Abstract
We report 16 bacitracin-resistant Streptococcus pyogenes isolates recovered from pharyngitis patients in Belgium, 14 of which belonged to a particular emm type (emm28). All 16 isolates were constitutively resistant to macrolides and carried erm(B). The emergence of a bacitracin-resistant S. pyogenes clone raises questions about the continued reliability of bacitracin susceptibility testing for S. pyogenes identification.
Streptococcus pyogenes, a Lancefield group A streptococcus, is a common pathogen in humans, causing tonsillo-pharyngitis and serious invasive infections, such as necrotizing fasciitis and toxic shock syndrome. The prevalence of S. pyogenes infections has increased drastically in the last decade (4, 8) and correlates with increasing resistance to macrolide and tetracycline groups of antibiotics, which act by disrupting prokaryotic protein synthesis (2, 3, 6). However, S. pyogenes has remained uniformly susceptible to antibiotics that disrupt cell wall synthesis: i.e., penicillins, glycopeptides, and bacitracin. In fact, susceptibility to bacitracin is one of the preliminary laboratory tests employed in the presumptive differentiation of S. pyogenes from other beta-hemolytic streptococci. We report here 16 bacitracin-resistant isolates recovered from tonsillo-pharyngitis patients as part of a national surveillance study conducted in Belgium during 2002. These isolates were investigated further for clonality, as well as resistance to macrolides and other antibiotic groups.
A total of 1,572 presumptive S. pyogenes isolates were collected from 10 Belgian provinces. Of these, 1,229 isolates were confirmed to be S. pyogenes on the basis of a battery of tests: beta-hemolysis on blood agar, Gram stain, catalase, pyrrolidonyl aminopeptidase (PYR), group A antigen, and the bacitracin disk diffusion test (0.4 U; Rosco, Taastrup, Denmark). All S. pyogenes isolates showed the expected results, except for 16 isolates that showed resistance to bacitracin (disk diffusion zone diameters of 0 mm). Macrolide resistance for these isolates was determined by the conventional double-disk diffusion test with erythromycin (78 μg) and clindamycin (25 μg) Neo-Sensitab disks (Rosco), and the results were interpreted as reported previously (7). MICs of erythromycin, clarithromycin (Abbott, Ottignies, Belgium), azithromycin (Pfizer, Groton, Conn.), clindamycin and telithromycin (Aventis, Romainville, France), penicillin and ciprofloxacin (Bayer AG, Leverkusen, Germany), and tetracycline were determined by the agar dilution method. The inoculum (104 CFU/spot) was incubated under aerobic conditions at 37°C for 18 to 24 h, and the results were interpreted according to National Committee for Clinical Laboratory Standards guidelines. For telithromycin, breakpoints of susceptibility and resistance were taken as ≤1 and ≥4 μg/ml, respectively. Unless specifically mentioned, antibiotics were purchased from Sigma Chemical Co. (St. Louis, Mo.). In addition, the presence of the macrolide resistance determinants erm(B), mef(A), and erm(A) was detected by PCR. Genomic DNA was extracted by the alkaline lysis method (0.25% sodium dodecyl sulfate, 0.05 N NaOH). PCR was performed with a DNA thermal cycler (9600 GeneAmp PCR system; Perkin-Elmer, Zaventem, Belgium). The primers described previously for erm(B) and mef(A) give PCR products of 639 and 348 bp (9), respectively, while a 590-bp product was obtained with the following primers for erm(A): 5′ CCCGAAAAATACGCAAAATTTCAT 3′ and 5′ CCCTGTTTACCCATTTATAAACG 3′ (G. Cornaglia, personal communication). The PCR mix and cycling conditions for erm(B), and mef(A) were described previously (2). For erm(A), each 50-μl PCR mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 0.01% gelatin, 200 μM deoxynucleoside triphosphates (dNTPs), 1.5 mM MgCl2, 20 pmol of primers, 0.45 U of SuperTaq polymerase (Enzyme Technologies Ltd., United Kingdom), and 2 μl of template DNA. The cycling conditions were an initial cycle of 5 min at 94°C; 35 cycles of 30 s of denaturation at 90°C, 60 s of annealing at 60°C, and 90 s of extension at 72°C; and finally 1 cycle of 5 min of elongation at 72°C. Positive controls used for erm(A), erm(B), and mef(A) were S. pyogenes strains UR1092, STP016, and STP046, respectively. Clonality was studied by pulsed field gel electrophoresis (PFGE) as well as emm typing as described previously (2, 5).
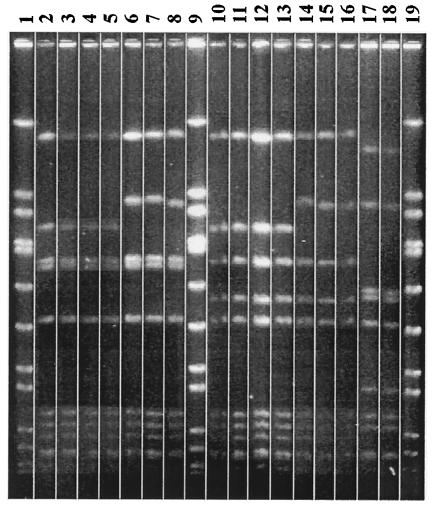
All 16 bacitracin-resistant S. pyogenes strains demonstrated constitutive resistance to erythromycin and clindamycin, explained by the uniform presence of the erm(B) gene. Neither mef(A) nor erm(A) was detected in any isolate. For these 16 isolates, the MICs at which 90% of the isolates tested are inhibited of erythromycin, clindamycin, clarithromycin, azithromycin, telithromycin, tetracycline, penicillin, and ciprofloxacin were >512, >512, 512, >512, 8, 0.125, 0.01, and 0.5 μg/ml, respectively. The PFGE clusters correlated completely with the emm typing results. Most interestingly, of the 16 isolates, 14 belonged to one PFGE cluster (10) and 1 particular emm type (emm28), and 2 isolates belonged to a distinct non-emm-typeable PFGE cluster (Fig. 1). Of the 14 clonal isolates, 13 were isolated from patients residing in the southern Belgian provinces of Hainaut, Luxembourg, and Liège, while 1 isolate was from the northern province of Vlaams-Brabant.
FIG. 1.
PFGE pattern of the bacitracin-resistant S. pyogenes. Lanes 2 to 8 and 10 to 16 correspond to cluster 1 (emm28), and lanes 17 and 18 correspond to cluster 2 (nontypeable isolates) Chromosomal DNA was digested with SmaI. Lanes 1, 9, and 19 correspond to control strain Staphylococcus aureus NCTC 8325, a reference type strain for SmaI digests. The PFGE profiles were obtained by analysis with the computer software Gelcompar, version 4.0 (Applied Maths, Kortrijk, Belgium).
So far, there has only been a single published report of a bacitracin-resistant S. pyogenes clone that was recovered from invasive infections (11), indirectly suggesting that bacitracin resistance could be related to invasiveness. However, our findings of bacitracin-resistant S. pyogenes from pharyngitis patients suggest that there is no link between bacitracin resistance and invasiveness. Interestingly, both the earlier report (11) and this study show that the majority of the bacitracin-resistant S. pyogenes isolates are clonal. Although no gene was ascribed to macrolide resistance in the previous study (11), macrolide resistance in our clone was explained by the presence of erm(B). Moreover, the emm28 bacitracin-resistant clone was concentrated in southern Belgian provinces. We are currently investigating whether the presence of such strains could be related to the use of non-prescription-based bacitracin-containing throat lozenges. Also, efforts to elucidate the mechanism of bacitracin resistance in the emm28 clone are under way. Since bacitracin acts by preventing dephosphorylation and recycling of a lipid carrier (undecaprenol pyrophosphate), resistance to bacitracin, although not definitively characterized, is believed to result from an overproduction of undecaprenol kinase encoded by the bacA gene (reviewed in reference 1). Regardless of the precise reason for the resistance, a further search for bacitracin-resistant S. pyogenes isolates warrants that a preliminary screening for S. pyogenes should not rely on susceptibility to bacitracin.
Acknowledgments
We thank the following Belgian centers for their participation in this study: AML BVBA, Antwerp; Laboratoire de Biologie Clinique et Hormonale-S.P.R.L., Couillet; Centraal Laboratorium, Hasselt; Medisch Centrum Huisarten, Leuven; Centre Hospitalier de L'Ardenne Laboratoire de Biologie Clinique et de Ria, Libramont; and Laboratoire Marchand, Liège.
REFERENCES
- 1.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 16:175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Descheemaeker, P., S. Chapelle, C. Lammens, M. Hauchecorne, M. Wijdooghe, P. Vandamme, M. Ieven, and H. Goossens. 2000. Macrolide resistance and erythromycin resistance determinants among Belgian Streptococcus pyogenes and Streptococcus pneumoniae isolates. J. Antimicrob. Chemother. 45:167-173. [DOI] [PubMed] [Google Scholar]
- 3.Jasir, A., A. Tanna, A. Noorani, A. Mirsalehian, A. Efstratiou, and C. Schalen. 2000. High rate of tetracycline resistance in Streptococcus pyogenes in Iran: an epidemiological study. J. Clin. Microbiol. 38:2103-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan, E. L. 1991. The resurgence of group A streptococcal infections and their sequelae. Eur. J. Clin. Microbiol. Infect. Dis. 10:55-57. [DOI] [PubMed] [Google Scholar]
- 5.Kaufhold, A., A. Podbielski, G. Baumgarten, M. Blokpoel, J. Top, and L. Schouls. 1994. Rapid typing of group A streptococci by the use of DNA amplification and non-radioactive allele-specific oligonucleotide probes. FEMS Microbiol. Lett. 119:19-25. [DOI] [PubMed] [Google Scholar]
- 6.Seppala, H., A. Nissinen, H. Jarvinen, S. Huovinen, T. Henriksson, E. Herva, S. E. Holm, M. Jahkola, M. L. Katila, and T. Klaukka. 1992. Resistance to erythromycin in group A streptococci. N. Engl. J. Med. 326:292-297. [DOI] [PubMed] [Google Scholar]
- 7.Seppala, H., A. Nissinen, Q. Yu, and P. Huovinen. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32:885-891. [DOI] [PubMed] [Google Scholar]
- 8.Stevens, D. L. 1995. Streptococcal toxic-shock syndrome: spectrum of disease, pathogenesis, and new concepts in treatment. Emerg. Infect. Dis. 1:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.York, M. K., L. Gibbs, F. Perdreau-Remington, and G. F. Brooks. 1999. Characterization of antimicrobial resistance in Streptococcus pyogenes isolates from the San Francisco Bay area of Northern California. J. Clin. Microbiol. 37:1727-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]