Abstract
The purpose of our study was the molecular characterization of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated in 21 hospitals in the Czech Republic in the period 2000-2002 and comparison with previous results from 1996-1997. Strains were analyzed by pulsed-field gel electrophoresis (PFGE) of SmaI digests and ribotyping of HindIII digests hybridized with a 16S-23S DNA probe. The prevalence of the most clinically important macrolide (ermA, ermB, ermC, and msrA) and aminoglycoside (aph3′, ant4′, and aac6′-aph2") resistance genes was evaluated as well. Selected isolates representative of each clonal type were analyzed by multilocus sequence typing and by a multiplex PCR method capable of identifying the structural type of the staphylococcal cassette chromosome mec (SCCmec) carried by the bacteria. Our results document the displacement of the Brazilian clone (ST239, SCCmec type IIIA, PFGE type B, ribotype H1) by a new clone that we named “Czech clone” (ST239, SCCmec type IIIA, PFGE type F, ribotype H6) and the maintenance of the Iberian clone (ST247, SCCmec type IA, PFGE type A, ribotype H2) exclusively in one hospital in the Czech Republic. In addition, we found a correlation between the distribution of aminoglycoside resistance genes and MRSA clonal types.
In the first unique study dealing with the characterization of methicillin-resistant Staphylococcus aureus (MRSA) in the Czech Republic by molecular typing, the prevalence of two multiresistant clones of MRSA that were particularly widely disseminated in the country was documented for the years 1996-1997 (19). One of these clones was the pandemic Iberian MRSA (10, 18, 29, 31), which represented 12% of the Czech MRSA isolates. The other distinct multiresistant clone, the Brazilian MRSA, widely spread in South America (3, 8, 34) and Portugal (4, 25), represented 80% of the Czech isolates of 1996-1997.
MRSA strains have acquired multiple resistance to a wide range of antibiotics, including aminoglycosides and macrolides (11). Three genes (aph3′, ant4′, and aac6′-aph2"), encoding three types of aminoglycoside-modifying enzymes, are of particular significance because they modify aminoglycosides of therapeutic importance, including kanamycin, tobramycin, and gentamicin, respectively (14). Macrolide resistance can be triggered by several mechanisms (16), the predominant one being target modification mediated by one or more erm genes encoding a 23S rRNA methylase, rendering the strain resistant to most macrolides, lincosamides, and streptogramin B compounds (MLSB) (33). Resistance to macrolide-streptogramin (MS) antibiotics resulting from the presence of macrolide efflux pumps in staphylococci (encoded by msrA) has also been documented (30).
In this paper, we focus on the characterization by different typing techniques of the following two collections of MRSA isolates from the Czech Republic: (i) 45 isolates collected during 2000-2001 exclusively from blood samples at 20 hospitals and (ii) 55 isolates collected during 2001-2002 from different clinical sources at three hospitals. In an attempt to determine the status of macrolide and aminoglycoside resistance among Czech MRSA isolates, we investigated the prevalence of the most clinically important macrolide (ermA, ermB, ermC, and msrA) and aminoglycoside (aph3′, ant4′, and aac6′-aph2") resistance genes.
MATERIALS AND METHODS
Hospitals.
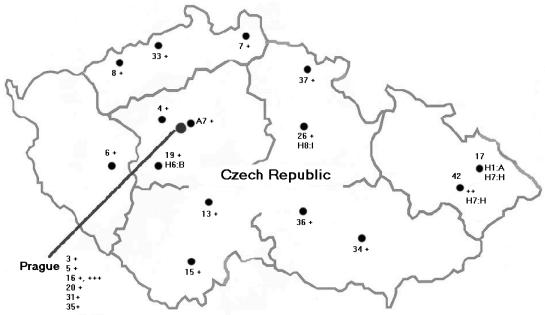
Nine large (>1,000 beds), 6 medium (between 500 and 1,000 beds), and 6 small (<500 beds) hospitals in the Czech Republic, including 7 teaching hospitals and 14 general hospitals, participated in this study (Table 1). A map of the Czech Republic with the locations of the 21 participating hospitals is shown in Fig. 1.
TABLE 1.
Hospital data and MRSA isolates from the Czech Republic collected during 2000-2002
| Hospital codea | City | Hospital type | No. of beds | % MRSA isolates (July 2000-March 2002) | No. of MRSA in this study |
|---|---|---|---|---|---|
| Collection 1 (invasive MRSA isolates) | |||||
| 3 | Prague | University-teaching | ≥1,000 | 16.7 | 9 |
| 4 | Kladno | General | 500-1,000 | 3.7 | 2 |
| 5 | Prague | University-teaching | ≥1,000 | 4.2 | 1 |
| 6 | Plzeň | University-teaching | ≥1,000 | 6.1 | 4 |
| 7 | Liberec | General | 500-1,000 | 4.3 | 1 |
| A7 | Brandys | General | <500 | Not known | 1 |
| 8 | Most | General | ≥1,000 | 6.3 | 1 |
| 13 | Tábor | General | <500 | 5.0 | 1 |
| 15 | České Budějovice | General | ≥1,000 | 2.0 | 2 |
| 17 | Ostrava | University-teaching | ≥1,000 | 9.2 | 1 |
| 19 | Příbram | General | 500-1,000 | 6.7 | 1 |
| 20 | Prague | University-teaching | <500 | 8.5 | 7 |
| 26 | Pardubice | General | 500-1,000 | 2.8 | 1 |
| 31 | Prague | General | 500-1,000 | 7.4 | 6 |
| 33 | Ústí nad Labem | General | ≥1,000 | 18.5 | 2 |
| 34 | Brno | University-teaching | ≥1,000 | 7.4 | 1 |
| 35 | Prague | General | <500 | 18.0 | 1 |
| 36 | Havličkuv Brod | General | <500 | 5.0 | 1 |
| 37 | Trutnov | General | <500 | 7.7 | 1 |
| 42 | Nový Jičín | General | <500 | 16.0 | 1 |
| Collection 2 (noninvasive MRSA isolates) | |||||
| 16 | Prague | University-teaching | ≥1,000 | 6.5 | 22 |
| 20 | Prague | University-teaching | <500 | 8.5 | 10 |
| 42 | Nový Jičín | General | <500 | 16.0 | 23 |
| Total | 100 |
Underlined hospital codes indicate hospitals that are included in both collections. Collection 1, 45 invasive isolates (from blood samples) collected at 20 hospitals; collection 2, 55 noninvasive isolates collected at three hospitals.
FIG. 1.
Map of the Czech Republic with locations of the 21 participating hospitals. Hospitals are designated with code numbers. The appearance of a particular clonal type is indicated by a symbol (+, Czech clone; ++, Iberian clone; +++, Brazilian clone) or by its molecular characteristics (ribotype:PFGE type).
Bacterial isolates.
For this study, we analyzed 100 single-patient MRSA isolates from the following two different collections: (i) 45 invasive isolates from blood samples obtained from 20 hospitals in the period from April 2000 through November 2001 (collection 1) and (ii) 55 noninvasive isolates recovered from April 2001 through March 2002 from different clinical sources at three hospitals (two located in Prague and one located in the eastern part of the country) (collection 2). Collection 2 isolates were recovered from wounds (42%), sputum (20%), catheters (11%), and other diverse clinical sites (27%). Of the 100 isolates, 14 (31%), 17 (38%), and 13 (29%) from collection 1 and 16 (29%), 12 (22%), and 5 (9%) from collection 2 were recovered from patients interned in intensive care units, surgical wards, and internal medicine wards, respectively.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed according to the National Committee for Clinical Laboratory Standards guidelines (21) for oxacillin, ciprofloxacin, clindamycin, erythromycin, fusidic acid, gentamicin, tobramycin, kanamycin, streptomycin, mupirocin, tetracycline, trimethoprim-sulfamethoxazole (SXT), chloramphenicol, rifampin, vancomycin, and teicoplanin. Susceptibility to vancomycin and teicoplanin was also determined according to the EARSS protocol (22). Production of β-lactamase was tested by nitrocefin (Oxoid, Hampshire, United Kingdom).
Identification of MRSA isolates.
S. aureus isolates were confirmed to be methicillin resistant by the MRSA-Screen slide latex agglutination kit for the rapid detection of PBP2a, according to the manufacturer's recommendations (Denka Seiken Co., Ltd., Tokyo, Japan).
Detection of enterotoxins A, B, C, D, and E, TSST-1, and exfoliative toxins A and B.
Enterotoxins (A, B, C, D, and E), toxic shock syndrome toxin 1 (TSST-1), and exfoliative toxins A and B were detected by the SET-RPLA, TST-RPLA, and EXT-RPLA kits, respectively (Denka Seiken Co., Ltd.), following the manufacturer's recommendations.
Preparation of whole-cell DNA for PCR.
One DNA disk prepared for pulsed-field gel electrophoresis (PFGE) as described by Chung et al. (7) was melted at 70°C. A volume of 180 μl of water was added, and the mixture was incubated at 95°C for 15 min and chilled on ice for 5 min. Two microliters of DNA was used as template.
PCR amplification of the mecA, aminoglycoside, and macrolide resistance genes.
Amplification of the mecA (19), aph3′, ant4′, aac6′-aph2" (36), ermA, ermB, ermC, and msrA (17) genes was performed as previously described.
Ribotyping, PFGE, MLST, and SCCmec typing.
Ribotyping of HindIII digests (19), PFGE of SmaI digests of chromosomal DNAs (7), and multilocus sequence typing (MLST) (12) were performed as previously described. The staphylococcal cassette chromosome mec (SCCmec) types were determined by a multiplex PCR strategy (24).
RESULTS
Antimicrobial susceptibility.
All 100 strains were resistant to oxacillin, which was confirmed by the presence of the mecA gene and production of PBP2a. The majority of the isolates tested were resistant to ciprofloxacin (99%), erythromycin (98%), tetracycline (97%), streptomycin (95%), rifampin (94%), gentamicin, kanamycin, tobramycin (93%), and clindamycin (86%), and a few of them showed resistance to SXT (10%), chloramphenicol (9%), mupirocin (4%), and fusidic acid (1%). All isolates were susceptible to vancomycin and teicoplanin (Table 2). Constitutive production of β-lactamase was found in 95% of the isolates.
TABLE 2.
Phenotypic and genotypic properties of 100 MRSA isolates from the Czech Republic
| Ribotype | PFGE type | No. of isolates | Clonal type | Antibiotic resistanceb (>50% of the isolates) | Presence of aminoglycosidesc
|
Presence of macrolidesc
|
MLST typed | ST | SCCmec type | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aac6′-aph2′′ | aph3′ | ant4′ | ermA | ermB | ermC | msrA | ||||||||
| Collection 1a | ||||||||||||||
| H1 | A1 | 1 | H1:A | O, E, G, K, S, TO, CLI, C, T, R | + | − | + | + | − | + | − | 3-3-1-12-4-4-16 | 247 | IA |
| H2 | A2 | 1 | H2:A | O, E, G, K, S, TO, | + | − | + | + | − | + | − | 3-3-1-12-4-4-16 | 247 | |
| Iberian | CLI, C, T, R | |||||||||||||
| H6 | F1 | 36 | H6:F Czech | O, E, G, K, S, TO, CLI, C, T, R | + | − (7 +) | −(1 +) | + (1 −) | − | + (3 −) | − | 2-3-1-1-4-4-3 | 239 | IIIA |
| F3 | 1 | O, E, G, K, S, TO, CLI, C, T, R | + | − | + | + | − | + | − | |||||
| F5 | 1 | O, E, G, K, S, TO, CLI, C, T, R | + | − | + | + | − | + | − | |||||
| F6 | 2 | O, E, G, K, S, TO, CLI, C, T, R | + | − | + | + | − | + | − | |||||
| H6 | B1 | 1 | H6:B | O, E, G, K, S, TO, CLI, C, T, R | + | − | − | + | − | − | − | 2-3-1-1-4-4-3 | 239 | IIIA |
| H7 | H2 | 1 | H7:H | O, E, CLI, C | + | − | − | − | − | + | − | |||
| H8 | I | 1 | H8:I | O, E, G, K, S, TO, CLI, C | + | + | − | + | − | + | − | 1-4-1-4-46-24-29 | 111 | I |
| Collection 2 | ||||||||||||||
| Hospital 16 | ||||||||||||||
| H1 | B1 | 3 | H1:B Brazilian | O, E, G, K, S, TO, CLI, SXT, C, T, R | + | + | − | + | − | + | − | 2-3-1-1-4-4-3 | 239 | IIIA |
| B5 | 1 | O, E, G, K, S, TO, SXT, C, T, F, CHL, R | + | + | − | + | − | − | − | |||||
| H6 | F1 | 14 | H6:F Czech | O, E, G, K, S, TO, CLI, C, T, R | + (1 −) | − | − (+ 1) | + | − | + (2 −) | − | 2-3-1-1-4-4-3 | 239 | IIIA |
| F2 | 2 | O, E, G, K, S, TO, CLI, C, T, Re | ± | − | − | + | − | + | − | |||||
| F3 | 1 | O, E, G, K, S, TO, CLI, C, T, R | + | − | − | + | − | + | − | |||||
| F5 | 1 | O, E, G, K, S, TO, CLI, C, T, R | + | − | − | + | − | + | − | |||||
| Hospital 20 | ||||||||||||||
| H6 | F1 | 10 | H6:F Czech | O, E, G, K, S, TO, CLI, C, T, R | + | − | −(4 +) | + | − | + (4 −) | − | 2-3-1-1-4-4-3 | 239 | IIIA |
| Hospital 42 | ||||||||||||||
| H2 | A1 | 1 | H2:A Iberian | O, E, G, K, S, TO, CLI, C, T, R | − | − | − | − | − | + | + (1 −) | 3-3-1-12-4-4-16 | 247 | IA |
| A2 | 19 | O, E, G, K, S, TO, CLI, C, T, R | + (1 −) | − | + | + (1 −) | − | − | − | |||||
| H7 | H1 | 3 | H7:H | O, E, C | − | − | − | − | − | + | + (1 −) | 7-6-1-5-8-8-6 | 22 | IV |
Collection 1, isolates collected exclusively from blood samples from 20 hospitals; collection 2, isolates collected from three different hospitals from a variety of clinical sources.
O, oxacillin; C, ciprofloxacin; CLI, clindamycin; E, erythromycin; T, tetracycline; G, gentamicin; K, kanamycin; S, streptomycin; TO, tobramycin; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol; R, rifampin; F, fusidic acid.
+, positive, −, negative; exceptions are indicated in parentheses.
The strains characterized by MLST were the following: CCM 7109 (ST22, SCCmec type IV, PFGE SmaI type H, ribotype HindIII H7), CCM 7110 (ST111, SCCmec type, PFGE SmaI type I, ribotype HindIII H8), CCM 7111 (ST239, SCCmec type IIIA, PFGE SmaI type B, ribotype HindIII H1), CCM 7112 (ST239, SCCmec type IIIA, PFGE SmaI type F, ribotype HindIII H6), CCM 7113 (ST239, SCCmec type IIIA, PFGE SmaI type B, ribotype HindIII H6), CCM 7114 (ST247, SCCmec type IA, PFGE SmaI type A, ribotype HindIII H1), and CCM 7115 (ST247, SCCmec type IA, PFGE SmaI type A, ribotype HindIII H2). CCM refers to the Czech Culture Collection of Microorganisms.
One isolate was susceptible to gentamicin, kanamycin, and tobramycin.
Production of enterotoxins, TSST-1, and exfoliative toxins A and B.
Representatives of each clonal type detected were tested for the production of enterotoxins A, B, C, D, and E, TSST-1, and exfoliative toxins A and B. Enterotoxin A was found in 16 isolates. The other toxins were not detected in any isolate.
Prevalence of macrolide resistance genes.
Among the 100 MRSA isolates screened for the presence of MLSB resistance genes, 99% contained one or more of the erm genes, which is consistent with the erythromycin resistance phenotype. The most prevalent erm gene was ermA, which was detected in 94% of the isolates. The msrA gene was detected in only two isolates, in association with ermC (Table 3).
TABLE 3.
Distribution of macrolide and aminoglycoside resistance genes in MRSA isolates from the Czech Republica
| No. of isolates with macrolide profile | Macrolide profile
|
No. of isolates with aminoglycoside profile | Aminoglycoside profile
|
|||||
|---|---|---|---|---|---|---|---|---|
| ermA | ermB | ermC | msrA | aac6′-aph2′′ | aph3′ | ant4′ | ||
| 30 | + | − | − | − | 54 | + | − | − |
| 0 | − | + | − | − | 1 | − | − | + |
| 3 | − | − | + | − | 0 | − | + | − |
| 0 | − | − | − | + | 28 | + | − | + |
| 0 | + | + | − | − | 12 | + | + | − |
| 64 | + | − | + | − | 0 | − | + | + |
| 0 | + | − | − | + | 0 | + | + | + |
| 0 | − | + | + | − | 5 | − | − | − |
| 0 | − | + | − | + | ||||
| 2 | − | − | + | + | ||||
| 1 | − | − | − | − | ||||
The numbers of isolates that carried each antibiotic resistance gene were as follows: macrolide genes, ermA, 94; ermB, 0; ermC, 69; mrsA, 2 (total, 100), aminoglycoside genes, aac6′-aph2′′, 94; aph3′, 12; ant4′, 29 (total, 100).
Prevalence of aminoglycoside resistance genes.
Among the 100 isolates tested, the aac6′-aph2" gene was the most frequently encountered aminoglycoside resistance gene (94%), and 40% of the isolates carried this gene in combination with one of the other aminoglycoside resistance genes. The ant4′ and aph3′ genes were present in 29 and 12% of the isolates, respectively (Table 3). Isolates with none of the three genes (n = 5) were fully susceptible to gentamicin, tobramycin, and kanamycin.
Clonal assignments. (i) Ribotypes.
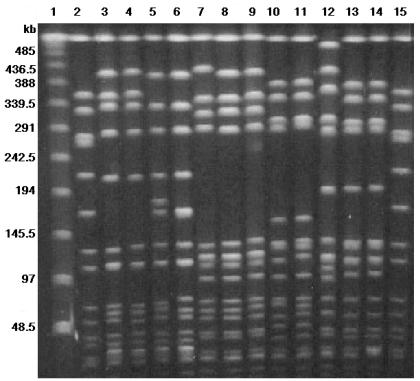
The 100 strains were classified into ribotypes on the basis of fingerprints produced after hybridization of HindIII digests with the 16S-23S probe. All ribotypes found in this study are indicated in Table 2 and shown in Fig. 2. The majority of the isolates from collection 1 (41 of 45 isolates; 91%) were included in ribotype H6, whereas four other ribotypes (H1, H2, H7, and H8) were represented by single isolates. For collection 2, most isolates also belonged to ribotype H6 (28 of 55 isolates; 51%), followed by ribotype H2 (20 of 55 isolates; 36%). Ribotypes H1 and H7 included four and three isolates, respectively. However, the distribution of ribotypes for collection 2 varied from hospital to hospital (Table 2). Ribotype H6 was represented by 100% of the isolates from hospital 20, 82% of the isolates from hospital 16, and none of the isolates from hospital 42. Ribotype H2 was found exclusively in hospital 42 and was represented by 87% of those isolates. Ribotype H1 was only present in hospital 16 and included 18% of those isolates. Ribotype H7 was detected in two hospitals only (hospitals 42 and 17) and was represented by 3 of 24 isolates (13%) and 1 of 2 isolates, respectively.
FIG. 2.
Ribotypes found among 100 MRSA clinical isolates from the Czech Republic. Lane 1, molecular size standard (lambda digest with StyI plus HindIII); lanes 3 to 5 and 7, ribotype H1; lanes 2 and 10, ribotype H2; lanes 6, 8, 9, 11, and 12, ribotype H6; lanes 13, 14, and 16, ribotype H7; and lane 15, ribotype H8.
(ii) PFGE types.
The 100 MRSA isolates were distributed into five PFGE types (Table 2). A major PFGE type, F, appeared among collection 1 isolates (89%) and also among collection 2 isolates from hospitals 16 (82%) and 20 (100%). We provisionally named this clone the Czech clone. PFGE type A, characteristic of the Iberian clone, was represented by two isolates in collection 1 (4%) and was found as a major PFGE type (87%) among collection 2 isolates from hospital 42. PFGE type B, characteristic of the Brazilian clone, was represented by a single isolate in collection 1 and by four isolates from hospital 16 in collection 2. PFGE type H was found in both collection 1 (one isolate) and collection 2 (three isolates from hospital 42). Sporadic PFGE type I was represented by a single isolate in collection 1. Figure 3 shows the major PFGE types found in this study as well as representatives of some international MRSA clones.
FIG. 3.
PFGE of SmaI macrorestriction fragments of MRSA clinical isolates from the Czech Republic and representatives of some international MRSA clones. Lane 1, lambda marker; lanes 2 and 15, NCTC8325; lanes 3 to 6, representatives of the Iberian clone (lane 3, PER34 [10]; lane 4, PL21 [19]; lane 5, 1NJ [this study]; lane 6, 2NJ [this study]); lanes 7 to 9, representatives of the Brazilian clone (lane 7, HU25 [34]; lane 8, KV173 [19]; lane 9, 2A8 [this study]); lanes 10 and 11, representatives of the Hungarian clone (lane 10, HUSA 304 [9]; lane 11, HU101 [23]); lane 12, representative of the new Czech MRSA clone, 2HK (this study); lanes 13 and 14, representatives of the major clone of Taiwan (lane 13, TAW3; lane 14, TAW 10 [1]).
(iii) MLST and SCCmec types.
MLST and SCCmec typing were applied to representatives of each of the clonal types identified in this study by PFGE and ribotyping. ST239 and SCCmec type IIIA are characteristic of both the Brazilian (PFGE type B, ribotype H1) and the Czech (PFGE type F, ribotype H6) clones. A single strain that was ST239 and SCCmec type IIIA also was HindIII ribotype H6, typical of the Czech clone, and PFGE pattern B, typical of the Brazilian clone. A clone isolated in hospital 42 only, distinguished by HindIII ribotype H2 and PFGE type A, belonged to ST247 and SCCmec type IA, which are typical of the Iberian clone.
All four strains of a sporadic clone that were resistant to erythromycin, clindamycin, and ciprofloxacin only belonged to ribotype H, PFGE type H, ST22, and SCCmec type IV.
The single isolate of the clone characterized by ribotype H8 and PFGE type I displayed SCCmec type I and belonged to ST111, which has so far been only detected in one MRSA strain (AB-903627/02) from Norway (http://www.mlst.net).
DISCUSSION
The mean proportion of MRSA blood isolates in the Czech Republic over the years 1999-2001 was reported to be 3 to 10% (22). However, MRSA prevalence varies substantially among Czech hospitals (2 to 18.5%) (Table 1), which may be due to differences in the hospital hygiene guidelines applied and/or antibiotic use. In an attempt to determine the evolution of MRSA clonal types in the Czech Republic from 1996-1997 to 2000-2002, 100 recent isolates were characterized by different molecular typing methods. The most surprising observation of this study was the high frequency of a clonal group defined by ST239, SCCmec type IIIA, PFGE type F, and ribotype H6 which was dominant (89%) among the isolates from blood samples (collection 1) and also among the isolates from two hospitals of collection 2 (82 and 100%) (Table 2). This clonal type was detected in 17 of the 21 hospitals included in this study (Fig. 1).
The Brazilian clone (ST239, SCCmec type IIIA, PFGE type B, ribotype H1), which in 1996-1997 was the major clone spread in two Prague and one Brno hospital (19), was no longer the dominant one in 2000-2002, being represented only by four isolates from the same hospital in collection 2. Interestingly, three of these isolates were collected from outpatients, which indicates that the Brazilian clone may still be present in the community. The Iberian clone (ST247, SCCmec type IA, PFGE type A, ribotype H2), represented by 12% of the isolates in 1996-1997 and detected in two hospitals, one in Prague and the other in Plzeò (19), was found to be the dominant clone in one of the hospitals of collection 2 (hospital 42) and was detected in one isolate of collection 1 collected at the same hospital. This hospital was located 350 km from Prague and 450 km from Plzeò. We can hypothesize that the blood isolates (collection 1), which are often unique isolates of the hospitals included in this study, might reflect the predominant clone of the hospital in which they were collected. What should be pointed out is the switch in the SXT resistance pattern from 90% SXT-resistant strains in 1996-1997 to only 10% in 2000-2002, which is consistent with the displacement of the SXT-resistant (Sxtr) Brazilian clone by the new SXT-susceptible (Sxts) Czech MRSA clone. A similar situation had been observed in a Portuguese hospital in which the Sxts Iberian clone was replaced by the Sxtr Brazilian clone (5). Although susceptible to SXT, the new Czech MRSA clone is multidrug resistant, showing resistance to penicillin, oxacillin, erythromycin, gentamicin, clindamycin, ciprofloxacin, and rifampin.
On the basis of identical MLST profiles (ST239) and SCCmec types (IIIA) between the Brazilian and the new Czech MRSA clones, we can hypothesize that the later clone might have evolved from the Brazilian clone, which was spread in large Czech cities in 1996-1997. The possible origin of the new Czech clone from the Brazilian one could be explained by rearrangement of chromosomal DNA. The precise mechanism of such rearrangement is still unclear, but some mutations must have taken place in, or close to, some of the ribosomal operons, because the HindIII ribotype of the Czech clone (H6) was different from that of the Brazilian one (H1). Interestingly, we found one isolate that showed HindIII profile H6, typical of the Czech clone, and PFGE profile B, characteristic of the Brazilian clone. This fact is also in agreement with our hypothesis that the Czech clone evolved from the Brazilian clone on Czech territory between 1996 and 2000. However, the biological properties that conferred a selective advantage to this new clone are still unknown, except for the production of enterotoxin A that seems to be associated with the new Czech clone (seven strains of nine strains tested expressed the toxin) and not with the Brazilian clone (all of four strains tested were negative). What is alarming is the production of enterotoxin A by most strains of the new Czech clone, to which the majority of the Czech MRSA isolates from 2000-2002 belonged: this enterotoxin with its superantigen properties might cause development of toxic shock syndrome, with possibly lethal outcomes (6).
The Hungarian (9) and Taiwan clones, which belong to ST239 and ST241 (single locus variant of ST239) (1, 23, 26, 27) and ribotypes H1 and H2, respectively, may also be progenitors of the Czech clone. Representatives of ST239 were also reported from other countries all over the world, including the countries neighboring the Czech Republic (Hungary, Poland, and Germany) (13, 23).
Some hospitals in Prague and one hospital in Plzeò have specialized units (hospital 3, transplantation unit; hospital 5, transplantation and burn units; hospital 20, trauma and neurology diagnosis units; hospital 6, transplantation unit) that receive patients from all over the country who are further transferred to regional hospitals. This fact might have played an important role in the massive dissemination of the new Czech MRSA clone all over the country in a relatively short period of time. The spread of MRSA strains could be traced based on movements of patients and healthcare staff between hospitals. The precise mechanism of the displacement of the predominant (80%) Brazilian clone (1996-1997) by the Czech clone before the year 2000 still remains unknown.
All of the four strains of a sporadic clonal type that were resistant to erythromycin, clindamycin, and ciprofloxacin only and were characterized by HindIII ribotype H7 and PFGE profile H shared the group ST22. PFGE type H seems to be identical to the PFGE type characteristic of EMRSA-15, which, together with EMRSA-16, is the most prevalent MRSA clone in hospitals in the United Kingdom and was also detected in northern Berlin, Germany (20, 28, 38). The strains from the United Kingdom and Germany showed the same antibiogram as clone H7:H (28, 38) but were producers of enterotoxin C (28), whereas clone H7:H produced enterotoxin A. Both clone H7:H and the German strains possess the ermC determinant (38), and two strains of clone H7:H possess the msrA gene as well. Clone ST22, H7:H, could have spread to the Czech Republic from the United Kingdom or from Germany.
We have studied the distribution of MLSB resistance determinants by PCR among the 100 MRSA isolates. Resistance to erythromycin was found for 98 isolates, and all of them contained one or more erm genes. When a single MLSB resistance determinant was present, the ermA gene was the most common (94%), followed by ermC (69%), whereas ermB was not detected in any isolate. Macrolide resistance due to msrA was rare (2%) and seems to be more frequent in coagulase-negative staphylococci than in S. aureus (17). Analysis of S. aureus strains isolated from blood in Denmark indicated that the ermA and/or ermC genes were responsible for erythromycin resistance in 98% of 428 isolates (37). A study involving 851 S. aureus isolates collected in 1997-1998 from 24 European university hospitals (33) and another one involving 294 S. aureus strains isolated in 1995 in French hospitals (17) concordantly concluded that the ermA gene was more common in MRSA isolates than in methicillin-susceptible S. aureus isolates. ermA occurs on the transposon Tn554 (35), which is present in the large majority of clinical MRSA isolates and absent from methicillin-susceptible S. aureus isolates (15), which may explain the high prevalence of the ermA gene among clinical MRSA isolates.
The frequency of genes encoding aminoglycoside-modifying enzymes was studied in the 100 MRSA isolates. The aac6′-aph2", ant4′, and aph3′ genes were present in 94, 29, and 12% of the isolates, respectively. These results confirmed those of Schmitz et al. (32), who documented that aac6′-aph2" has been the gene most frequently found in MRSA strains isolated in Europe. However, ant4′ was more prevalent in Japanese isolates (14). The major MRSA clonal type found in Japan (2) is very distinct from the ones found in European countries, which might explain the predominance of the ant4′ gene in Japanese isolates (14) compared with the higher prevalence of aac6′-aph2" in Europe. Among the 100 MRSA isolates included in this study, five were susceptible to gentamicin, kanamycin, and tobramycin and did not carry any of the three aminoglycoside determinants. However, two of these isolates were resistant to streptomycin, which could be explained by the presence of the resistance gene str or chromosomal mutations (strA) (32).
When we combined data of aminoglycoside and macrolide resistance genes with clonal types, we observed that the different clonal types found in this study could be distinguished by the presence or absence of these resistance genes (Table 2). The Iberian clone could be distinguished from the Brazilian clone by the presence of ant4′ and the absence of aph3′, whereas the new Czech MRSA clone seldom carried even one of these two genes. None of the aminoglycoside resistance genes was detected in isolates belonging to the minor clone characterized by PFGE type H, ribotype H7, ST22, and SCCmec type IV, which also had a different aminoglycoside resistance gene profile. Regarding macrolide resistance genes, the Iberian clone could be distinguished from the Brazilian clone and the new Czech clone by the absence of ermC. However, the Brazilian and the Czech clones have identical macrolide resistance gene profiles.
In summary, our results, based on two distinct collections of invasive and noninvasive MRSA isolates, clearly document the displacement of the Brazilian clone (ST239, SCCmec type IIIA, PFGE type B, ribotype H1) by the new Czech clone (ST239, SCCmec type IIIA, PFGE type F, ribotype H6) and the maintenance of the Iberian clone (ST247, SCCmec type IA, PFGE type A, ribotype H2) exclusively in one hospital in the Czech Republic. In addition, we found a correlation between the distribution of aminoglycoside resistance genes and the different MRSA clonal types.
Acknowledgments
This study was supported by grant 310/01/1363 of the Grant Agency of the Czech Republic awarded to O. Melter. Support was also received from the Fundação para a Ciência e Tecnologia, Lisbon, Portugal, within project POCTI/1999/ESP/34872 awarded to H. de Lencastre. M. Aires de Sousa was supported by grant BD/13731/97, PRAXIS XXI, from the Fundação para a Ciência e Tecnologia, Lisbon, Portugal, and by grant 310/01/1363 of the Grant Agency of the Czech Republic.
We are grateful to the Czech EARSS participants for collecting staphylococcal isolates. We thank P. Petras for the detection of staphylococcal toxins, E. Kodytková and H. Leitnerová for excellent assistance, and A. Gabrielová for providing data and additional isolates. Some observations were made possible by the information available at the MLST database (www.mlst.net) hosted by The Imperial College and the University of Bath.
REFERENCES
- 1.Aires de Sousa, M., M. I. Crisóstomo, I. Santos Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2002. Frequent recovery of a single clonal type of multidrug resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., H. de Lencastre, I. Santos Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6:253-258. [DOI] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., M. Miragaia, I. Santos Sanches, S. Ávila, I. Adamson, S. T. Casagrande, M. C. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America, 1996-1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aires de Sousa, M., I. Santos Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amorim, M. L., M. Aires de Sousa, I. S. Sanches, R. Sá-Leão, J. M. Cabeda, J. M. Amorim, and H. de Lencastre. 2002. Clonal and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus (MRSA) from a Portuguese hospital over time. Microb. Drug Resist. 8:301-309. [DOI] [PubMed] [Google Scholar]
- 6.Bargs, N. L., and T. Harris. 1997. Toxin-mediated syndromes, p. 527-545. In K. B Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 7.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 8.Corso, A., I. Santos Sanches, M. Aires de Sousa, A. Rossi, and H. de Lencastre. 1998. Spread of a dominant methicillin-resistant multiresistant Staphylococcus aureus (MRSA) clone in Argentina. Microb. Drug Resist. 4:277-288. [DOI] [PubMed] [Google Scholar]
- 9.de Lencastre, H., E. P. Severina, H. Milch, M. Konkoly Thege, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 3:289-296. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez, M. A., H. de Lencastre, J. Liñares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duval, J. 1985. Evolution and epidemiology of MLS resistance. J. Antimicrob. Chemother. 16(Suppl. A):137-149. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ida, T., R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga, and M. Inoue. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. Maslow, A. McGeer, D. E. Low, and R. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 16.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lina, G., A. Quaglia, M. E. Reverdy, R. Leclercq, F. Vandenesch, and J. Etienne. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mato, R., I. Santos Sanches, M. Venditti, D. J. Platt, A. Brown, and H. de Lencastre. 1998. Spread of the multiresistant Iberian clone of methicillin resistance Staphylococcus aureus (MRSA) to Italy and Scotland. Microb. Drug Resist. 4:107-112. [DOI] [PubMed] [Google Scholar]
- 19.Melter, O., I. Santos Sanches, J. Schindler, M. Aires de Sousa, R. Mato, V. Kovarova, H. Zemlickova, and H. de Lencastre. 1999. Methicillin-resistant Staphylococcus aureus clonal types in the Czech Republic. J. Clin. Microbiol. 37:2798-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, P. C. L., and J. A. Lindsay. 2002. Molecular characterization of the dominant UK methicillin-resistant S. aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 51:516-521. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 1995. Performance standards for antimicrobial disk susceptibility test. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.National Institute of Public Health and the Environment. 2001. European antimicrobial resistance surveillance system annual report. National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
- 23.Oliveira, D. C., I. Crisóstomo, I. Santos Sanches, P. Major, C. R. Alves, M. Aires de Sousa, M. K. Thege, and H. de Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 36:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira, D., I. Santos Sanches, R. Mato, M. Tamayo, G. Ribeiro, D. Costa, and H. de Lencastre. 1998. Virtually all methicillin-resistant Staphylococcus aureus (MRSA) infections in the largest Portuguese teaching hospital are caused by two internationally spread multiresistant strains: the “Iberian” and the “Brazilian” clones of MRSA. Clin. Microbiol. Infect. 4:373-384. [DOI] [PubMed] [Google Scholar]
- 26.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 28.O′Neill, G. L., S. Murchan, A. Gil-Setas, and H. M. Aucken. 2001. Identification and characterization of phage variants of a strain of epidemic methicillin-resistant S. aureus (EMRSA-15). J. Clin. Microbiol. 39:1540-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, R. B., A. M. Tennenberg, W. Eisner, J. Hargrave, L. M. Drusin, R. Yurt, and B. N. Kreiswirth. 1998. Outbreak in a New York city teaching hospital caused by the Iberian epidemic clone of MRSA. Microb. Drug Resist. 4:175-183. [DOI] [PubMed] [Google Scholar]
- 30.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of ATP-binding transport super-gene family. Mol. Microbiol. 4:1207-1214. [DOI] [PubMed] [Google Scholar]
- 31.Sanches, I. S., M. Ramirez, H. Troni, M. Abecassis, M. Pádua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitz, F. J., A. C. Fluit, M. Gondolf, R. Beyrau, E. Lindenlauf, J. Verhoef, H. P. Heinz, and M. E. Jones. 1999. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43:253-259. [PubMed] [Google Scholar]
- 33.Schmitz, F. J., R. Sadurski, A. Kray, M. Boos, R. Geisel, K. Köhrer, J. Verhoef, and A. C. Fluit. 2000. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45:891-894. [DOI] [PubMed] [Google Scholar]
- 34.Teixeira, L., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. S. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tillotson, L. E., W. D. Jenssen, L. Moon-McDermott, and D. T. Dublin. 1989. Characterization of a novel insertion of the macrolide-lincosamide-streptogramin B resistance transposon Tn554 in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 33:541-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanhoof, R., C. Godard, J. Content, H. J. Nyssen, and E. Hannecart-Pokorni. 1994. Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. J. Med. Microbiol. 41:282-290. [DOI] [PubMed] [Google Scholar]
- 37.Westh, H., D. M. Hougaard, J. Vuust, and V. T. Rosdahl. 1995. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob. Agents Chemother. 39:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witte, W., M. Enright, F. J. Schmitz, C. Cuny, C. Braulke, and D. Heuck. 2001. Characteristics of a new epidemic MRSA in Germany ancestral to United Kingdom EMRSA15. Int. J. Med. Microbiol. 290:677-682. [DOI] [PubMed] [Google Scholar]