Abstract
Eukaryotic cells arose from an ancient endosymbiotic association of prokaryotes, with plant cells harboring 3 genomes as the remnants of such evolution. In plant cells, plastid and mitochondrial DNA replication [organelle DNA replication (ODR)] occurs in advance of the subsequent cell cycles composed of nuclear DNA replication (NDR) and cell division. However, the mechanism by which replication of these genomes with different origins is coordinated is largely unknown. Here, we show that NDR is regulated by a tetrapyrrole signal in plant cells, which has been suggested as an organelle-to-nucleus retrograde signal. In synchronized cultures of the primitive red alga Cyanidioschyzon merolae, specific inhibition of A-type cyclin-dependent kinase (CDKA) prevented NDR but not ODR after onset of the cell cycle. In contrast, inhibition of ODR by nalidixic acid also resulted in inhibition of NDR, indicating a strict dependence of NDR on ODR. The requirement of ODR for NDR was bypassed by addition of the tetrapyrrole intermediates protoporphyrin IX (ProtoIX) or Mg-ProtoIX, both of which activated CDKA without inducing ODR. This scheme was also observed in cultured tobacco cells (BY-2), where inhibition of ODR by nalidixic acid prevented CDKA activation and NDR, and these inhibitions were circumvented by Mg-ProtoIX without inducing ODR. We thus show that tetrapyrrole-mediated organelle–nucleus replicational coupling is an evolutionary conserved process among plant cells.
Keywords: Cyanidioschyzon merolae, retrograde signal, tobacco BY-2
The origin of eukaryotic cells is still unclear, but it is now generally accepted that mitochondria and plastids arose from endosymbiosis of rickettsia-like α-proteobacteria and cyanobacteria-like photosynthetic bacteria, respectively (1). Many of the genes of these endosymbionts, including those for DNA replication and maintenance of genomic integrity, were subsequently lost or transferred to the nuclear genome [endosymbiotic gene transfer (EGT)]. According to current dogma, the transfer of genes of endosymbiotic origin to the nucleus that are required for organelle DNA replication (ODR) has resulted in loss of the independence of the cell cycles of the endosymbionts and in their integration into the eukaryotic control system that is mediated largely by cyclins and cyclin-dependent kinase (CDK). However, coordination of cell-cycle events such as DNA replication would have been essential for establishing integrity of eukaryotic cells, at least during the early stages of endosymbiotic association. Although there is little evidence for discrete cell-cycle control of ODR in animal and fungal cells, studies with algae and flowering plants have shown that ODR precedes the subsequent cell proliferation cycles composed of nuclear DNA replication (NDR) and cell division (2–4).
C. merolae is an unicellular red alga, each cell of which contains 1 plastid, 1 mitochondrion, and 1 nucleus. The advantage of studying nuclear and organelle proliferation cycles in C. merolae is that the cultures can be highly synchronized by exposure of the cells to controlled dark–light cycles, with the overall cell cycle being arrested at G1 phase in the dark (5). Initiation of the cell cycle by illumination in combination with microscopic quantification of organelle DNA content revealed that ODR always occurs before NDR (6). After completion of both ODR and NDR, division of the plastid, mitochondrion, and nucleus occurs sequentially and is followed by cell division (7). The complete nucleotide sequences of the 3 genomes of C. merolae have been determined (8–11), with the result that many tools are now available or are under development for genomics analysis. In this study, we have identified the chemical signal that coordinates ODR and NDR in C. merolae and have shown that this mechanism is also working in flowering plants.
Results and Discussion
ODR Precedes NDR in the C. merolae Cell Cycle.
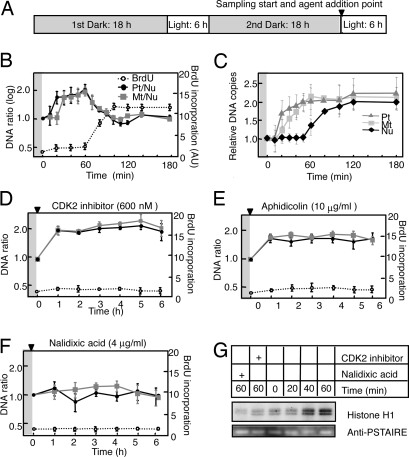
A synchronizing cultivation condition for C. merolae has been established (12), with the optimal cycle found to be 6-h light/18-h dark (Fig. S1). Microscopic observations revealed a highly synchronous G1–S phase transition after the onset of the second period of illumination. To examine the timing of ODR and NDR, we used quantitative PCR (qPCR) analysis with primers targeted to specific regions of each genome to monitor changes in the copy ratio of organelle DNA to nuclear DNA. The plastid/nucleus (Pt/Nu) and mitochondrion/nucleus (Mt/Nu) ratios of DNA copy number increased from 1 to ≈2 during the first 60 min after the onset of illumination (Fig. 1 A and B), indicating that the DNA content of the plastid and mitochondrion doubled during this period. Both Pt/Nu and Mt/Nu ratios returned to ≈1 during the subsequent 40 min, corresponding to the occurrence of NDR. We also observed ODR and NDR by directly counting the DNA contents by fluorescence microscopy after staining with the DNA-specific fluorochrom DAPI using a video-intensified microscope photon-counting system (VIMPCS). The results were consistent with the qPCR results and a previous report (6), indicating the reliability of the qPCR-based calculation. The timing of NDR was further confirmed by monitoring the BrdU incorporation reflecting the de novo DNA synthesis. Incorporation of BrdU was detected from 60 to 100 min after the onset of illumination (Fig. 1C), representing synthesis of a new nuclear genome. Given that the plastid and mitochondrial genomes are much smaller than the nuclear genome, BrdU incorporation during ODR was not evident in this assay. Based on these results, we thus routinely used the synchronized culture system and the qPCR-based method for subsequent experiments. In the absence of the second illumination, Pt/Nu and Mt/Nu ratios remained constant, indicating that neither ODR nor NDR was occurring (data not shown).
Fig. 1.
Dynamics of nuclear, plastid, and mitochondrial DNA replication in C. merolae. (A) Protocol for synchronization of the cell cycle in C. merolae by exposure to light–dark cycles. Arrowhead indicates the reagent addition to the culture medium and the initiating time point of cell sampling. (B and D–F) Changes in the Pt/Nu and Mt/Nu ratios of DNA copy number as determined by qPCR and in BrdU incorporation [arbitrary units (AU)] during the second period of illumination (mean ± SD; n = 3). (C) Changes in the DNA copy numbers of plastid, mitochondrion, and nucleus. DNA amounts were determined by measuring the fluorescence intensity of DAPI-stained DNA by VIMPCS and expressed as relative copy numbers normalized to the G1 state (mean ± SD; n = 30). Cells were incubated in the absence (B and C) or presence of the indicated concentrations of CDK2 inhibitor (D), aphidicolin (E), or nalidixic acid (F). Gray and white backgrounds correspond to dark and light conditions, respectively. (G) Activity of CDKA-type kinase with histone H1 as substrate at the indicated times after the dark/light shift for cells exposed to aphidicolin, CDK2 inhibitor, or nalidixic acid; quantitative data are presented in Fig. S3. Immunoblot analysis with antibodies to PSTAIRE, a cyclin-binding motif specific for CDKA-type kinases, is also shown for the various conditions.
ODR Is Essential to Activate Type A CDK (CDKA) and NDR.
In other eukaryotic cells, activation of a G1 cyclin–CDK complex is essential for initiation of NDR. This complex phosphorylates replication and other proteins such as the retinoblastoma protein (13–15). Analysis of the genomic sequence of C. merolae has revealed 3 candidate CDK genes, with 1 of the gene products (CME119C or CDKA; http://merolae.biol.s.u-tokyo.ac.jp) being orthologous to Cdc2 of fission yeast and CDKA;1 of Arabidopsis (16). Given that a candidate gene for a G1 cyclin, a unique E-type cyclin (CML219C), has been identified in the genome of C. merolae, it would seem reasonable to assume that the G1–S phase transition is similarly regulated in this alga. We therefore initially hypothesized that ODR is also under the control of CDKA activity in C. merolae. We examined this possibility by determining the effect of addition of a CDK2 inhibitor (17), which was expected to be specific for CDKA, to the culture immediately before the onset of illumination. We found that ODR was not affected by CDKA inhibition, whereas NDR was completely blocked, as revealed by the qPCR and VIMPCS analyses and the BrdU incorporation assay (Fig. 1D and Fig. S2a). Specific inhibition of CDKA activity by the CDK2 inhibitor was confirmed by direct monitoring of CDKA activity (Fig. 1G and Fig. S3). Addition of the CDK2 inhibitor to the culture either 30 min or 1 h before the onset of illumination gave essentially the same results (data not shown). These observations indicate that ODR is regulated independently of CDKA. When another type of CDK inhibitor, roscovitine, was used in place of the CDK2 inhibitor, ODR was also inhibited and NDR (Fig. S4a). Because roscovitine inhibits various types of CDKs, CDKs other than CDKA could be involved in the ODR initiation, where further analyses should be required to understand the underlying details. We next examined the effects of the addition of various other inhibitors of cellular processes to the culture medium immediately before light onset. Aphidicolin, a specific inhibitor of nuclear DNA polymerase α (18), inhibited NDR but not ODR or CDKA activity (Fig. 1 E and G and Fig. S2b). In contrast, nalidixic acid, which inhibits organelle DNA gyrase and therefore ODR (19), blocked not only ODR but also CDKA activation and NDR (Fig. 1 F and G and Fig. S2c). Another organelle DNA gyrase inhibitor, novobiocin, had similar effects on ODR and NDR (Fig. S4b). These data strongly indicate that ODR occurs independently of both CDKA activation and subsequent NDR, whereas NDR requires ODR and subsequent CDKA activation. Cycloheximide, an inhibitor of cytoplasmic translation, blocked both ODR and NDR (Fig. S4c), indicating that de novo protein synthesis is required at least for ODR.
Tetrapyrrole Signal Activates CDKA and NDR.
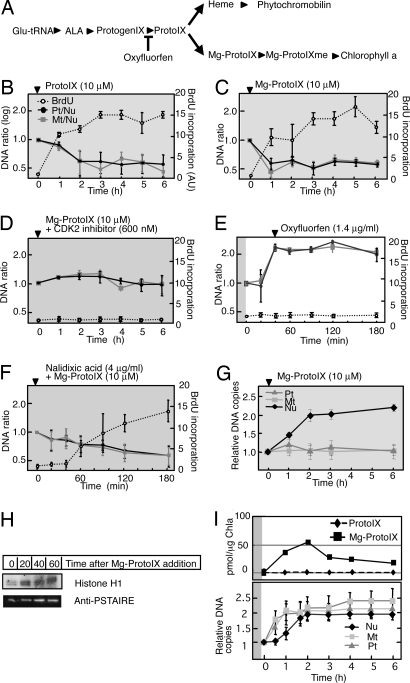
We next hypothesized that ODR and NDR are controlled independently, and that a signal generated as a result of ongoing or completed ODR leads to activation of CDKA and thereby triggers NDR. It was shown that plastid and mitochondrial status are relayed to the nucleus by tetrapyrrole molecules, resulting in transcriptional regulation of genes important for organelle function (20–22). We therefore examined whether such retrograde signaling mediated by tetrapyrrole molecules also might play a role in DNA replicational coupling. We added intermediates of chlorophyll or heme biosynthesis, including protoporphyrin IX (ProtoIX), Mg-ProtoIX, Mg-ProtoIX monomethyl ester (Mg-ProtoIXme), and hemin (an oxidized form of heme that is reduced to heme in vivo) (Fig. 2A), to synchronized cultures in the dark, given that these compounds produce reactive oxygen species in the presence of light, possibly complicating the results. Both Pt/Nu and Mt/Nu ratios decreased from 1 to ≈0.5 after the addition of ProtoIX (Fig. 2B) or Mg-ProtoIX (Fig. 2C), whereas addition of Mg-ProtoIXme or hemin had no effect (Fig. S4 d and e). Results of the VIMPCS (Fig. 2G and Fig. S2d) and BrdU incorporation (Fig. 2 B and C) analyses also indicated that ProtoIX and Mg-ProtoIX induce NDR in the absence of both light and prior ODR. Given that ProtoIX is the immediate precursor of Mg-ProtoIX, the latter is likely the effecter molecule in vivo. Direct monitoring of CDKA activity suggested that the effect of Mg-ProtoIX on NDR was mediated through the activation of CDKA (Fig. 2H and Fig. S3). Consistent with this idea, synchronous addition of CDK2 inhibitor and Mg-ProtoIX prevented the activation of NDR by Mg-ProtoIX (Fig. 2D). We also examined the effect of oxyfluorfen, which inhibits the biosynthesis of ProtoIX from protoporphyrinogen IX (23) (Fig. 2A). The addition of oxyfluorfen 40 min after the onset of illumination, around the time of completion of ODR, resulted in inhibition of NDR (Fig. 2E and Fig. S2e), consistent with the proposed role of Mg-ProtoIX as a signaling metabolite. Mg-ProtoIX addition in the dark in combination with nalidixic acid or novobiocin could induce NDR, indicating that Mg-ProtoIX cancelled the effect of ODR inhibition on NDR and that these inhibitors were not inhibitory for NDR itself (Fig. 2F and Fig. S4f, and Fig. S2f). Mg-ProtoIX addition in combination with oxyfluorfen could induce NDR, indicating that these inhibitors were not deleterious to the cell (Fig. S4g). To substantiate the tetrapyrrole molecules as the signaling metabolite, we measured intracellular accumulation of ProtoIX and Mg-ProtoIX through the G1 and S phases. As shown in Fig. 2I, transient accumulation of Mg-ProtoIX was observed correlating with the ODR occurrence, whereas ProtoIX remained at an undetectable level. The correlation of ODR and Mg-ProtoIX accumulation was also observed when the synchronized cells were cultivated in the presence of 2% CO2 (Fig. S5) under which condition the timing of ODR and the subsequent NDR delays 1.5 h after the dark to light shift. These results are consistent with the role of Mg-ProtoIX as the replicational signal.
Fig. 2.
Effects of tetrapyrroles and an inhibitor of ProtoIX biosynthesis on DNA replication in C. merolae. (A) Tetrapyrrole biosynthetic pathway. The step inhibited by oxyfluorfen is indicated. Glu-tRNA, ALA, and ProtogenIX denote glutamyl-tRNA, α-aminolevulinic acid, and protoporphyrinogen IX, respectively. (B–F) Changes in the Pt/Nu and Mt/Nu ratios of DNA determined by qPCR and in BrdU incorporation during incubation of cells with ProtoIX (B), Mg-ProtoIX (C), Mg-ProtoIX and CDK2 inhibitor (D), oxyfluorfen (E), or Mg-ProtoIX and nalidixic acid (F) (mean ± SD; n = 3). (G) Changes in DNA contents of plastid, mitochondrion, and nucleus determined by VIMPCS analysis during incubation of cells with Mg-ProtoIX, expressed as relative copy numbers normalized to the G1 state (mean ± SD; n = 30). Arrowhead indicates the reagent addition to the culture medium. All reagents were added at the end of the second dark period, with the cells subsequently maintained in the dark, with the exception of oxyfluorfen, which was added 40 min after the onset of the second light period. Gray and white backgrounds correspond to dark and light conditions, respectively. (H) Activity of CDKA-type kinase with histone H1 as substrate and immunoblot analysis with anti-PSTAIRE for cells exposed to Mg-ProtoIX. Quantitative data for kinase activity are presented in Fig. S3. (I) Changes in ProtoIX and Mg-ProtoIX accumulation in synchronized culture cells. (Upper) Time course of ProtoIX and Mg-ProtoIX accumulation in cells. The tetrapyrrole amounts were normalized by chlorophyll a content. Experiments were reproduced twice with biologically independent conditions, and the averaged value was plotted in the graph. (Lower) Changes in relative DNA amounts determined by VIMPCS (mean ± SD; n = 30).
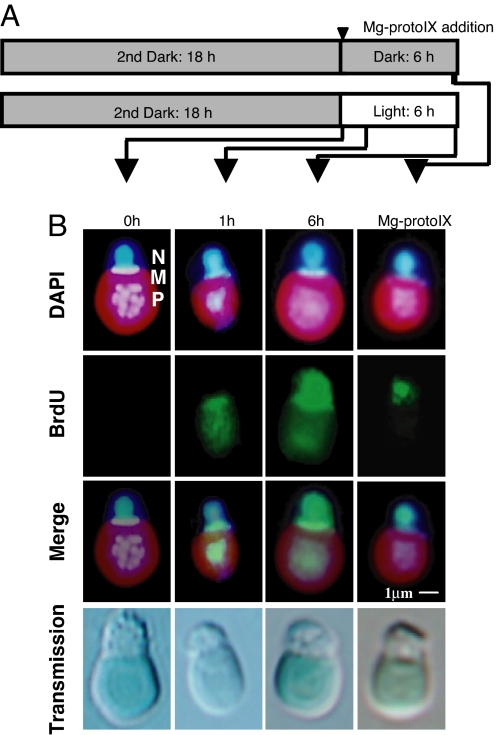
The occurrence of ODR and NDR was also analyzed more directly by immunocytofluorescence staining of BrdU incorporation. We added BrdU to the culture medium after synchronization and either before the dark–light shift or before addition of Mg-ProtoIX (Fig. 3A). Consistent with the qPCR data, BrdU signals were detected only in the plastid and mitochondrion at 1 h after the onset of illumination, whereas they were detected both in these organelles and the nucleus at 6 h (Fig. 3B). In contrast, BrdU signals were detected only in the nucleus 6 h after Mg-ProtoIX addition in the dark.
Fig. 3.
Immunofluorescence detection of BrdU incorporation in organelles and the nucleus in C. merolae. (A) Protocol for synchronization of the cell cycle showing the time of Mg-ProtoIX addition (arrowhead) and time points for sampling (arrows). (B) Immunostaining of fixed cells. BrdU (green) was visualized with antibodies to BrdU and Alexa Fluor 488-conjugated secondary antibodies. DNA (blue) was visualized by staining with DAPI. Stained organelles are indicated as nucleus (N), mitochondrion (M), and plastid (P). The BrdU and DNA images are also shown merged. (Scale bar: 1 μm.)
Common Tetrapyrrole Signal in a Flowering Plant Cell Cycle.
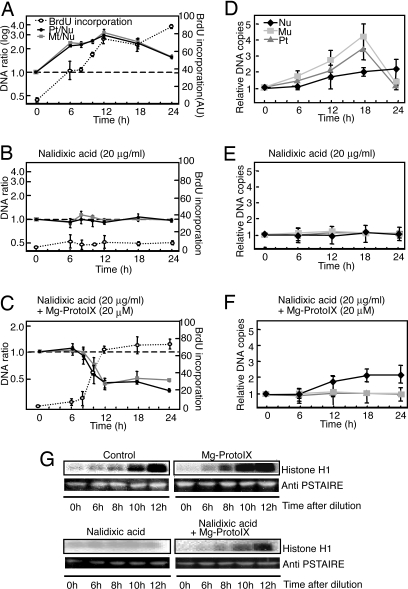
In flowering plants, it was suggested that ODR is independent of the cell-cycle events governing NDR (24, 25), but involvement of ODR in NDR control is not well understood. Thus, we examined the role of ODR in NDR in this study. During the formation of organs such as leaves and roots, amplification of organelle DNAs (ODR) occurs before multiple rounds of the cell cycle (3, 6). These events are apparently reproducible in unsynchronized tobacco BY-2 cell cultures (6). Therefore, we examined the ODR and NDR coupling mechanism by using this system with similar methodology as in C. merolae (Fig. 4). After subcultivation of stationary-phase cells by dilution, ODR occurred during the first phase, and subsequently NDR was activated (Fig. 4 A and D), which is consistent with a previous report (6). By assaying CDKA activity, we found that CDKA was gradually activated after dilution, and this activation was hastened by addition of Mg-ProtoIX at the time of dilution (Fig. 4G). This CDKA activation and NDR were completely inhibited by the addition of nalidixic acid that inhibits ODR (Fig. 4 B, E, and G and Fig. S6), and Mg-ProtoIX could circumvent ODR and induce CDKA activation and NDR (Fig. 4 C, F, and G and Fig. S6). Novobiocin in place of nalidixic acid gave similar results (Fig. S7 A and B). Although BY-2 cells are analogous to root cells and do not develop chlorophyll-containing chloroplasts, chlorophyll intermediates were detected in nonphotosynthetic root tissues (26), which is consistent with a general, photosynthesis-independent role of chlorophyll biosynthesis intermediates in plant cell-cycle events. In BY-2 cells, the strict dependence of ODR on a light signal has apparently been lost and might have been superseded by other, possibly hormonal signals, whereas the downstream tetrapyrrole signal has been conserved throughout evolution. The loss of light dependence of ODR, and the light-independent accumulation of the resultant tetrapyrrole signal as well, might be explained by the fact that multicellular plants contain tissues, such as roots, that are never exposed to light but nevertheless require coordinated ODR and NDR in flowering plants. Our results suggest that tetrapyrrole-mediated CDK activation is a commonly found mechanism in red and green plants with the primary plastids.
Fig. 4.
Effects of Mg-ProtoXI in tobacco BY-2 cells. Cells were sampled at the indicated times after dilution, and reagents were added at the time of dilution. (A–C) Changes in Pt/Nu and Mt/Nu ratios of DNA copy number as determined by qPCR and BrdU incorporation (mean ± SD; n = 3). Each sample was pulse-labeled for 30 min with 100 μM BrdU and 10 μM 5-fluorodeoxyuridine. (D–F) Changes in DNA copy numbers in plastid, mitochondrion, and nucleus determined by VIMPCS analysis, expressed as relative copy numbers normalized to the state at the subcultivation (mean ± SD; n = 10). (G) Activity of CDKA-type kinase with histone H1 as substrate and immunoblot analysis with anti-PSTAIRE at the indicated times after dilution. Quantitative data for kinase activity are presented in Fig. S6.
Implications.
Given that the prokaryotic progenitors of plastids and mitochondria would have had their own cell cycles before the endosymbiotic events, coupling of genome replication among these new organelles and the nucleus would have been essential for ensuring the integrity of the eukaryotic cell. In plant cells, tetrapyrrole biosynthesis is achieved through the interaction of multiple organelles including plastids and mitochondria. These metabolic connections may thus have given rise to the tight linkage among multiple replication cycles. Thus far, tetrapyrrole molecules have been suggested to mediate organelle-to-nucleus retrograde signaling to coordinate nuclear gene expression (20–22). However, our findings suggest that tetrapyrrole signaling could be a mechanism for coordinating the cell cycles as well, which may include fine-tuning of the nuclear gene expression in various contexts. During this study, we did not detect any difference in the dynamics of plastid and mitochondrial DNA replication. However, these 2 organelles have distinct origins, and elucidation of the mechanism responsible for their replicational coupling may provide fundamental insights into plant and other eukaryotic cells. Presently, we have little information on the underlying signal transduction mechanism. However, examining the effects of mutations that could affect biosynthesis of tetrapyrrole intermediates, such as gun mutations of Arabidopsis (20), may help us to understand the relationship between ODR and the tetrapyrrole signal. With respect to the mechanism for the CDKA activation by Mg-ProtoIX, there are a number of possibilities, such as activation of the cyclin E expression or inhibition of a specific CDKA inhibitor. Although we cannot yet discern these alternative hypotheses, it is evident from our study that a cytosolic receptor perceives the tetrapyrrole signal and regulates the CDKA activation to initiate NDR. Future biochemical identification of the receptor and the extremely simple genome content of C. merolae with little redundancy should facilitate the elucidation of the complete pathway, which will pave the way for a mechanistic understanding of tetrapyrrole-controlled NDR in the flowering plant system.
Materials and Methods
Materials and Culture Conditions.
Cells of C. merolae 10D were cultured and their growth was synchronized as described (5), with minor modifications (SI Text). Tobacco (Nicotiana tabacum) BY-2 cells were cultivated in modified Murashige and Skoog (MS) medium at 27 °C and 130 rpm with a gyratory shaker (Bioshaker BR-160 LF, Taitec) in the dark and subcultured by a 1 in 20 dilution each week (27).
qPCR Analysis of DNA Replication.
C. merolae total DNA was isolated by the hot phenol method and purified by using DNase-free ribonuclease A (Nippongene). For BY-2, total DNA was isolated by using a DNeasy plant mini kit (Qiagen). An MX 3000P instrument (Stratagene) was used for qPCR with FullVelocity SYBR Green QPCR Master Mix (Stratagene). Copy numbers of plastid and mitochondrial genomes were normalized compared with the nuclear genome, yielding Pt/Nu and Mt/Nu ratios. Means ± SD of values of these ratios were obtained from 3 biologically independent sets of DNA preparations, and each experimental value represents the mean of replicates. See Table S1 for primer sequences used.
Microfluorometry.
C. merolae and BY-2 cells were fixed by 1% glutaraldehyde and 3.7% formaldehyde, respectively. Fixed cells were stained with DAPI at the concentration of 2 μg/mL. The intensity of fluorescence of DAPI-stained DNA was quantified by using a VIMPCS as described (28), with minor modifications (SI Text).
Quantitative BrdU Incorporation Assay.
Total DNA (5 μg) from BrdU-treated cells were isolated and purified as described was applied to a nylon membrane filter with a dot blotter (Bio-Dot; Bio-Rad), after which the membrane was incubated for 1 h with antibodies to BrdU (Sigma). BrdU incorporation into DNA was detected with ECL Western blot analysis detection reagents (GE Healthcare).
Assay of CDKA Activity.
Precipitation of CDKA and assays of kinase activity were performed as described (29) with minor modifications (SI Text).
Quantification of Tetrapyrrole Intermediates.
Tetrapyrrole intermediates were quantified as in SI Text.
For more details and additional methods see SI Text.
Supplementary Material
Acknowledgments.
We thank T. Nagata (University of Tokyo, Tokyo, Japan) for BY-2 cells; N. Mochizuki (Kyoto University, Kyoto, Japan) for tetrapyrroles; and N. Mochizuki, M. Umeda, M. Hanaoka, and A. Weber for comments on the manuscript. This work was supported by Grant-in-Aid for Creative Scientific Research 16GS0304 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804270105/DCSupplemental.
References
- 1.Leister D. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene. 2005;354:110–116. doi: 10.1016/j.gene.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 2.Stolarsky L, Walfield AM, Brich RA, Hershberger CL. Light-stimulated synthesis of chloroplast DNA. Biochim Biophys Acta. 1976;425:438–450. doi: 10.1016/0005-2787(76)90008-3. [DOI] [PubMed] [Google Scholar]
- 3.Rose RJ, Cran DG, Possingham JV. Changes in DNA synthesis during cell growth and chloroplast replication in greening spinach leaf disks. J Cell Sci. 1975;12:27–41. doi: 10.1242/jcs.17.1.27. [DOI] [PubMed] [Google Scholar]
- 4.Blamire J, Flechtner VR, Sager R. Regulation of nuclear DNA replication by the chloroplast in Chlamydomonas. Proc Natl Acad Sci USA. 1974;71:2867–2871. doi: 10.1073/pnas.71.7.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki K, et al. Behavior of mitochondria, chloroplasts, and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur J Cell Biol. 1994;63:280–288. [PubMed] [Google Scholar]
- 6.Sakai A, Takano H, Kuroiwa T. Organelle nuclei in higher plants: Structure, composition, function, and evolution. Int Rev Cytol. 2004;238:59–117. doi: 10.1016/S0074-7696(04)38002-2. [DOI] [PubMed] [Google Scholar]
- 7.Kuroiwa T. The primitive red algae: Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. BioEssays. 1998;20:344–354. [Google Scholar]
- 8.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 9.Ohta N, et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2003;10:67–77. doi: 10.1093/dnares/10.2.67. [DOI] [PubMed] [Google Scholar]
- 10.Ohta N, Sato N, Kuroiwa T. Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 1998;26:5190–5198. doi: 10.1093/nar/26.22.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishida K, et al. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci USA. 2003;100:2146–2151. doi: 10.1073/pnas.0436886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekholm SV, Reed SI. Regulation of G1 cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;6:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 14.Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:272–274. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 16.De Clercq A, Inzé D. Cyclin-dependent kinase inhibitors in yeast, animals, and plants: A functional comparison. Crit Rev Biochem Mol Biol. 2006;41:293–313. doi: 10.1080/10409230600856685. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura K, Yokoyama N, Igarashi I. Cyclin-dependent kinase inhibitors block erythrocyte invasion and intraerythrocytic development of Babesia bovis in vitro. Parasitology. 2007;134:1347–1353. doi: 10.1017/S0031182007002831. [DOI] [PubMed] [Google Scholar]
- 18.Itoh R, et al. Aphidicolin uncouples the chloroplast division cycle from the mitotic cycle in the unicellular red alga Cyanidioschyzon merolae. Eur J Cell Biol. 1996;71:303–313. [PubMed] [Google Scholar]
- 19.Itoh R, et al. DNA gyrase involvement in chloroplast-nucleoid division in Cyanidioschyzon merolae. Eur J Cell Biol. 1997;73:252–258. [PubMed] [Google Scholar]
- 20.Nott A, Jung H, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 21.Strand A, et al. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 22.Mense SM, Zhang L. Heme: A versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinase. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, et al. Toxicity of 40 herbicides to the green alga Chlorella vulgaris. Ecotoxicol Environ Saf. 2002;51:128–132. doi: 10.1006/eesa.2001.2113. [DOI] [PubMed] [Google Scholar]
- 24.Heinhorst S, Cannon G, Weissbach A. Plastid and nuclear DNA synthesis are not coupled in suspension cells of Nicotiana tabacum. Plant Mol Biol. 1985;4:3–12. doi: 10.1007/BF02498710. [DOI] [PubMed] [Google Scholar]
- 25.Heinhorst S, Cannon G, Weissbach A. Chloroplast DNA synthesis during the cell cycle in cultured cells of Nicotiana tabacum: Inhibition by nalidixic acid and hydroxyurea. Arch Biochem Biophys. 1985;239:475–479. doi: 10.1016/0003-9861(85)90714-3. [DOI] [PubMed] [Google Scholar]
- 26.Walter G, Shalygo NV. Location and fate of protoporphyrin IX accumulated in etiolated leaves and roots of Zea mays L. and Pisum sativum L. J Photochem Photobiol B. 1997;40:175–182. [Google Scholar]
- 27.Nagata T, Kumagai F, Hasezawa S. The origin and organization of cortical microtubules during the transition between M and G1 phases of the cell cycle as observed in highly synchronized cells of tobacco BY-2. Planta. 1994;193:567–572. [Google Scholar]
- 28.Kuroiwa T, Nakamura S, Hizume M, Toh-e A, Miyakawa I, Sando N. Cytological characterization of NOR in the bivalent of Saccharomyces cerevisiae. Exp Cell Res. 1986;165:199–206. doi: 10.1016/0014-4827(86)90544-6. [DOI] [PubMed] [Google Scholar]
- 29.Samiei M, Daya-Makin M, Clark-Lewis I, Pelech SL. Platelet-activating factor and thrombin-induced stimulation of p34cdc2 cyclin histone H1 kinase activity in platelets. J Biol Chem. 1991;266:14889–14892. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.