36), and preliminary studies suggest that many USA300 and -400 isolates harbor this virulence determinant (CDC unpublished observations) in addition to the more recent SSCmec IV.
Among the five PFTs associated predominantly with health care-related infections (i.e., USA100, -200, -500, -600, and -800), USA100 was by far the most common (44% of all U.S. ORSA isolates examined). U.S. isolates of this PFT carried SCCmec II. This PFT was previously designated as the New York/Japan clone (1). Interestingly, several other previously described global lineages that are common as causes of health care-associated infections (e.g., the Brazilian clone and EMRSA15) were not found among U.S. isolates.
Most of the ORSA PFTs also contained OSSA isolates, suggesting that each of these lineages had independently acquired mecA. Five types of SCCmec have been identified. Type I (34 kb) was detected in the first ORSA strain isolated in 1961 in the United Kingdom (strain NCTC 10442) (31); type II (52 kb) was identified in an ORSA strain isolated in 1982 in Japan (strain N315) (30); type III (66 kb) was identified in an MRSA strain isolated in 1985 in New Zealand (strain 82/2082) (31); type IV (20 to 24 kb) was identified in two community-acquired ORSA strains (38) and in isolates of the Pediatric clone from Poland and Portugal (45); and a new type 2 was identified in three community onset ORSA strains from Adelaide, Australia (45). SCCmec II and III contain transposon Tn554, which encodes erythromycin and spectinomycin resistance (31). SCCmec I and IV carry no other resistance genes (31, 38).
In conclusion, results of SmaI PFGE typing, corroborated with those of limited MLST and spa typing, allowed us to identify genetic backgrounds and delineate the major U.S. ORSA lineages within a national database generated from PFGE fingerprinting and epidemiologic data from the CDC and U.S. state health laboratory investigations. PFGE has proven to be more discriminating than MLST for monitoring the spread of ORSA isolates in the United States, although periodic typing of selected isolates using MLST will be critical for assessing major changes in the lineages over time.
Acknowledgments
We thank Mark Enright for providing the MLST data on several of the isolates cited in this paper; we also thank Molly Kellum, Bette Jensen, Glennis Westbrock, and Loretta Carson for pulsed-field electrophoresis typing, Stephen Weber for assistance with SCCmec analysis, Matthew Kuehnert and Jeff Hageman for providing isolates, and J. Kamile Rasheed and Leslie McGee for comments on the manuscript.
The use of trade names is for identification purposes only and does not constitute endorsement by the Public Health Service or the U.S. Department of Health and Human Services.
REFERENCES
- 1.Aires de Sousa, M., H. de Lencastre, I. Santos Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6:253-258. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., M. Miragaia, I. Santos Sanches, S. Ávila, I. Adamson, S. T. Casagrande, M. C. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aucken, H. M., M. Ganner, S. Murchan, B. D. Cookson, and A. P. Johnson. 2002. A new UK strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-17) resistant to multiple antibiotics. J. Antimicrob. Chemother. 50:171-175. [DOI] [PubMed] [Google Scholar]
- 4.Bannerman, T. L., G. A. Hancook, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1997. Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morb. Mortal. Wkly. Rep. 46:765-766. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2001. Methicillin-resistant Staphylococcus aureus skin or soft tissue infections in a state prison—Mississippi, 2000. Morb. Mortal. Wkly. Rep. 50:919-922. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2002. Public health dispatch: vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2003. Public health dispatch: outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections—Los Angeles County, California, 2002-2003. Morb. Mortal. Wkly. Rep. 52:88. [PubMed] [Google Scholar]
- 11.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin, and the Vancomycin-ResistantStaphylococcus aureus Investigative Team. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 12.Charlebois, E. D., D. R. Bangsberg, N. J. Moss, M. R. Moore, A. R. Moss, H. F. Chambers, and F. Perdreau-Remington. 2002. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin. Infect. Dis. 34:425-433. [DOI] [PubMed] [Google Scholar]
- 13.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, and the Multilaboratory Project Collaborators: I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leão, I. Santos Sanches, J. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 14.Crisostoma, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 9:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A. Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neil, M. J. Struelens, and The European Study Group on Epidemiological Markers of the ESCMID. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deplano, A., W. Witte, W. J. van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 17.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 18.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 19.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enright, M. C., D. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, F. C. Tenover, and the Vancomycin-Intermediate Staphylococcus aureus Epidemiology Study Group. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2000. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 23.Gillet, Y., B. Issartel, P. Vanhems, J. Fournet, G. Lina, M. Bes, F. Vandenesch, Y. Piémont, N. Brousse, D. Floret, and J. Etienne. 2002. Association between Staphylococcus aureus strains carrying the gene for the Panton-Valentine leukocidin and highly-lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753-759. [DOI] [PubMed] [Google Scholar]
- 24.Gomes, A. R., I. Santos Sanches, M. Aires de Sousa, E. Castaneda, and H. De Lencastre. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Colombian hospitals: dominance of a single unique multidrug-resistant clone. Microb. Drug Resist. 7:23-32. [DOI] [PubMed] [Google Scholar]
- 25.Groom, A. V., D. H. Wolsey, T. S. Naimi, K. Smith, S. Johnson, D. Boxrud, K. A. Moore, and J. E. Cheek. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 286:1201-1205. [DOI] [PubMed] [Google Scholar]
- 26.Haley, R. W., N. B. Cushion, F. C. Tenover, T. L. Bannerman, D. Dryer, J. Ross, P. J. Sanchez, and J. D. Siegel. 1995. Eradication of endemic methicillin-resistant Staphylococcus aureus infections from a neonatal intensive care unit. J. Infect. Dis. 171:614-624. [DOI] [PubMed] [Google Scholar]
- 27.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 28.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-146. [DOI] [PubMed] [Google Scholar]
- 29.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 30.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karchmer, A. W. 2000. Nosocomial bloodstream infections: organisms, risk factors, and implications. Clin. Infect. Dis. 31(Suppl. 4): S139-S143. [DOI] [PubMed] [Google Scholar]
- 33.Kim, M., C. H. Pai, J. H. Woo, J. S. Ryu, and K. Hiramatsu. 2000. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38:3879-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, T. F., M. E. Kellum, S. S. Porter, M. Bell, and W. Schaffner. 2002. An outbreak of community-acquired foodborne illness caused by methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 8:82-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 36.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 37.Lindenmayer, J. M., S. Schoenfeld, R. O'Grady, and J. K. Carney. 1998. Methicillin-resistant Staphylococcus aureus in a high school wrestling team and the surrounding community. Arch. Intern. Med. 158:895-899. [DOI] [PubMed] [Google Scholar]
- 38.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chontrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maguire, G. P., A. D. Arthur, P. J. Boustead, B. Dwyer, and B. J. Currie. 1996. Emerging epidemic of community-acquired methicillin-resistant Staphylococcus aureus infection in the Northern Territory. Med. J. Aust. 164:721-723. [DOI] [PubMed] [Google Scholar]
- 40.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. E. Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 43.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., vol. 20, no. 2. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 44.O'Brien, F. G., J. W. Pearman, M. Gracey, T. V. Riley, and W. B. Grubb. 1999. Community strain of methicillin-resistant Staphylococcus aureus involved in a hospital outbreak. J. Clin. Microbiol. 37:2858-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 47.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 48.Panlilio, A. L., D. H. Culver, R. P. Gaynes, S. Banerjee, T. S. Henderson, J. S. Tolson, and W. J. Martone. 1992. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975-1991. Infect. Control Hosp. Epidemiol. 13:582-586. [DOI] [PubMed] [Google Scholar]
- 49.Phillips, S., and R. P. Novick. 1979. Tn554—a site-specific repressor-controlled transposon in Staphylococcus aureus. Nature 278:476-478. [DOI] [PubMed] [Google Scholar]
- 50.Ploy, M. C., C. Grélaud, C. Martin, L. de Lumley, and F. Denis. 1998. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet 351:1212. [DOI] [PubMed] [Google Scholar]
- 51.Roberts, R. B., H. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, and A. Tomasz, and the MRSA Collaborative Study Group.1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 52.Roberts, R. B., M. Chung, H. de Lencastre, J. Hargrave, A. Tomasz, D. P. Nicolau, J. F. John, Jr., O. Korzeniowski, and the Tri-State MRSA Collaborative Study Group. 2000. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey, and Pennsylvania. Microb. Drug Resist. 6:245-251. [DOI] [PubMed] [Google Scholar]
- 53.Rosdahl, V. T., and A. M. Knudsen. 1991. The decline of methicillin resistance among Danish Staphylococcus aureus strains. Infect. Control Hosp. Epidemiol. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 54.Richardson, J. F., and S. Reith. 1993. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 25:45-52. [DOI] [PubMed] [Google Scholar]
- 55.Sa-Leao, R., I. Santos-Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salgado, C. D., B. M. Farr, and D. P. Calfee. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin. Infect. Dis. 36:131-139. [DOI] [PubMed] [Google Scholar]
- 57.Santos-Sanches, I., M. Ramirez, H. Troni, M. Abecassis, M. Padua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simor, A. E., M. Ofner-Agostini, E. Bryce, A. McGeer, S. Paton, and M. R. Mulvey. 2002. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: results of 5 years of national surveillance, 1995-1999. J. Infect. Dis. 186:652-660. [DOI] [PubMed] [Google Scholar]
- 60.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, and W. R. Jarvis. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 61.Struelens, M. J., R. Bax, A. Deplano, W. G. Quint, and A. Van Belkum. 1993. Concordant clonal delineation of methicillin-resistant Staphylococcus aureus by macrorestriction analysis and polymerase chain reaction genome fingerprinting. J. Clin. Microbiol. 31:1064-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and the CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teixeira, L., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. S. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hebert, B. Hill, and R. Hollis. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C. M. O'Hara, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. ElSolh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voss, A., D. Milatovic, C. Wallrauch-Schwarz, V. T. Rosdahl, and I. Braveny. 1994. Methicillin-resistant Staphylococcus aureus in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 13:50-55. [DOI] [PubMed] [Google Scholar]
- 68.Wei, M. Q., and W. B. Grubb. 1992. Typing of Australian methicillin-resistant Staphylococcus aureus strains by pulsed-field gel electrophoresis. J. Med. Microbiol. 37:187-191. [DOI] [PubMed] [Google Scholar]
- 69.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]