Abstract
Background/Aims
To specify roles of HNF4α in mouse liver development, we have analyzed the ex vivo morphogenetic potential of HNF4α-null embryonic hepatic cells.
Methods
Using mice with floxed or deficiency alleles of HNF4α, hepatic cells lacking this transcription factor were explanted into primary culture and derived into cell lines.
Results
Contrary to behavior in vivo where HNF4α-null liver cells fail to show normal polarity and epithelialization, e18.5 hepatic cells in primary culture from mutant embryos show restoration of apical expression of tight junction protein-1 and of transcripts for E-cadherin. Clones of control and HNF4α -null cell lines were indistinguishable, even when differentiation of bile canalicular formation was induced. HNF4α-null and control cell lines showed similar potential to colonize livers of the murine ALB-uPA/SCID model of liver regeneration, but null cells formed only bile ducts and not clusters of hepatocytes. Finally, analysis of mutant embryonic livers revealed a transcriptional signature consistent with a stress response, which could underlie the morphogenetic defects observed in vivo.
Conclusions
We conclude that the lack of epithelialization characteristic of the HNF4α-null embryonic liver is due, at least in part, to non-cell autonomous defects, and that null cells do not suffer intrinsic defects in polarization.
Keywords: Hepatocytes, Cholangiocytes, Liver repopulation, Cell polarization, Stress response
1. Introduction
Hepatocyte nuclear factor 4α (Hnf4α, NR2A1) is a member of the nuclear receptor super-family of transcription factors. Mutations in the HNF4α gene can induce maturity-onset diabetes of the young (MODY1 [1,2]) while mutations in HNF4α binding sites in the HNF1α and Factor IX genes are responsible for a subtype of MODY3 and hemophilia Leyden, respectively [3,4]. There is therefore considerable interest in determining the in vivo functions and gene targets of this transcription factor.
Most genes known to be controlled by HNF4α are involved in lipid, carbohydrate or amino acid transport and metabolism (see www.sladeklab.ucr.edu for list), indicating a central role in energy homeostasis. Invalidation of the HNF4α gene in mouse results in a lethal failure in the metabolic functions of the visceral endoderm [5] while conditional knockout of the HNF4α gene in the post-natal liver results in dyslipidemia and numerous other metabolic defects [6–8].
While the role of HNF4α in metabolism is undisputed, recent data suggest a wide role in hepatocyte biology. Indeed, HNF4α protein was detected at nearly half of all transcribed hepatic gene promoters [9], but it could bind directly or indirectly, and act as an activator or repressor, depending on the context. A role for HNF4α in hepatic morphogenesis was suggested because its deficiency in embryonic mouse liver resulted in abnormal tissue architecture and a lack of appropriate cell–cell contacts [10], while its over-expression induced cell polarity in F9 embryonic carcinoma [11], H5 hepatoma [12] and even NIH3T3 fibroblast [10] cells.
To determine the level at which absence of HNF4α interferes with hepatic morphogenesis, we examined properties of HNF4α-deficient embryonic liver cells in primary culture, and as BMEL (Bipotential Mouse Embryonic Liver) cell lines, analyzed both in vitro, and in vivo in a model of competitive liver regeneration. We show that HNF4α-null hepatic cells do not possess intrinsic deficiencies in epithelialization, for they can polarize, differentiate as hepatocytes and cholangiocytes in vitro and colonize mouse Alb-uPA/SCID livers as polarized cholangiocytes.
2. Materials and methods
2.1. Preparation of primary cultures from e18.5 day embryonic liver
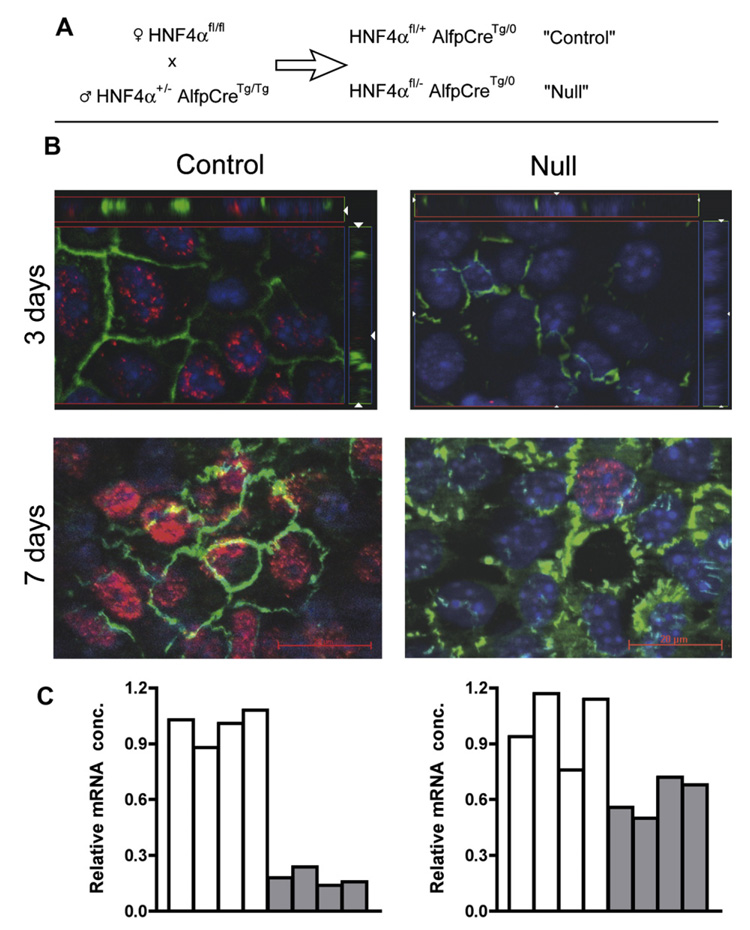
Animal care followed institutional guidelines. Individual livers of e18.5 control and HNF4α-null mice (as depicted in Fig. 1A) were diced and digested in Liberase Blendzyme 3 (25 µg/ml; Roche Diagnostics, Meylan, France) for 30 min at 37 °C. Cells were cultured on collagen-coated dishes in William’s E medium with antibiotics and 10% FCS. PCR genotyping was on a tail sample from each embryo. Cells were immunostained as in [13].
Fig. 1.
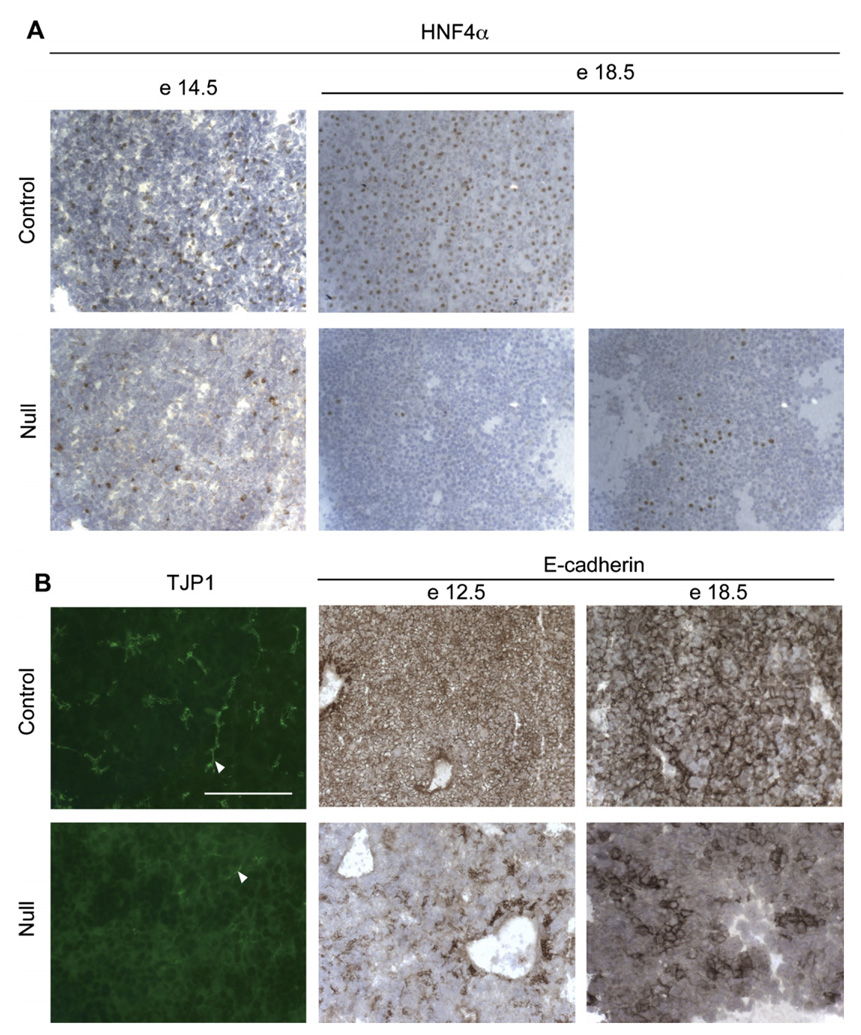
Short-term ex vivo culture of hepatic cells from e18.5 (embryonic day) HNF4α-null liver restores polarity and partially rescues expression of the E-cadherin gene. (A) Breeding scheme used to generate null (HNF4αfl/− AlfpCreTg/0) and littermate control (HNF4aαfl/+ AlfpCreTg/0). (B) Double immunofluorescence for tight junction protein 1 (TJP1/ZO1, green), and HNF4α (red) with nuclei visualized by DAPI (blue) staining, carried out at 3 and 7 days of culture. The “3 days” photomicrographs have bars to show vertical distribution of the fluorescence signals through the culture (z-stack). Immunostaining for HNF4α is intense in the control cells at 7 days, and a single stained nucleus has been captured for the null culture. (C) E-Cadherin transcripts, for control (open bars) and HNF4α-null (filled bars) e18.5 livers (left) and e18.5 three day cultures of hepatic cells (right).
2.2. Isolation and characterization of BMEL cells from livers of HNF4αfl/fl mice
Mice homozygous for allele HNF4α tm1.1Gonz(HNF4αfl/fl) in which exons 4 and 5 are flanked by loxP sites (described in [6]) were generated. Embryonic livers (e14.5) were removed and treated as described [14] to obtain BMEL cell lines. Embryo tails were genotyped by PCR as in [6].
2.3. Disruption of the HNF4α gene in BMEL cell lines by Cre-mediated recombination
Cloned BMEL lines were transfected with expression vectors (10 µg) encoding eGFPnlsCre-fusion protein or eGFP-alone, driven by the elongation factor1α promoter, and a selectable neomycin resistance cassette. Cells (3 × 106) were electroporated and selected in G418 (1.2 mg/ml). Isolated colonies were picked and expanded, DNA isolated, and analyzed for deletion efficiency. Control and null lines from the same clone of origin were compared. Three independent lines gave identical results; data from only one (26A1) are presented.
2.4. Test of BMEL cell lines for functional bile canaliculi
Dense BMEL cell cultures had three successive feedings with 0.04% Matrigel, then were given fluorescein diacetate (FDA) [15]. This color-less compounds penetrates into cells where cytoplasmic esterases cleave the diacetate to yield fluorescein. The treated cultures were immediately used for time-lapse fluorescence and phase-contrast image acquisition at 37 °C using a 10× objective and a Zeiss Axiovert 200M microscope.
2.5. Embryos for immunohistochemistry and RNA isolation
Staged embryos were collected and samples for immunohistochemistry frozen in OCT. Immunocytochemistry employed standard protocols using antibodies anti-HNF4α (A445), rat monoclonal anti-ZO1 (TJP1) and rabbit polyclonal anti-E-cadherin (Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were from Santa Cruz, Caltagor (Invitrogen, Carlsbad, CAl) or Molecular Probes (Invitrogen). Total RNA was isolated using Trizol reagent, treated with DNAse I and re-purified using Quiagen RNeasy columns (Quiagen, Courtaboeuf, France). For cell lines RNA was isolated directly using the R Neasy mini kit (Quiagen).
2.6. Immunofluorescent localization of proteins in cultured cells
Cells were cultured on collagen-I-coated culture wells, fixed with ice-cold methanol for 10 min or with methanol for 1 min, then 10 min with 3% paraformaldehyde, washed and incubated with anti-bodies. Bound antibody was detected with Texas-Red (Santa Cruz) or Alexa Fluor 594 (Molecular Probes)-conjugated secondary antibodies. Slides were analyzed on a Zeiss Axioplan 2 microscope fitted with a Zeiss ApoTome. Images were acquired using an Axiocam Hrc digital camera and processed using the Zeiss Axiovision 4.2 program.
2.7. BMEL cell injection into Alb-uPA/SCID mice
Pools of 3 clones of control or null cells were used. Experiments were conducted as described in [16]; 0.5 × 106 cells were injected into the spleen of anesthetized 4-week-old mice. Mice were sacrificed three weeks later and livers frozen in OCT. Serial sections were analyzed by immunohistochemistry using antibodies as detailed in [16]. BMEL (control and HNF4α-null) cells were detected using an anti-GFP antibody (Molecular Probes). Serial sections (10 µm) were stained using antibodies to bile duct-specific markers CK19 (Troma 3), a gift from R. Kemler (Max-Planck Institute of Immunobiology, Freiburg, Germany), CK7 (Progen, Heidelberg, Germany) or HNF1β (a gift from M. Yaniv, Institut Pasteur) or for the marker of epithelial polarity E-cadherin (Transduction Laboratories). Three mice were injected with control cells and nine mice with HNF4α-null cells.
2.8. Disruption of the HNF4α gene in the embryonic liver
HNF4α+/− mice were crossed with the AlfpCre transgenic line [17] and bred to produce HNF4α+/− AlfpCreTg/Tg males that were crossed with HNF4αfl/fl females in timed pregnancies. Genotyping was by PCR as described [6].
2.9. Real-time PCR quantitation of relative mRNA concentration
Reverse transcription reactions were performed using the cDNA archive kit (Applied Biosystems, Foster City CA) as recommended. Real-time PCR measurements were on an ABI sequence detector 7700 using ABI reagents. For SYBR green quantitation (Apoa1, Apoa4, Afp, CK19, c-kit, Hnf6, and Hnf1β) primers were designed using ABI Primer Express software and in all cases flank exons. Full primer lists are available on request. For Taqman quantitation (18s, Cdh1, Pck1, Tat, Mat1a, Osmr, Vegfc, Epo, Fah, Fos, Ddit3) primers and probes were purchased as Gene Expression Assay kits directly from ABI. The Osm assay was designed by ABI. PCR was performed in duplicate or triplicate on RT reactions and relative quantities calculated from a standard curve. Values were corrected to 18s and are expressed as concentration relative to the appropriate control. Histograms are of individual livers, or means of primary cultures from three livers. Statistics were performed using non-paired t-tests assuming equal variance.
3. Results
3.1. HNF4α-null hepatic cells in primary culture show apical expression of TJP1
Mice carrying null (−) or floxed (fl) alleles of HNF4α [6] were crossed with AlfpCre transgenic mice where Cre recombinase is active in the hepatic field beginning at e10 [17,18] to obtain HNF4α deficiency (Fig. 1A).
We prepared primary cultures from e18.5 HNF4α-null (HNF4α fl/−;AlpfpCreTg/0) and littermate control (HNF4α fl/+;AlpfpCreTg/0) livers and analyzed them after 3 and 7 days. In control cultures, expression of HNF4α was observed throughout, especially after 7 days. As shown in Fig. 1B, both control and HNF4α-deficient cultures showed membrane localized TJP1. Thus, HNF4α deficiency does not interfere with simple polarization of hepatic epithelial cells, unlike the in vivo findings of Parviz et al. [10] and Battle et al. [21]. We confirmed that E-cadherin expression is severely repressed in e18.5 hepatic tissue, but expression is substantially restored in the primary cultures. These results suggest that the deficient liver could suffer indirect effects (Fig. 1C).
3.2. HNF4α-null BMEL cells are polarized
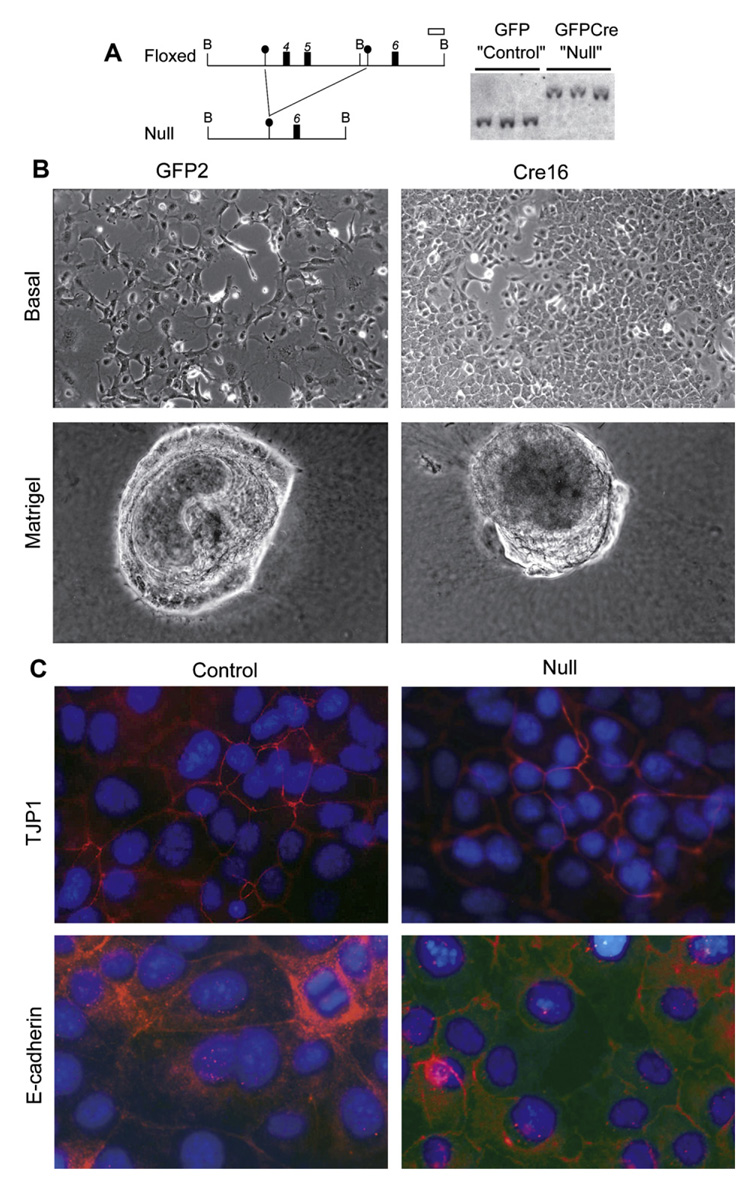
We isolated a series of bipotential mouse embryonic liver (BMEL) cell lines, using e14.5 livers from homozygous HNF4α floxed embryos. As previously shown [14], in basal culture conditions BMEL cells express liver-enriched transcription factors HNF4α, HNF1α and HNF3α but not liver-specific functions such as albumin or ApoA1. Cells cultured as aggregates express characteristic hepatocyte markers while culture in Matrigel™ induces hepatocyte and bile-duct markers. All HNF4αfl/fl cell lines tested showed the hallmarks of bipotentiality. They were transfected with vectors encoding a Cre recombinase-GFP-fusion protein, or only GFP. Stably transfected cells were selected for drug resistance conferred by the plasmid and isolated colonies expanded. HNF4α-null genotypes were verified by Southern blotting (Fig. 2A).
Fig. 2.
Morphology and cell polarity of bipotential mouse embryonic liver (BMEL) cell lines grown in basal or differentiation-inducing culture conditions is independent of HNF4α genotype. (A) Structure of the floxed and null alleles of HNF4α and Southern blot analysis of HNF4α-floxed BMEL cell lines stably transfected with vectors expressing GFP (green fluorescent protein) alone (GFP, Control) or a GFPnlsCre-fusion protein (GFPCre, null). Positions of BamHI sites (B), loxP sites (●), exons (■) and the probe (□) are represented. The HNF4α-floxed and -null alleles yield bands of 3.4 and 5.6 Kb, respectively. (B) Phase-contrast micrographs of cells grown under basal (proliferation) conditions and in Matrigel. (C) Immunofluorescent staining of tight junction protein 1 (TJP1) and E-cadherin.
Fig. 2B shows that control and HNF4α-null BMEL cells were indistinguishable in morphology: both presented mixed palmate/epithelial morphology at low density, and regular epithelial monolayer sheets at confluence. When grown on Matrigel, both types of cells formed aggregates with cystic structures, implying presence of epithelial cell polarity. Immunostaining for TJP1 and E-cadherin (Fig. 2C) showed similar expression and localization patterns in control and HNF4α-null BMEL cells, even though amounts were lower in mutant cells. Thus, clonal BMEL cell lines show indistinguishable growth and morphological properties in the presence or absence of HNF4α.
3.3. HNF4α-null BMEL cells can differentiate as hepatocytes in vitro
To judge differentiation as hepatocytes in culture, we employed the criterion of formation of bile canaliculi, a hepatocyte-specific structure specialized in bile secretion at the apical pole. In cultured hepatic cell lines, bile canaliculi form mainly as pockets between adjacent hepatocytes [15].
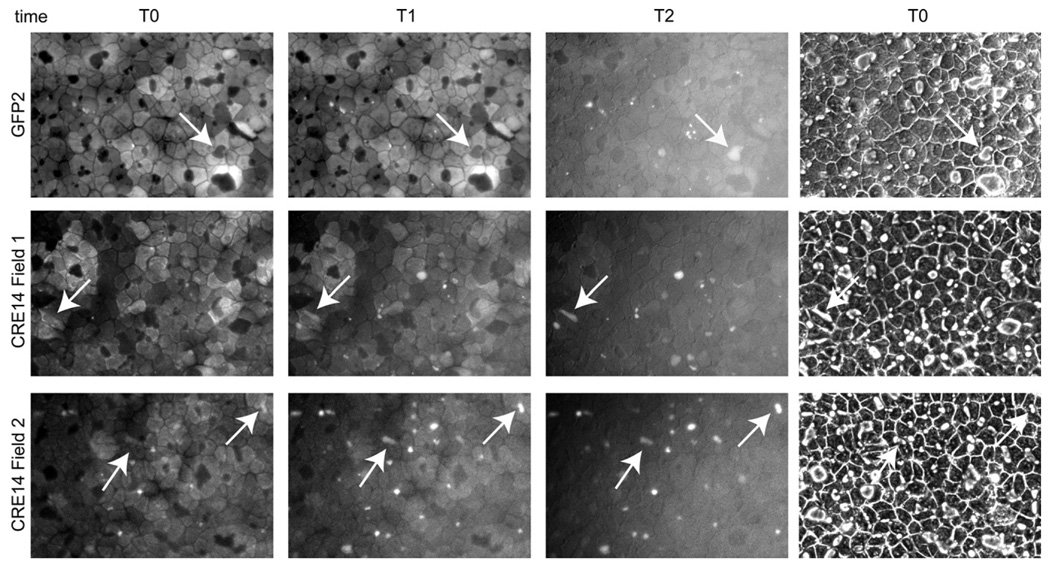
Under basal conditions, BMEL cells exhibit simple polarity of epithelial cells. If diluted Matrigel is added to a culture [13], the cells undergo morphogenesis to form bile canaliculi. To demonstrate functionality of the canaliculi, FDA (fluorescein diacetate) was given to the cells. The arrays of photos of Fig. 3 are each of a single field, at different times after addition of FDA: immediately after addition the cytoplasm is fluorescent, and thereafter the fluorescein is secreted via the apical surface and fills the bile canalicular “pockets”. In all cases, including the HNF4α-null cells, numerous active canaliculi were identified by the FDA test.
Fig. 3.
BMEL cells, whether they can express functional HNF4α or not, have the potential to form functional bile canalilculi. Frames illustrate loading of bile canalicular pockets with fluorescein during 0–12 min at 37 °C after addition of FDA, using the null clone Cre14 and control GFP2. At time 0 fluorescence is in the cytoplasm, and progressively disappears as the vesicles become fluorescent. Individual fields at successive times, with arrows identifying the evolution of fluorescence of bile canaliculi, that can be seen as spaces between the confluent cells in the phase contrast image (right). Images at T0 were captured as rapidly as possible after FDA was added to the cultures. T1 was 1 min later for Cre14 (Fields 1 and 2), and 2 min later for GFP2. T2 corresponds to 2 min for Cre14 and to 11 min for GFP2. (The rates of exchange differ from one field to another in a given dish; the differences in timing are not considered significant.)
3.4. HNF4α-null cells participate in liver regeneration, differentiating as bile ducts but not hepatocytes
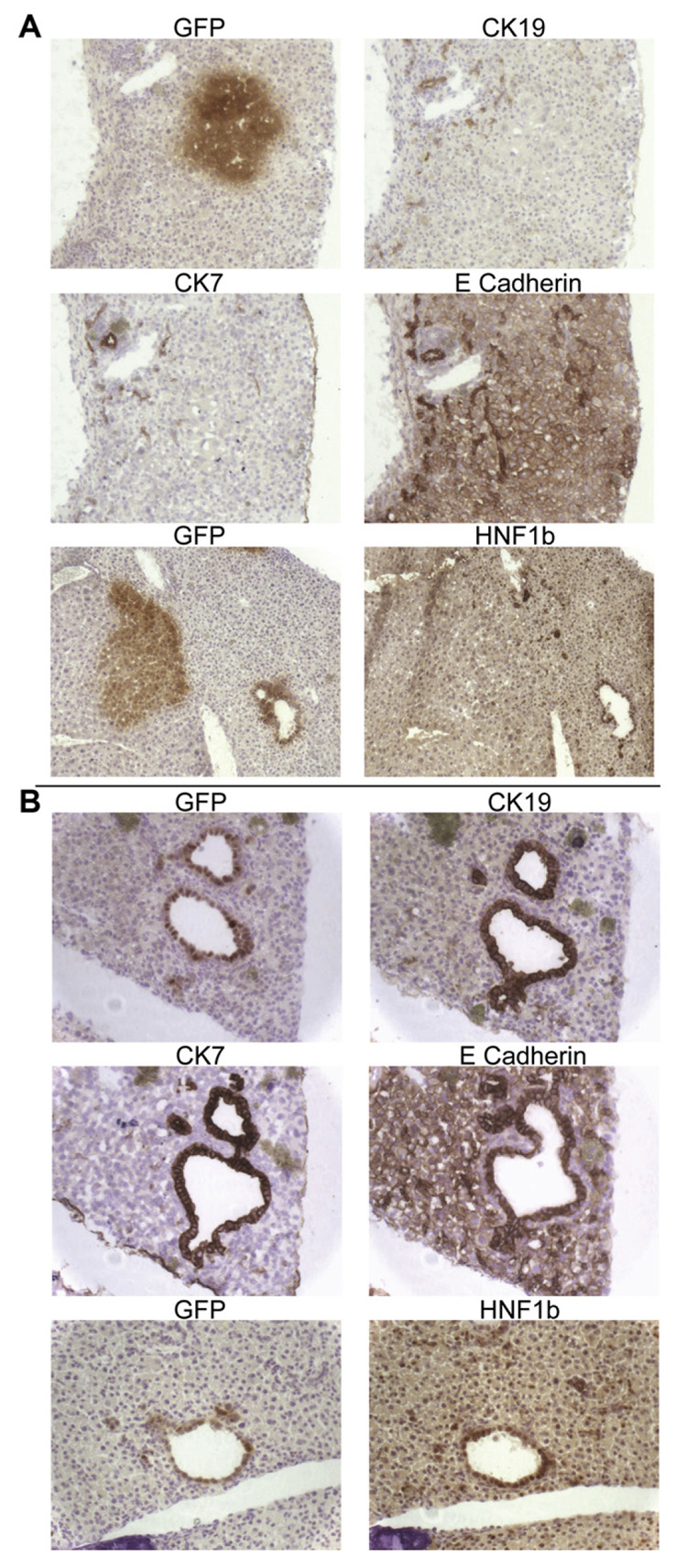
A stringent test of the differentiation potential of hepatic cells is their ability to participate in tissue repair. We employed competitive liver regeneration, using the Alb-uPA/SCID mouse model, where it was previously shown that BMEL cells form clusters of hepatocytes as well as bile ducts and can constitute up to 5% of the liver mass [16]. In this test, BMEL control cells indeed differentiated as hepatocytes and bile ducts (Fig. 4A), and expressed the appropriate markers: heaptocytes showed membrane-bound E-cadherin and nuclear HNF1β, but did not express the bile duct-specific markers CK7 and CK19. As expected, bile ducts stained for E-cadherin and for HNF1β (Fig. 4A).
Fig. 4.
Control and HNF4α-null BMEL cells participate in liver regeneration in the Alb-uPA/SCID mouse, forming polarized, differentiated bile ducts. (A) Liver sections of Alb-uPA/SCID (Albu-min-urokinase Plasminogen Activator trangene/Severe Combined Immunodeficiency) mice injected with HNF4α control BMEL cells. Immunohistochemistry (brown) of serial adjacent sections for GFP and for the bile duct-specific cytokeratins CK19 and CK7, and for E-cadherin, expressed in both hepatocytes and cholangiocytes. Two sections from a different area, stained, respectively, with GFP, highlighting both a cluster of BMEL hepatocytes and a bile duct. HNF1β nuclear staining is observed as expected in hepatocytes and especially in bile duct cells. (B) Alb-uPA/SCID mice transplanted with HNF4α-null BMEL cells. Four serial sections showing BMEL derived bile ducts stained with GFP, CK19, CK7 and E-cadherin. Two serial sections stained, respectively, with GFP and HNF1β.
In mice transplanted with HNF4α-null BMEL cells, injected cells participated only in reconstituting the bile duct cell lineage: no hepatocyte clusters were observed. HNF4α-null BMEL derived bile ducts were well differentiated and expressed CK7, CK19, and HNF1β and correctly localized E-cadherin (Fig. 4B). Thus, HNF4α is dispensable in vivo for morphogenesis of polarized bile ducts and for bile duct cell differentiation. The absence of clusters of GFP-positive hepatocytes in mice injected with the HNF4α-null BMEL cells could indicate inability of the deficient cells to differentiate into hepatocytes. Alternatively, the deficient cells differentiated as hepatocytes in vivo could have failed to thrive in the environment of competitive regeneration because HNF4α-target genes critical for hepatocyte metabolism were not optimally expressed. The fact that null cells are competent to form functional bile canaliculi in vitro (Fig. 3) leads us to favor the second alternative.
3.5. Quantitative PCR transcript analysis of embryonic livers and primary hepatic cultures indicates no loss of cell type identity
Fig. 5 shows a comparison of transcript levels for markers of cholangiocytes, and hepatocytes, including in the latter category some basal measurements for glucocorticoid inducible enzymes that show steadily increasing expression levels during liver development and are particularly abundant in adult hepatocytes. Results from embryonic livers at e14.5, 16.5 and 18.5 (Fig. 5A), and 3-day-old primary cultures from e18.5 livers (Fig. 5B) are shown. Among these functions, the promoters/enhancers of TAT, Pck1 and especially ApoAI are all known to harbor functional HNF4α binding sites, so diminished expression in the deficient hepatocytes is anticipated.
Fig. 5.
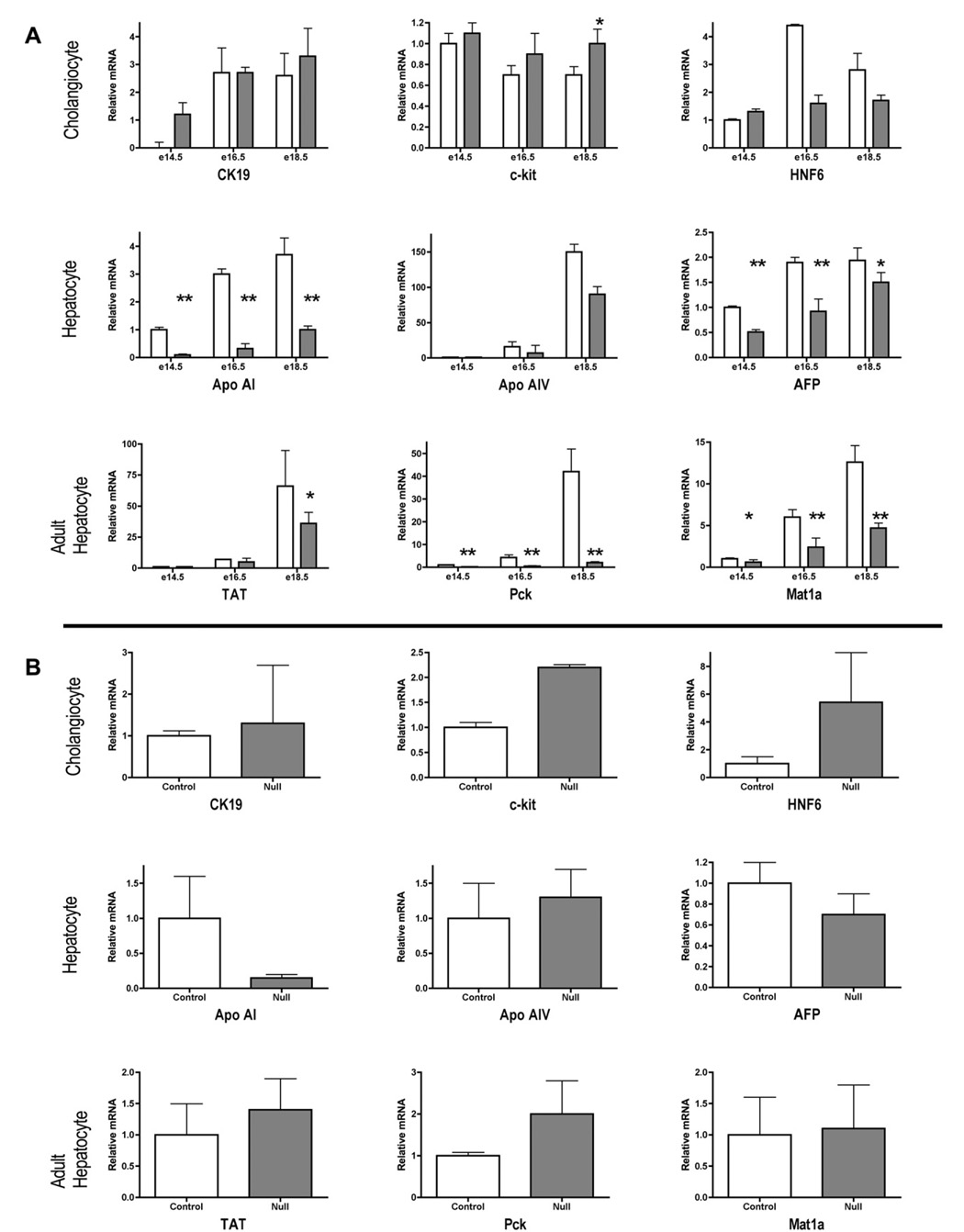
Gene expression patterns in HNF4α-null and control liver and primary hepatic cell cultures. Steady-state RNA concentrations in: (A) control (open bars) and HNF4α-null (filled bars) embryonic livers during development, and (B) 3 day primary cultures from e18.5 embryonic livers, as measured by real-time PCR. Cholangiocyte markers include a cytokeratin (CK19), a surface receptor (c-kit) and a transcription factor (HNF6). Hepatocyte markers include a serum protein (AFP, alpha-feto protein), genes required for lipid transport (Apoa1, apolipoprotein; Apoa4), and amino acid (Mat1a, methionine adenosyl transferase; TAT; tyrosine aminotransferase) and carbohydrate (Pck1, phospho(enol)pyruvate carboxykinase) metabolism. Values were corrected to 18s and are expressed relative to the mean value for control livers at e14.5 (±standard deviation, n = 4), or relative to control livers (standard deviation, n =≥3) in primary culture. Histograms are means of ≥3 livers or cultures from ≥3 different livers. Statistical comparisons of the effect of genotype on RNA concentrations in embryonic liver tissue were performed using unpaired, 2-tailed t-tests assuming equal variance. *≤60.05, ** ≤0.01.
As shown in Fig. 5A, for control and null livers of embryos of all three ages analyzed, the three cholangiocyte markers CK19, c-kit and HNF6 were expressed at similar levels. In contrast, all of the heaptocyte markers (except ApoA4, see [10]) showed a significant depression of transcript levels in the deficient tissue.
The same functions (Fig. 5B). were then examined in 3-day-old primary cultures prepared from control and null e18.5 embryos Here, as in the tissue, cholangiocyte markers were expressed at normal levels or were upregulated. Concerning hepatocyte functions, the known HNF4α dependent ApoAI gene was depressed as in the liver tissue, while others were comparable to (Apo-AIV and AFP) or even enhanced compared to the corresponding tissue (TAT, pck1 and Mat 1a). A similar re-expression of E-cadherin transcripts in primary cultures of HNF4α-deficient embryonic liver cells was documented in Fig. 1.
We conclude that in situ, HNF4α-deficient hepatocytes suffer a generalized depression of hepatocyte transcript levels.
3.6.The HNF4α-null allele presents the same characteristics as described [10]
Since the work here makes use of an HNF4α deficiency allele that differs from that of Parviz et al. [10], we verified that both alleles provoke the same phenotype, using the crosses depicted in Fig. 1A to generate deficient embryos. Fig. 6A shows immune-staining of HNF4α of e14.5 and e18.5 embryonic liver, revealing a progressive loss of HNF4α positive nuclei in the null tissue, as previously reported. Clonal proliferation of positive nuclei can occasionally be observed at e18.5 in null livers (Fig. 6A and see also [19]).
Fig. 6.
Disruption of allele HNF4αtm1.1Gonz in the developing mouse liver leads to defects in hepatic morphogenesis. Breeding scheme used to generate null (HNF4αfl/− AlfpCreTg/0) and littermate control (HNF4αfl/+ AlfpCreTg/0) embryos is depicted in Fig. 1A. (A) Immunohistochemistry for HNF4α on embryonic mouse liver (10 µm sections) at e14.5 and e18.5 stages of development. HNF4α positive nuclei stain brown. In occasional 18.5 day samples, clonal proliferation of cells that have escaped gene inactivation was observed. (B) Immunocytology of TJP1 (bar = 100 µm) and E-cadherin in control and HNF4α-null mouse liver at e12.5 (10×) and e18.5 (20×).
Finally, in confirmation of the results of [10], the deficient livers examined at E18.5 failed to show appropriate expression of TJP1 (Fig. 6B): in control liver the TJP1 staining outlines the canalicular network while in mutant liver it is mainly cytoplasmic. E-Cadherin staining of e12.5 and 18.5 livers is shown. At e12.5 for both genotypes, regions of intense E-cadherin staining represent ductal plate areas around veins, but only the controls show homogeneous staining of hepatoblasts. As development progressed, the differences between wild-type and deficient tissues intensified, as expected from increasing complexity of the hepatic transcription factor network during liver development [20] as well as from the stage dependent enhancement of hepatocyte metabolic enzyme expression (Fig. 5A). In wild-type embryos, all hepatocytes showed membrane E-cadherin staining while only small clusters of positive cells were visible in the mutant tissue.
Furthermore, Q-PCR analysis of liver-specific genes previously described to be either unaffected or under-expressed in the null tissue show the same patterns of transcript accumulation in the two types of HNF4α deficiency alleles (Fig. 5 and data in [10,21]). We conclude that the two HNF4α deficiency alleles confer the same phenotype.
3.7.Transcript analysis reveals the existence of a stress response in null livers
The differences in behavior of HNF4α -null embryonic liver cells in situ and in culture is consistent with the idea that non-cell autonomous effects, due to the absence of HNF4α, could be responsible for the severe in vivo phenotype. We therefore analyzed transcripts whose mis-expression could confer pathology on the HNF4α-deficient tissue. Genes were chosen to test for expression of essential metabolic functions of the hepatocyte, a generalized stress response, and molecules involved in signaling among hepatic cell types.
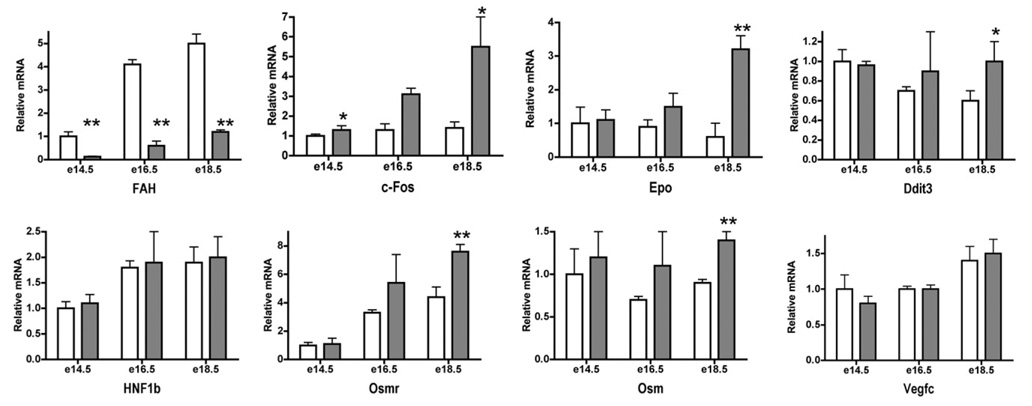
We began by analysis of FAH, an hepatocyte-enriched enzyme of tyrosine metabolism, whose absence causes accumulation of toxic intermediates in deficient mouse liver and a non-specific block to liver differentiation in embryos [22–24]. In addition, deficient mouse liver shows profound ultrastructural abnormalities of the ER and Golgi [25]. FAH transcript levels were significantly depressed at all times (Fig. 7), indicating that it could be one of the many metabolically critical targets of HNF4α. At later times, c-Fos, an immediate early marker of oxidative stress, is enhanced in livers of HNF4α-null embryos, while increased levels of Epo are indicative of hypoxia. These stress responses are pathway-specific as we detected no differences in expression of the DNA-damage-induced gene Ddit3.
Fig. 7.
Gene expression patterns in HNF4α-null embryonic liver indicate multiple metabolic defects and late-onset cellular responses to stress and hypoxia. Steady-state RNA concentrations in control (open bars) and HNF4α-null (filled bars) embryonic livers during development as measured by real-time PCR. First row: an essential metabolic enzyme of the liver (FAH, fumerylacetoacetate hydrolase) and markers of cellular stress (c-Fos), hypoxia (Epo, Erythropoietein) and DNA damage (Ddit3, DNA damage inducible). Second row: markers of the major cell lineages in the developing liver: hepatoblasts (HNF1β, Osmr, oncostatin M receptor), hematopoietic stem cells (Osm) and angioblasts (Vegfc, vascular endothelial growth factor). Values were corrected to 18s and are expressed relative to the mean value for control livers at e14.5 (±standard deviation. n = 4). See legend to Fig. 5 for statistical analysis.
We saw no major differences in expression of markers of the major cell lineages in the developing liver (Fig. 7): hepatoblasts (HNF1β, Osmr), hematopoietic stem cells (Osm) or angioblasts (Vegfc), consistent with the data of [10], which showed that disruption of the HNF4α gene in the developing liver does not alter the cellular composition.
Taken together our data indicate that while HNF4α-null hepatic cells are competent to undergo epithelialization, loss of HNF4α-controlled metabolic functions in vivo may induce a stress response that interferes with liver morphogenesis.
4. Discussion
It is not a simple matter to elucidate the developmental role of a transcription factor, because its targets may include genes not only directly implicated in morphogenesis, but also genes required to insure an environment that is permissive for morphogenesis. We have attempted to discriminate between these possibilities concerning the role of HNF4α in liver development. Although it has been described [10,21] that this transcription factor is necessary for hepatic morphogenesis and cellular polarity, we hypothesized that absence of HNF4α, a pivotal factor in specialized metabolism of the hepatocyte, could lead to a stress response of the tissue, similar to the phenotype associated with deficiency of the hepatocyte-specific enzyme FAH [22–24]. Our first prediction was that stressed HNF4α-deficient hepatic cells should present a normalized phenotype in primary culture, where culture medium would provide essential nutrients and dilute soluble toxic metabolites. Further, hepatic bipotential cell lines rendered deficient in HNF4α compared to their controls could provide a test for traits that are direct consequences of HNF4α deficiency.
HNF4α-deficient embryos present abnormal liver architecture, highlighted by absence of cell polarization and deficiency in E-cadherin expression [10]. However, in primary culture, the hepatic cells acquire rapidly the normal apical localization of tight junction protein TJP1. Furthermore, E-cadherin transcripts, severely repressed in vivo, are increased to about 60% of the values of control liver cultures. Together, these results indicate that lack of epithelialization in vivo is not a direct effect of the absence of HNF4α. We next established BMEL cell lines from embryos containing floxed HNF4α alleles, induced deletion and examined morphogenetic potential of the resulting cells. By all criteria, the null and floxed control cell lines were indistinguishable in vitro. Pools of clones of both genotypes were injected into Alb-PA/SCID mice to test their capacity to participate in liver regeneration. Control cells were identified as hepatocyte clusters and bile ducts, while the null cells were found only as bile ducts. If this result were due to inability of HNF4α-deficient cells to differentiate as hepatocytes, they should also fail to form hepatocytes in vitro. When we tested capacity of the control and null BMEL cells to form functional bile canaliculi, a highly specialized apical membranous structure, the cell lines of the two genotypes were indistinguishable. We conclude that HNF4α-deficient cells remain competent to differentiate as hepatocytes in vitro.
These data suggest that the observed in vivo defects in hepatocyte polarity are non-cell autonomous. The loss of expression of metabolic genes controlled by HNF4α could be the mechanism underlying this phenotype. Indeed, it is known that phenotypes described for HNF4α-deficient adult liver, including perturbations of bile acid homeostasis [6] as well as cholesterol depletion, can induce loss of hepatocyte polarity [26]. We propose that HNF4α-null embryonic liver suffers metabolic disturbances which indirectly cause a block to differentiation. In these livers we observe enhanced expression of c-fos and Epo transcripts, indicative of a stress response.
Our observation that HNF4α is not necessary for epithelialization of hepatic precursors seems inconsistent with the demonstration that forced expression of this factor promotes epithelialization in various cell lines [10–12]. However, while over-expression assays have been widely used to identify candidate functions for gene products, results obtained by targeted mutagenesis in the mouse may show that conclusions drawn from such experiments require reassessment. For example, the initial analysis by over-expression of bHLH myogenic factors indicated that they were “determination” genes, competent to effect myogenic conversion of recipient cells [27]. It is now appreciated that family members fulfill distinct roles and no single factor is sufficient for normal muscle formation in the embryo [28,29]. Hence, a given factor may be sufficient in a forced expression assay but not necessary in vivo.
Our data broadly caution the use of knockout phenotype, taken alone, as instructive of gene function, especially in tissues that fulfill a metabolic function. We conclude with a quotation from a 1993 review paper of the search for a liver differentiation gene to explain the hepatic phenotype of a deletion allele of lethal albino mice, later revealed to be due to absence of the liver-specific metabolic enzyme FAH: “It is unlikely that lethal albino mice will be the last case in which absence of an enzyme of intermediary metabolism will lead to the disturbance of a differentiation process” [30].
Acknowledgements
We thank all members of the Plateforme d’Imagerie Dynamique, Pasteur Institute, and particularly Emmanuelle Perret for their help with image acquisition, Francois Tronche and Lionel Gresh for the Alfp-Cre mice, Francey Sladek and Doris Cassio for HNF4α and ZO1 antibodies, respectively, Margaret Buckingham, and all members of the Unité de Génétique de la Différenciation for advice and constructive criticism. This work was supported by the Action Thématique Concertée Biotherapies Program of the IN-SERM, by the Minisère de l’Education National, de l’Enseignement Supérieur et de la Recherche, and by the Grand Programme Horizontal 7 (Stem Cells) of the Institut Pasteur, which also supported H.S.-M., and by an agreement with Baylor College of Medicine/Institut Pasteur, which in turn was supported by Grant 1 U01 DK63588-01 from the National Institutes of Health (coordinator G. Darlington)
Footnotes
NIH funded study (Grant 1 U01 DK63588-01). The authors declare that they do not have anything to disclose regarding funding from industries or conflict of interest with respect to this manuscript.
References
- 1.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 2.Hansen SK, Parrizas M, Jensen ML, Pruhova S, Ek J, Boj SF, et al. Genetic evidence that HNF-1alpha-dependent transcriptional control of HNF-4alpha is essential for human pancreatic beta cell function. J Clin Invest. 2002;110:827–833. doi: 10.1172/JCI15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gragnoli C, Lindner T, Cockburn BN, Kaisaki PJ, Gragnoli F, Marozzi G, et al. Maturity-onset diabetes of the young due to a mutation in the hepatocyte nuclear factor-4 alpha binding site in the promoter of the hepatocyte nuclear factor-1 alpha gene. Diabetes. 1997;46:1648–1651. doi: 10.2337/diacare.46.10.1648. [DOI] [PubMed] [Google Scholar]
- 4.Reijnen MJ, Sladek FM, Bertina RM, Reitsma PH. Disruption of a binding site for hepatocyte nuclear factor 4 results in hemophilia B Leyden. Proc Natl Acad Sci USA. 1992;89:6300–6303. doi: 10.1073/pnas.89.14.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm:tetraploid rescue of Hnf-4(−/−) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- 6.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Repa JJ, Inoue Y, Hayhurst GP, Gonzalez FJ, Mangelsdorf DJ. Identification of a liver-specific uridine phosphorylase that is regulated by multiple lipid-sensing nuclear receptors. Mol Endocrinol. 2004;18:851–862. doi: 10.1210/me.2003-0285. [DOI] [PubMed] [Google Scholar]
- 8.Inoue Y, Hayhurst GP, Inoue J, Mori M, Gonzalez FJ. Defective ureagenesis in mice carrying a liver-specific disruption of hepatocyte nuclear factor 4alpha (HNF4alpha). HNF4alpha regulates ornithine transcarbamylase in vivo. J Biol Chem. 2002;277:25257–25265. doi: 10.1074/jbc.M203126200. [DOI] [PubMed] [Google Scholar]
- 9.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 11.Chiba H, Gotoh T, Kojima T, Satohisa S, Kikuchi K, Osanai M, et al. Hepatocyte nuclear factor (HNF)-4alpha triggers formation of functional tight junctions and establishment of polarized epithelial morphology in F9 embryonal carcinoma cells. Exp Cell Res. 2003;286:288–297. doi: 10.1016/s0014-4827(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 12.Spath GF, Weiss MC. Hepatocyte nuclear factor 4 provokes expression of epithelial marker genes, acting as a morphogen in dedifferentiated hepatoma cells. J Cell Biol. 1998;140:935–946. doi: 10.1083/jcb.140.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fougere-Deschatrette C, Imaizumi-Scherrer T, Strick-Marchand H, Morosan S, Charneau P, Kremsdorf D, et al. Plasticity of hepatic cell differentiation: bipotential adult mouse liver clonal cell lines competent to differentiate in vitro and in vivo. Stem Cells. 2006;24:2098–2109. doi: 10.1634/stemcells.2006-0009. [DOI] [PubMed] [Google Scholar]
- 14.Strick-Marchand H, Weiss MC. Inducible differentiation and morphogenesis of bipotential liver cell lines from wild-type mouse embryos. Hepatology. 2002;36:794–804. doi: 10.1053/jhep.2002.36123. [DOI] [PubMed] [Google Scholar]
- 15.Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, et al. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J Cell Biol. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strick-Marchand H, Morosan S, Charneau P, Kremsdorf D, Weiss MC. Bipotential mouse embryonic liver stem cell lines contribute to liver regeneration and differentiate as bile ducts and hepatocytes. Proc Natl Acad Sci USA. 2004;101:8360–8365. doi: 10.1073/pnas.0401092101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellendonk C, Opherk C, Anlag K, Schutz G, Tronche F. Hepatocyte-specific expression of Cre recombinase. Genesis. 2000;26:151–153. doi: 10.1002/(sici)1526-968x(200002)26:2<151::aid-gene17>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 18.Coffinier C, Gresh L, Fiette L, Tronche F, Schutz G, Babinet C, et al. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development. 2002;129:1829–1838. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- 19.Stanulovic VS, Kyrmizi I, Kruithof-de Julio M, Hoogenkamp M, Vermeulen JL, Ruijter JM, et al. Hepatic HNF4alpha deficiency induces periportal expression of glutamine synthetase and other pericentral enzymes. Hepatology. 2007;45:433–444. doi: 10.1002/hep.21456. [DOI] [PubMed] [Google Scholar]
- 20.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, et al. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci USA. 2006;103:8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trigg MJ, Gluecksohn-Waelsch S. Ultrastructural basis of biochemical effects in a series of lethal alleles in the mouse. Neonatal and developmental studies. J Cell Biol. 1973;58:549–563. doi: 10.1083/jcb.58.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruppert S, Kelsey G, Schedl A, Schmid E, Thies E, Schutz G. Deficiency of an enzyme of tyrosine metabolism underlies altered gene expression in newborn liver of lethal albino mice. Genes Dev. 1992;6:1430–1443. doi: 10.1101/gad.6.8.1430. [DOI] [PubMed] [Google Scholar]
- 24.Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- 25.Kelsey G, Ruppert S, Beermann F, Grund C, Tanguay RM, Schutz G. Rescue of mice homozygous for lethal albino deletions: implications for an animal model for the human liver disease tyrosinemia type 1. Genes Dev. 1993;7:2285–2297. doi: 10.1101/gad.7.12a.2285. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Boyer JL. The maintenance and generation of membrane polarity in hepatocytes. Hepatology. 2004;39:892–899. doi: 10.1002/hep.20039. [DOI] [PubMed] [Google Scholar]
- 27.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 28.Tajbakhsh S, Buckingham M. The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr Top Dev Biol. 2000;48:225–268. doi: 10.1016/s0070-2153(08)60758-9. [DOI] [PubMed] [Google Scholar]
- 29.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, et al. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 30.Kelsey G, Schutz G. Lessons from lethal albino mice. Curr Opin Genet Dev. 1993;3:259–264. doi: 10.1016/0959-437x(93)90032-k. [DOI] [PubMed] [Google Scholar]