Abstract
Chemotherapeutic alkylating agents, such as bifunctional nitrogen mustards and cisplatins, generate interstrand DNA crosslinks that inhibit cell proliferation by arresting DNA transcription and replication. A synthetic N4C-ethyl-N4C interstrand crosslink between opposing cytidines mimics the DNA damage produced by this class of clinically important compounds, and can be synthesized in large quantities to study the repair, physical properties, and structures of these DNA adducts. The X-ray structure of a DNA duplex d(CCAAC*GTTGG)2 containing a synthetic N4C-ethyl-N4C interstrand crosslink between the cytosines of the central CpG step (*) has been determined at 1.65 Å resolution. This structure reveals that the ethyl crosslink in the CpG major groove does not significantly disrupt the B-form DNA helix. Comparison of the N4C-ethyl-N 4C crosslinked structure with the structure of an uncrosslinked oligonucleotide of the same sequence reveals that the crosslink selectively stabilizes a pre-existing alternative conformation. The conformation preferred by the crosslinked DNA is constrained by the geometry of the ethyl group bridging the cytosine amines. Characteristics of the crosslinked CpG step include subtle differences in the roll of the base pairs, optimized Watson-Crick hydrogen bonds, and loss of a divalent cation binding site. Given that the N4C-ethyl-N4C crosslink stabilizes a pre-existing conformation of the CpG step, this synthetically accessible substrate presents an ideal model system for studying the genomic effects of covalently coupling the DNA strands, independent of gross alterations in DNA structure.
Keywords: DNA damage, DNA repair, alternative conformations, interstrand crosslink
DNA crosslinking agents, including bischloroethylnitroso ureas, cisplatins, and nitrogen mustards such as melphalan and oxazaphosphorine are widely-prescribed as chemotherapeutic treatments for cancers. These chemotherapeutics exert their toxic effects by forming interstrand DNA crosslinks, which block the essential processes of transcription and replication by preventing separation of the DNA strands (reviewed in (1, 2)). In addition, unsolicited DNA interstrand crosslinks may arise naturally from unavoidable exposure to endogenous compounds such as the products of lipid peroxidation (3), or environmentally derived compounds such as formaldehyde (4). Cells have developed a variety of mechanisms, including homologous recombination and nucleotide excision repair (NER), to identify and repair interstrand DNA crosslinks. Accordingly, enhanced DNA repair plays a major role in the resistance of certain cancers to chemotherapy, for example melphalan-resistant multiple myeloma (5) or oxazaphosphorine-resistant medulloblastoma (6). Given that failed cancer treatment often results from drug resistance, a clear understanding of the molecular mechanisms involved in the recognition and repair of DNA interstrand crosslinks would facilitate the development of more effective chemotherapeutic agents.
Interstrand crosslinks are only a minor percentage (1-5%) of the adducts formed by most of DNA alkylating agents, although interstrand DNA crosslinks are by far the most toxic (7, 8), and therefore the most physiologically relevant adduct for investigation. The low availability of defined DNA crosslinks is a roadblock to studying their structure and repair. Using an approach in which the crosslink is introduced during solid phase DNA synthesis, short DNA duplexes containing N4C-alkyl-N4C interstrand crosslinks have been synthesized in quantities sufficient for detailed structural and mechanistic investigation (9-12). These synthetic N4C-alkyl-N4C interstrand crosslinks provide a well-defined mimic of the DNA damage introduced by therapeutic DNA alkylating agents.
DNA interstrand crosslinks produced by different chemical agents induce a variety of structural changes in DNA (2). This structural diversity could lead to differences in the mechanism and effectiveness of repair, and hence to the physiological activity of the DNA crosslinking agent. The relationship between structure and repair of interstrand crosslinked DNA was investigated by studying two different orientations of the same N4C-ethyl-N4C interstrand crosslink, connecting cytidines in a single CpG versus GpC step of double-stranded DNA (13). Both crosslink orientations were repaired efficiently in wild-type and homologous-recombination deficient E. coli. However, a functional NER pathway was required for the crosslinked CpG step to be repaired efficiently, whereas the crosslinked GpC step was repaired efficiently regardless of the status of the NER pathway. Proton chemical shifts coupled with distance-restrained molecular dynamic regularization established that the structure of a DNA duplex containing the N4C-ethyl-N4C crosslink at a CpG step remains undistorted. In contrast, an oligonucleotide in which the orientation of the crosslink was reversed to a GpC step behaves poorly in NMR experiments. Moreover, atomic force microscopy experiments demonstrated that longer DNAs containing the crosslinked GpC step are more flexible than the CpG counterpart. The correlated differences between the structure and repair of the two crosslink orientations suggest that undistorted DNA crosslinks are recognized and repaired in a different manner from those lesions that grossly alter the overall DNA conformation.
Given the importance of correlating the structures of damaged DNAs with their mechanisms of repair, here we present a high resolution view of an important model system for studying DNA repair, an oligonucleotide containing a synthetic N4C-ethyl-N4C intrastrand crosslink at a CpG step. The X-ray structure of a palindromic DNA oligonucleotide,d(CCAAC*GTTGG) 2, was determined at 1.65 Å resolution (*, crosslink). Comparison with an atomic (1.0 Å) resolution structure of the uncrosslinked DNA counterpart (14) reveals that conformational details, which could mark the crosslinked DNA duplex for recognition by DNA repair factors, instead pre-exist in the unmodified structure.
MATERIALS AND METHODS
Crosslinked DNA Synthesis and Crystallization
The DNA decamer containing the N4C-ethyl-N4C interstrand crosslink was synthesized, purified using strong anion-exchange HPLC, and analyzed using mass spectrometry using previously described procedures (12). The purified crosslinked DNA was then desalted using C-18 Sep-Pak cartridges (Waters), and dissolved in a buffer containing 22 mM ammonium acetate, 11 mM tris-hydroxymethyl-aminomethane (Tris)-HCl pH 8.0, and 11 mM calcium acetate. Crystals were grown using the sitting drop vapor diffusion method at 4 °C, in which 3 μL of a reservoir solution containing 27% 2-3 methyl-pentadiol, 115 mM calcium acetate, and 10 mM Tris-HCl pH 8.0 was added to an equal volume of DNA solution, and the mixture was equilibrated against 700 μL of the reservoir solution. Crystals first appeared after two weeks. Prior to data collection, crystals were flash-cooled to liquid nitrogen temperatures using 35% MPD as a cryoprotectant.
X-ray Data Collection and Structure Determination
X-ray data from a single crystal were collected at a wavelength of 1.54 Å, using a Rigaku rotating anode generator and Raxis-IV image plate detector at the Johns Hopkins School of Medicine (Table 1). The space group and unit cell dimensions were C2, a=31.0 Å, b=24.9 Å, c=34.4 Å, β=114.5°, with one DNA strand per asymmetric unit of the crystal. Since this crystal form was isomorphous with the uncrosslinked oligonucleotide of identical sequence (14), the structure was solved by difference Fourier methods starting with the coordinates of conformation ‘A’ of the uncrosslinked counterpart (PDB code 1EN8).
Table 1.
Crystallographic data and refinement statistics†
| Resolution Limit (Å) | 20.00-1.65 |
| Redundancy | 4.0 |
| Completeness (%) | 96.6 (79.4) |
| Rsym‡ (%) | 4.6 (4.7) |
| I/σ(I) | 38.2 (25.8) |
| Number of DNA Atoms | 203 |
| Number of Water Atoms | 87 |
| Number of Calcium Ions | 3 |
| RMSD Bond Lengths | 0.01 Å |
| RMSD Bond Angles | 1.13° |
| Average Temperature Factor | 14.1 Å2 |
| Rcryst§ | 18.2% |
| Rfree§ | 20.8% |
Values in parenthesis are for the highest resolution shell, 1.65-1.71Å.
where Ii is an intensity I for the ith measurement of a reflection with indices hkl and <I> is the weighted mean of all measurements of I.
for the working set of reflections, Rfree is Rcryst for 5.3% of the reflections excluded from the refinement.
A difference Fourier electron density map revealed well-defined electron density for the two methylene carbons of the alkyl interstrand crosslink (Figure 1), which are related by the twofold symmetry axis of the crystal. The methylene carbon was placed manually using the program O (15). The topology and parameter files for the crosslinked cystosine were generated using XPLO2D of the Uppsala Software Factory, then modified manually for chemically reasonable restraints (16). The structure was refined using CNSsolve v.1.1 (17). Three water molecules were reassigned as Ca+2 ions based on coordination geometry and unusually low temperature factors. Final refinement statistics are given in Table 1. The SigmaA-estimated coordinate error of the structure is 0.15 Å (18). Global helical parameters were calculated using the program Curves v5.1 (19), and figures were made using the Pymol molecular graphics system (htttp://www.pymol.org).
Figure 1.
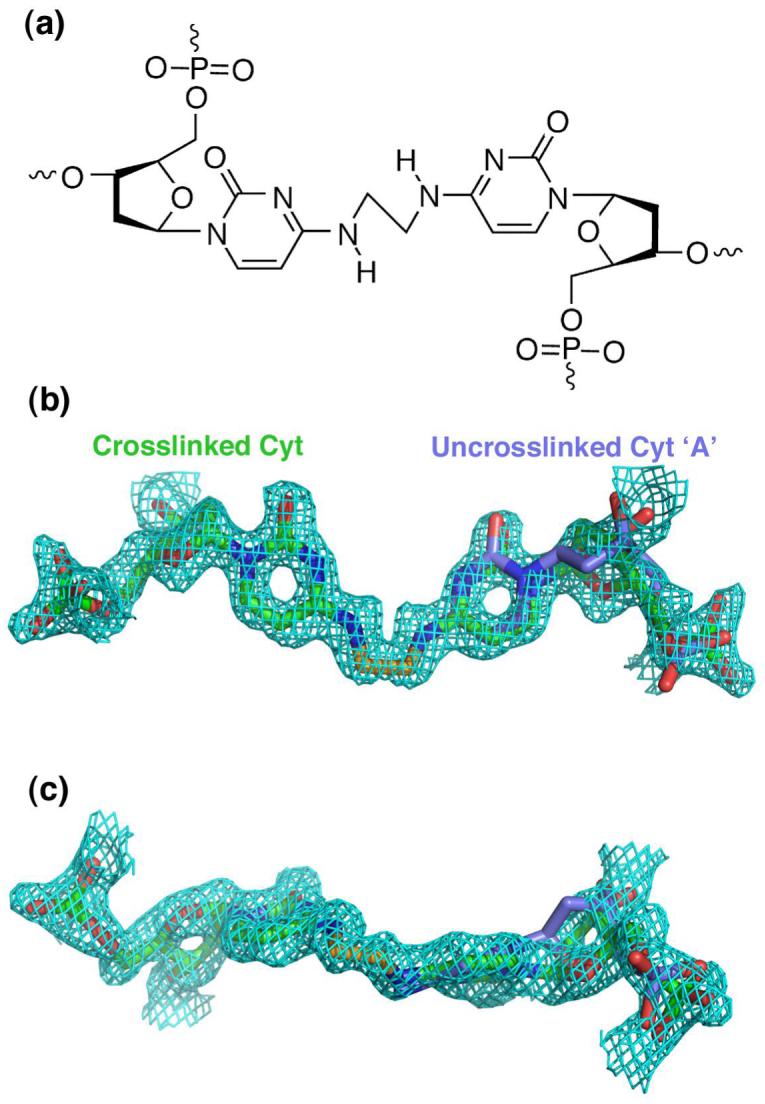
(a) Chemical structure of the N4C-ethyl-N 4C crosslinked cytidines. (b) A 2|Fo| - |Fc| composite omit electron density map, shown at 1 contour level around the crosslinked cytidines. For comparison, conformation ‘A’ of the uncrosslinked DNA structure (colored blue) is overlaid on one of the two crosslinked cytidines (colored green), which are related by crystallographic symmetry. (c) Same as in (b), but rotated 90° about the horizontal axis.
RESULTS
Overall Structure
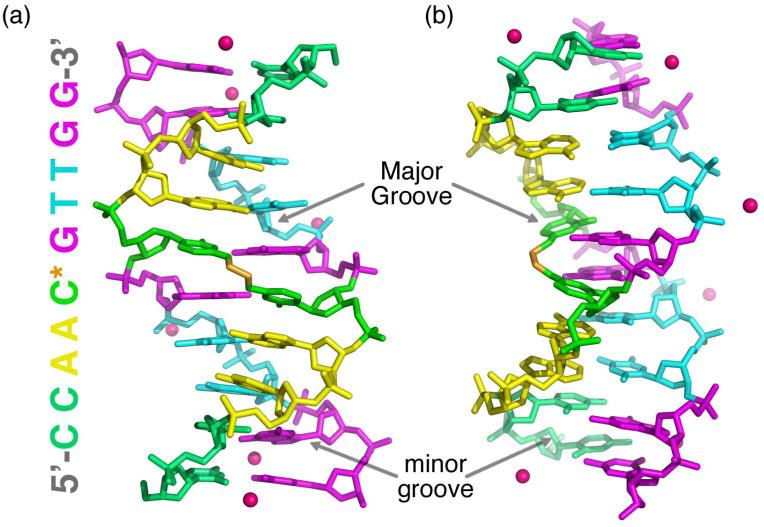
The X-ray structure of the N4C-ethyl-N4C interstrand crosslinked DNA decamer of sequence d(CCAAC*GTTGG) 2 was determined by difference Fourier analysis starting with the coordinates of the uncrosslinked DNA counterpart (14). The structure of the crosslinked DNA was refined to a final Rcryst of 18.7% and Rfree of 20.2% at 1.65 Å resolution, including 87 water molecules and three Ca+2 ions (Table 1). Since the palindromic strands of the DNA duplex are related by crystallographic symmetry, the two halves of the DNA duplex share identical conformations (Figure 2).
Figure 2.
Overall X-ray structure of a DNA duplex d(CCAAC*GTTGG)2 with the N4C-ethyl-N 4C interstrand crosslink between the cytosines of the central CpG step (*).Cyt, green; Thy, cyan; Gua, magenta; Ade, yellow. The ethyl crosslink is colored gold. The two strands of the double helix are related by crystallographic symmetry. The orientation in (b) is rotated 90° about the vertical axis relative to (a). The DNA sequence is shown to the left.
Overall, the N4C-ethyl-N 4C interstrand crosslink is accommodated in the major groove of an undistorted B-form DNA helix (Figure 2). The average twist, rise, and sugar pucker of the crosslinked duplex are 35°, 3.32 Å, and C2′-endo, respectively, compared with the standard B-DNA twist, rise, and sugar pucker of 36°, 3.37 Å, and C3′-exo/C2′-endo (20). A locally overwound, helical twist (40°) at the CpG step is compensated by underwound flanking ApC and GpT steps (26°). This slight deviation from the ideal B-form prototype is unlikely to result from the N4C-ethyl-N4C interstrand crosslink, since it is shared by its uncrosslinked DNA counterpart (Table 2).
Table 2.
Comparison of helical parameters
| (a) Helical parameters with similarity between crosslinked and uncrosslinked -CG- conformations | |||
|---|---|---|---|
| Crosslinked Cyt·Gua | Uncrosslinked Cyt·Gua, Minor Conformation | Uncrosslinked Cyt·Gua, Major Conformation | |
| Rise (Å) | 3.4 | 3.5 | 3.5 |
| Twist (°) | 40 | 45 | 40 |
| Propeller twist (°) | -21 | -26 | -5 |
| Buckle (°) | -5 | -5 | 3 |
| Phase (Sugar pucker) (°) | 52 (C4′-exo) | 24 (C3′-endo) | 152 (C4′-endo) |
| Shift (Å) | 0.5 | 0.45 | 0.1 |
| Slide (Å) | 0 | 0 | 0.5 |
| Tilt (°) | 7.0 | 9.8 | 1.8 |
| (b) Helical parameters with differences between crosslinked and uncrosslinked -CG- conformations | |||
|---|---|---|---|
| Crosslinked Cyt·Gua | Uncrosslinked Cyt·Gua, Minor Conformation | Uncrosslinked Cyt·Gua, Major Conformation | |
| Roll (°) | 12.7 | -2.2 | -0.2 |
| Shear (Å) | 0.1 | -0.3 | 0.2 |
| Stretch (Å) | 0.0 | 0.5 | 0.2 |
| Stagger (Å) | 0.0 | 0.2 | -0.4 |
| (c) Distances between atoms of the Cyt·Gua base pair (Å) | |||
|---|---|---|---|
| Crosslinked Cyt·Gua | Uncrosslinked Cyt·Gua, Minor Conformation | Uncrosslinked Cyt·Gua, Major Conformation | |
| Cyt-N4:Cyt-N4 | 2.8 | 3.1 | 3.8 |
| Cyt-O2:Gua-N2 | 2.9 | 3.1 | 3.3 |
| Cyt-N3:Gua-N1 | 2.9 | 3.1 | 3.3 |
| Cyt-N4:Gua-N6 | 2.9 | 3.0 | 3.3 |
Structure of the N4C-ethyl-N4C Crosslink
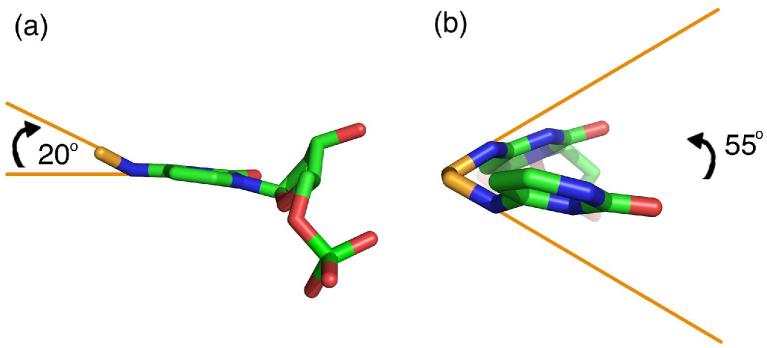
The exocyclic amines (N4) of the central cytosines are covalently linked by the two methylene carbons (CX) of the intervening ethyl group (Figure 3). Each of these carbons replace one of hydrogen atom of the amino group, which would normally be presented in the major groove of the DNA duplex for recognition by protein molecules or interaction with solvent. Based on the observation of optimal hydrogen bond distances between the cytosine-N4 and guanine-O6 atoms (Table 2(c)), the other hydrogen atom of the amino group presumably remains undisturbed by the crosslink. Conventionally, the cytosine exocyclic amine and its attached hydrogen atoms are considered to be sp2-hybridized, and coplanar with the base (21, 22). However, each methylene carbon in the N4C-ethyl-N4C crosslink is slightly rotated out of the plane of the cytosine (20° from base plane, 0.45 Å shortest distance from CX to base plane calculated using Geomcalc (23)) (Figure 3(a)). This rotation allows the N4-CX—CX-N4 torsion of the N4C-ethyl-N4C group to adopt a staggered, rather than unfavorably overlapped, conformer, with a 55° angle between N4 atoms when observed down the CX—CX bond (Figure 3(b)).
Figure 3.
Close views of the crosslinked cytosines, shown as ball-and-stick diagrams and colored by atom: ethyl crosslink, gold; carbon, green; nitrogen, blue; oxygen, red. (a) Viewed along the plane of the base. (b) View in the orientation of a Newman projection, along the CX-CX bond.
The rotation of the carbons of the ethyl crosslink out of the planes of the cytosine bases raises the possibility that the exocyclic amines are predominantly in the the sp3- as opposed to the presumed sp2-hybridized state. Although the linker CX carbon and base N4 nitrogen would display similar positions if sp3-hybridized rather than sp2-hybridized, several observations suggest that the exocyclic N4 amine is at least partially sp2-hybridized. First, no hydrogen bond donors, such as bound solvent molecules or other nucleotide atoms, are observed in the vicinity of the amine. This suggests the N4 electrons are delocalized and unavailable as hydrogen bond acceptors. Second, an sp3-hybridized state of the exocyclic N4 amine would require the N4-H---C4 hydrogen bond angle (105°) to deviate by 75° from the linear ideal, notably less than the most acute angle observed in a survey of two-center hydrogen bonds (132°) (24). In the sp2-hybridized state, the N4-H---C4 hydrogen bond angle (160°) would deviate only 20° from linearity, and would be within the most probably range of hydrogen bond angles (24). Thus, despite the deviation of the methylene CX carbon from the base plane, a predominately sp2-hybridized geometry of the cytosine exocyclic amine is consistent with the observed structure and the increased thermal stability of the N4C-ethyl-N4C interstrand crosslinked CpG steps (12).
Comparison with NMR Structural Analysis
The sequence of the six nucleotides surrounding the crosslinked CpG step of the X-ray structure is identical to an N4C-ethyl-N4C interstrand crosslinked oligonucleotide previously studied by nuclear magnetic resonance (NMR) experiments and distance-restrained molecular dynamics (respectively d(CCAAC*GTT GG)2 and d(CGAAC*GTT CG)2, sequence differences underlined) (13). Although the RMSD between corresponding atoms is relatively large (2.2 Å RMSD), it is within the range of RMSDs observed for other structures determined by both NMR and X-ray crystallography (25). In general, the overall features of the two structures are similar, with the crosslinked CpG step embedded within an overall B-form DNA duplex. Several helical parameters of the crosslinked CpG step are shared between the X-ray and NMR structures, including a relatively large propeller twist between the bases (-21°), and a C4′-exo sugar pucker for the crosslinked cytidine. Based on the NMR structure alone, these slightly unusual helical parameters were suggested to result from the N4C-ethyl-N 4C interstrand crosslink (13). However, comparison with the X-ray structure of the uncrosslinked double stranded DNA of the same sequence d(CCAACGTTGG)2 (14) reveals that these characteristics are also observed in one alternative conformation of the uncrosslinked DNA, as shown in Table 2(a) and described below.
Similarities with the Uncrosslinked DNA Structure
We chose to study the DNA sequence d(CCAACGTTGG)2, because the structure of this decamer has been thoroughly investigated in the uncrosslinked form (14, 26). This allows the crosslinked and uncrosslinked structures to be compared to understand the influence of the N4C-ethyl-N4C interstrand crosslink on the DNA conformation. Two 1.0 Å resolution structures of the uncrosslinked d(CCAACGTTGG)2 oligonucleotide are available, determined in the presence of either Mg+2 or Ca+2 divalent cations. Although the space group and unit cell parameters of the d(CCAACGTTGG)2 structures are isomorphous, the hydration and location of bound Ca+2 versus Mg+2 counterions induce slight structural differences. In the presence of Ca+2 ions, the central cytidine adopts two alternative conformations of nearly equivalent occupancy, designated ‘A’ and conformation ‘B’ (14). In the presence of Mg+2 ions (14, 26), a single, intermediate conformation of the cytidine is observed, consistent with previous observations that oligonucleotide structures are influenced by the type of counterion (14, 27, 28). Here, we compare the details of the N4C-ethyl-N4C crosslinked oligonucleotide structure, which was determined in the presence of Ca+2 ions, with the Ca+2-containing crystal form of the uncrosslinked oligonucleotide (Figure 4).
Figure 4.
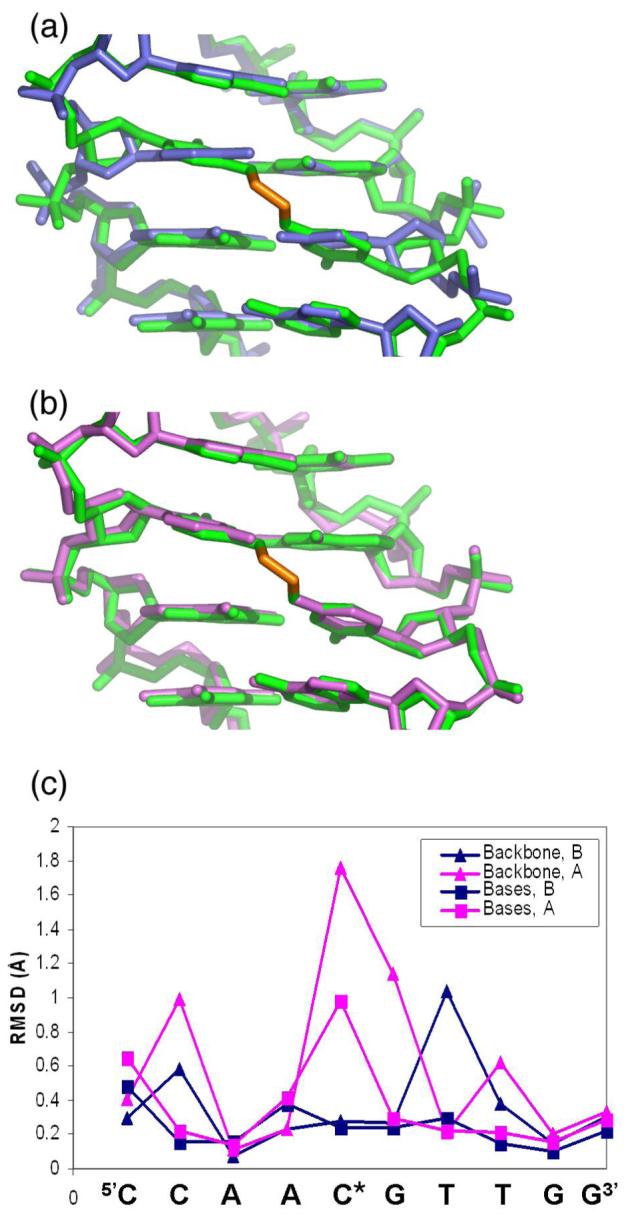
Comparison of the N4C-ethyl-N4C interstrand crosslinked site (green) with (a) conformation A (blue) or (b) conformation B (magenta) of the uncrosslinked structure. (c) RMSD between backbone (-▴-) or base atoms (-■-) of each nucleotide in the crosslinked structure with the uncrosslinked conformation A (blue) or conformation B (magenta).
Introducing the two carbon linker selectively stabilizes alternative conformation B of the cytidine, with no evidence for conformation A (Figure 1, Figure 4). Overall, the RMSDs between all non-hydrogen atoms of the interstrand crosslinked DNA and the uncrosslinked conformations A or B are 0.5 Å or 0.3 Å, respectively. For comparison, the RMSD between the uncrosslinked A and B conformations is 0.5 Å, similar to the differences between the uncrosslinked conformation A and the crosslinked structure. Further inspection of the RMSDs between each nucleotide at the modified CpG step showed that the RMSDs between the crosslinked structure and the uncrosslinked conformation B are remarkably slight (0.27 and 0.24 Å, for backbone or base atoms, respectively)(Figure 4(c)). In contrast, the largest differences between the crosslinked structure and the uncrosslinked conformation A are observed at the modified CpG step, regardless of whether backbone or base atoms are compared (1.76 and 1.0 Å, respectively). Accordingly, a difference omit electron density map for the backbone of the crosslinked cytidine shows no trace of conformation A in the interstrand crosslinked DNA helix (Figure 1). Instead, the conformation of the interstrand crosslinked cytidine closely matched the conformation of uncrosslinked cytidine ‘B’ (Figure 4(b)).
Comparison of the structures reveal that the gauche conformer of the ethyl crosslink is likely to explain the preference of the crosslinked CpG step to adopt conformation B of the uncrosslinked structure. In conformation B of the uncrosslinked structure, the exocyclic amines of the cytosines in the CpG step are 0.4 Å closer than those of conformation A. The N4 atoms of conformation B are within van der Waals contact (3.1 Å), whereas the N4 atoms of conformation A are slightly separated (3.8 Å). The observed gauche conformer of the N4C-ethyl-N4C crosslink, with the N4 atoms adjacent (Figure 3(b) and Table 2(c)), constrains the cytidines to adopt conformation B. An extended anti conformer would enable the N4C-ethyl-N4C crosslink to bridge the greater distance between the N4 atoms of conformation A. However, the anti conformer would drastically distort the positions of the linker CX carbons out of the planes of the cytosine bases, which would enforce predominately sp3-hybridization on the exocyclic N4 amino group and disrupt the N4-H---O6 hydrogen bond of the base pair. Instead, the methylene carbons of the interstrand crosslink are likely to be constrained to the planes of the cytosines by delocalization of the N4 electrons.
As expected given the similarity of the crosslinked CpG site and the uncrosslinked conformation B, several helical parameters of these sites are also similar (Table 2(a)). In particular, the cytosines of the crosslinked base pairs and the uncrosslinked conformation B are both highly propeller twisted (-26° and -21°, respectively), which accounts for the tight packing of the exocyclic amines between the consecutive cytosines. In contrast, conformation A of the uncrosslinked CpG step has a low propeller twist (-5°), contributing to the greater distance between the cytosine exocyclic amines in the major groove. In addition, the shift, slide, and tilt parameters of the crosslinked base pair closely matches those of the uncrosslinked B conformation, as opposed to the A conformation (Table 2(a)). The sugar puckers of both the crosslinked cytosine and the uncrosslinked B conformation are unusual, with low phase angles (52° and 24°, respectively) representative of A-form rather than B-form duplexes. Consistently, a similar, C4′-exo sugar pucker of the crosslinked cytosine base was also observed by NMR methods (13). Thus, far from distorting the DNA helix, the interstrand crosslink selectively stabilizes a pre-existing alternative conformation of the d(CCAACGTTGG)2 oligonucleotide.
Differences from the Uncrosslinked DNA Structure
Despite the many similarities between the N4C-ethyl-N4C crosslinked and conformation B of the uncrosslinked DNAs, a few, slight differences in the DNA conformation are evident (Table 2(b)). Namely, the crosslinked CpG step displays a large positive roll, which contributes to the proximity of the cytosine amines in the major groove, at the cost of opening the base pairs in the minor groove. Another notable difference is that the bases in the crosslinked pair are positioned ideally across the helical axis with no shear, stretch or stagger, whereas the bases of the uncrosslinked conformations are slightly out-of-alignment. This straightening optimizes the Watson-Crick hydrogen bonds in the base pairs of the CpG step (Table 2(c)), which may contribute to the increased thermal stability of DNA duplexes containing the N4C-ethyl-N4C interstrand crosslink at the CpG step (12).
Comparison of Bound Calcium Ions
Four hydrated calcium ions were assigned in the uncrosslinked d(CCAACGTTGG)2 structure based on coordination geometry and low temperature factors. Although the concentration of Ca2+ ions in the crystallization conditions of the crosslinked and uncrosslinked structures is similar (∼10 mM), only three of the four bound Ca2+ ions were observed in the interstrand crosslinked structure (Supplementary Figure S1). These three Ca2+ ions occupy nearly identical locations in the interstrand crosslinked structure compared with the non-crosslinked structure, and are labelled consistently by numbers 111, 113, and 114. The geometry of the coordination spheres around the three Ca2+ ions are also preserved between the structures. Calciums 111 and 114 mediate intermolecular contacts between distinct DNA helices in the crystal lattice, and calcium 113 directly coordinates the O6 and N7 atoms in the major groove of the terminal guanines. Thus, none of the three bound Ca2+ ions or their coordinated water molecules directly contact the alkylated cytidine.
A fourth Ca2+ ion (labelled calcium 112) is bound in the minor groove of the uncrosslinked structure between the Ade:Thy base pairs and the central Cyt:Gua base pair, which would be modified by the crosslink (Figure 5). In conformation A, one water molecule from the coordination sphere of this fully hydrated Ca2+ ion forms a direct hydrogen bond with the deoxyribose-O4′ atom of the central cytidine (2.8 Å H2O-O4′ distance). However, the position of this water molecule is incompatible with the unusual, A-form sugar pucker of conformation B, due to steric overlap between the water oxygen and the C1′-H atoms (3.0 Å H2O-C1′ distance, compared with 4.2 Å predicted van der Waals distance including the C1′-hydrogen with appropriate bond geometry). Hence, calcium 112 is likely to be partially occupied in the uncrosslinked structure, in a manner that is correlated with the alternative cytidine conformations.
Figure 5.
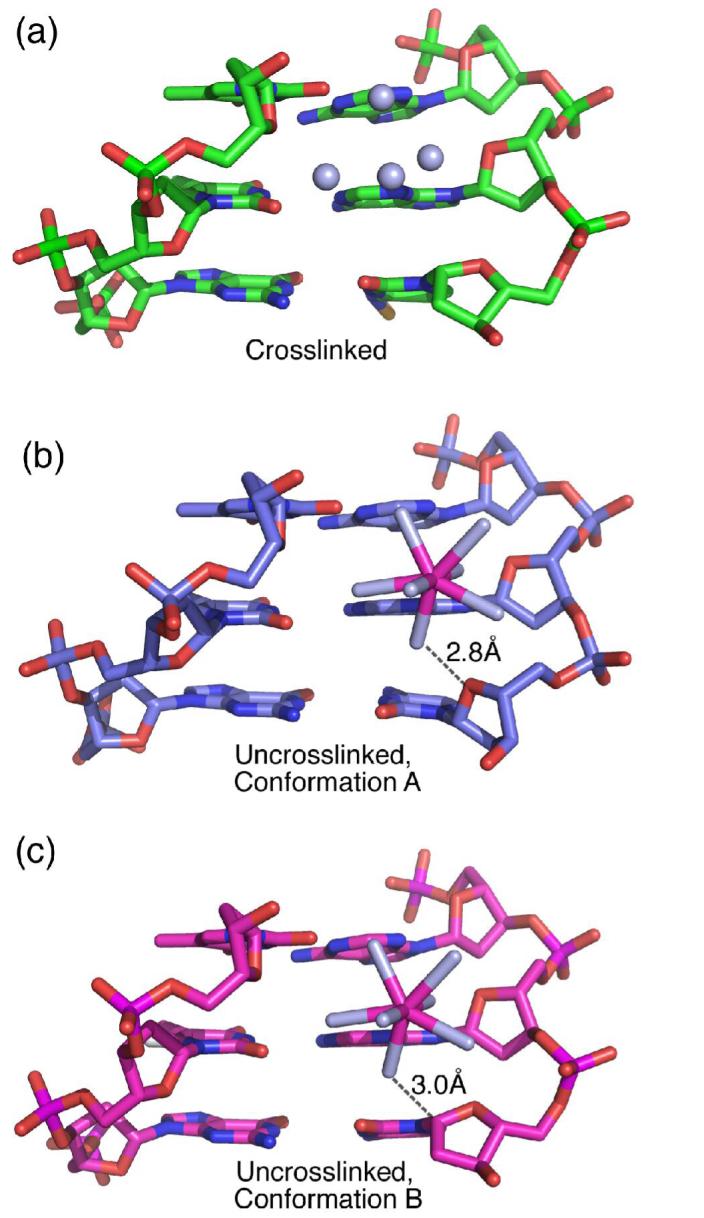
Comparison of the calcium 112 binding sites in the (a) crosslinked, (b) uncrosslinked conformation A, and (c) uncrosslinked conformation B. The hydrated calcium is depicted with a ball-and-stick representation, with calcium 112 colored pink and water ligands colored light blue. The four water molecules that replace the hydrated calcium in the crosslinked structure are shown as spheres. The distances are indicated between the hydrated calcium and either the deoxyribose-O4′ or C1′ atoms of conformations A or B, respectively.
The sugar pucker of the crosslinked cytidine is similar to that of the uncrosslinked conformation B, and accordingly would sterically conflict with the hydrated calcium 112 (2.8 Å predicted H2O-C1′ distance). Instead of a hydrated Ca2+ ion, a constellation of bound water molecules is observed in the crosslinked structure (Figure 5). Although four of these water molecules in the crosslinked structure correspond to four water ligands of calcium 112 in the uncrosslinked structure, these water molecules interact with atoms in the unmodified thymidine nucleotides rather than with the crosslinking site. No unaccounted electron density is observed near the position of calcium 112 in the uncrosslinked structure. The four bound water molecules lack the preferred coordination geometry of calcium, which ranges from six to eight ligands with calcium-oxygen ligand distances of ∼2.4 Å (29). Moreover, the refined temperature factors of the assigned water molecules are appropriate for oxygen (B-value of 19-28 Å2 compared with B-values of 1-5 Å2 when Ca2+ were inappropriately assigned as H2O). Thus, introduction of the N4C-ethyl-N 4C interstrand crosslink eliminates a divalent cation binding site that would otherwise be presented by the uncrosslinked oligonucleotide.
DISCUSSION
Stabilized Alternative Conformations
When compared with the structure of an uncrosslinked DNA oligonucleotide of the same sequence, the synthetic N4C-ethyl-N4C interstrand crosslink at the CpG step does not distort the B-form DNA helix, but instead fully adopts one alternative conformation of the undamaged DNA. Similarly, pre-existing alternative conformations of proteins are often stabilized during RNA recognition (30, 31). In particular, association with RNA selectively stabilizes a subset of alternative conformations displayed on the RNA-interaction surface of the unliganded protein (hnRNP A1 (30) or U2AF65 (31)). This work provides complementary evidence that alternative conformations observed in X-ray structures often play important biological roles in chemical modification and ligand binding, and should not be overlooked.
Conformation of the Crosslinked CpG Site
The relatively short, interstrand, N4C-ethyl-N4C crosslink forces the alkylated cytidines to adopt a single alternative conformation (‘B’) of the uncrosslinked duplex, due to an unusually large negative propeller twist that minimizes the distance between the exocyclic N4 amines. The X-ray structure reveals that a locally overwound twist and unusual C4′-exo sugar pucker at the CpG site also are observed in one conformation of the DNA in the absence of the crosslink, clarifying a previous conjecture based on NMR analysis that the crosslink induced these features. This conformation of the crosslinked deoxyribose interferes with association of a hydrated Ca+2 ion, which otherwise is observed in the minor groove of the other uncrosslinked conformation (‘A’). Unique features of the crosslinked CpG site include an increased roll of the base pair that brings together the cytosine amines, and optimized hydrogen bonds between the paired cytosine and guanine bases.
Locally, the N4-CX—CX-N4 bond of the N4C-ethyl-N4C crosslink adopts a staggered, gauche conformer, with the CX methylene carbons slightly displaced from the plane of the base. Although the geometry of the Watson-Crick hydrogen bond and lack of hydrogen bond donors near the crosslinked N4 amine are consistent with sp2-hybridized bond character, the deviation of the CX carbon from the base plane suggests partial sp3-hybridization. Conventionally, hydrogen atoms bonded to exocyclic amino groups of nucleotide bases are assumed to be coplanar with the base atoms (21, 22), due to conjugation of the lone pair electrons of the nitrogen with the neighboring π electron cloud of the base. However, ab initio quantum chemical calculations and surveys of hydrogen bonds in small molecule and macromolecular structure databases indicate that in some cases, the hydrogen atoms bonded to amino groups of nucleoside bases may adopt nonplanar, sp3-hybridized geometry (32-34). In the absence of high resolution neutron diffraction studies (35), direct experimental evidence for nonplanar exocyclic amino groups in nucleic acids is minimal. Even in atomic resolution X-ray structures (<1.0 Å resolution), hydrogen atoms are difficult to place due to low atomic scattering factors and generally more dynamic positions. The methylene carbons that replace the hydrogen atoms in the N4C-ethyl-N4C crosslink are clearly observed in the electron density (Figure 1), providing rare experimental evidence for nonplanarity of an exocyclic amino group belonging to a nucleic acid base. This observation reinforces the importance of considering nonplanar amino groups when analyzing the hydrogen bond interactions of nucleic acids with proteins or small molecule ligands.
Comparison with Chemotherapeutic Interstrand DNA Crosslinks and Significance for DNA Repair
The majority of DNA interstrand crosslinks induced by antitumor compounds that have been studied structurally thus far are highly distorted, including interstrand crosslinks of cis-diamminedichloroplatinum (cisplatin, DDP), nitrogen mustards, and psoralen (reviewed in (2)). For example, molecular dynamics and electrophoretic mobility experiments show that DNAs crosslinked with the nitrogen mustard mechlorethamine at 5′-GNC-3′ sites are locally distorted and bent by the lesion (36). The anticancer compound DDP forms interstrand crosslinks between guanines of 5′-GC-3′ sequences that significantly deform the DNA helix (37, 38). Although DDP reacts with the guanine N7 atoms, which are normally located in the major groove of B-form DNA, the highly distorted DNA conformation places this crosslink in the DNA minor groove and extrudes the cytosines from the dramatically bent and underwound DNA site, as shown by X-ray (39) and NMR structural analysis (40, 41), as well as by electrophoretic mobility and chemical footprinting experiments (42). Psoralens, which are natural DNA crosslinkers used to treat skin diseases (43), intercalate preferentially at AT-rich sites and produce interstrand crosslinks between thymidines when irradiated with ultraviolet light (44). NMR structures of 4′-hydroxymethyl-4,5′,8-trimethylpsoralen (45, 46) or 4′-aminomethyl-4,5′,8-trimethylpsoralen (47) interstrand crosslinked DNA oligonucleotides show that the intercalated and crosslinked site is locally distorted and significantly underwound compared with the canonical B-form. These interstrand crosslinked structures differ remarkably from the largely unaltered, B-form structure of the N4C-ethyl-N 4C crosslinked CpG site.
Since psoralen interstrand crosslinked DNA is chemically stable and relatively easy to synthesize, many studies on the repair of interstrand crosslinks in defined DNA substrates have employed the psoralen interstrand crosslink (2). However, in some cases, antitumor compounds result in interstrand crosslinks that minimally perturb the DNA structure. For example, the antibiotic mitomycin C (MMC) from Streptomyces caespitosus is an important treatment for a variety of cancers (48-50). MMC forms interstrand crosslinks between the exocyclic N2 amines of guanines, which slightly widen the DNA minor groove (51) but lacks detectable effect on the overall DNA conformation or bending (52). Although interstrand DNA crosslinks triggered by MMC are a major cause of cell death, these crosslinks occur with low frequency (1 per 20,000 base pairs) (53, 54). The UvrABC NER pathway in E. coli repairs site-specific interstrand crosslinks of a related but more active compound, mitomycin A, particularly in the context of supercoiled plasmid DNA (55). However, the low abundance of the MMC interstrand crosslinks presents a difficulty for studying the repair mechanism of MMC lesions in defined DNA sites of known sequence. Overall, the X-ray structural analysis presented here and previous NMR experiments (13) suggest that N4C-ethyl-N4C interstrand crosslinked CpG steps would provide a synthetic model for studying repair of interstrand crosslinks of defined sequences in the absence of gross distortions of the DNA helix, such as those produced by MMC. Accordingly, both the alkyl and MMC interstrand crosslinks of CpG sites are repaired by the UvrABC pathway in E. coli (13, 56).
Remarkably, our high resolution X-ray structure reveals that the N4C-ethyl-N4C interstrand crosslink does not even minimally alter the structure of the uncrosslinked d(CCAACGTTGG)2 oligonucleotide; rather, one pre-existing conformation of the DNA is selectively stabilized by the interstrand crosslink. Moreover, comparison of the N4C-ethyl-N4C interstrand crosslinked DNA with the DNA-bound structure of the nucleosome core (57) shows that the presence of two methylene carbons deep in the major groove are unlikely to interfere with the DNA contacts of the histone proteins, which are primarily formed with the phosphodiester backbone. Consequently, the N4C-ethyl-N4C crosslinked CpG site lacks explicit chemical handles for recognition by DNA repair factors. Nevertheless, the covalent crosslink between the DNA strands would block DNA replication or transcription. Thus, the synthetic, N4C-ethyl-N4C interstrand CpG crosslink provides a unique opportunity to study the repair of covalently linked DNA strands in the absence of other prominent features of damaged DNA. Together with studies on the structure and repair of the reversed, N4C-ethyl-N 4C interstrand GpC crosslink, which significantly disrupts the DNA helix unlike the minimal effects of the CpG crosslink (13), this work contributes to a growing arsenal of well-characterized synthetic alkyl interstrand crosslinks that can be used to address the detailed structural and mechanistic relationships of DNA repair.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to L.M. Amzel, D.J. Leahy, and C. Wolberger for advice and generous access to X-ray equipment.
Abbreviations
- DDP
diamminedichloroplatinum
- MMC
mitomycin C
- NER
nucleotide excision repair
- PDB
Protein Data Bank
- Tris
tris-hydroxymethyl-aminomethane
Footnotes
M.C.S. and S.R.P. contributed equally to this work. This research was supported by a grant from the National Cancer Institute (CA082785). M.C.S. and S.R.P. were supported in part by a training grant from the National Cancer Institute (CA009110).
Atomic coordinates and structure factors are available in the RCSB Protein Data Bank (2OKS).
REFERENCES
- (1).Scharer OD. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- (2).Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Niedernhofer LJ, Daniels JS, Rouzer CA, Greene RE, Marnett LJ. Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J Biol Chem. 2003;278:31426–33. doi: 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]
- (4).Mathison BH, Harman AE, Bogdanffy MS. DNA damage in the nasal passageway: a literature review. Mutat Res. 1997;380:77–96. doi: 10.1016/s0027-5107(97)00128-0. [DOI] [PubMed] [Google Scholar]
- (5).Chen Q, Van der Sluis PC, Boulware D, Hazlehurst LA, Dalton WS. The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood. 2005;106:698–705. doi: 10.1182/blood-2004-11-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dong Q, Johnson SP, Colvin OM, Bullock N, Kilborn C, Runyon G, Sullivan DM, Easton J, Bigner DD, Nahta R, Marks J, Modrich P, Friedman HS. Multiple DNA repair mechanisms and alkylator resistance in the human medulloblastoma cell line D-283 Med (4-HCR) Cancer Chemother Pharmacol. 1999;43:73–9. doi: 10.1007/s002800050865. [DOI] [PubMed] [Google Scholar]
- (7).Vogel EW, Barbin A, Nivard MJ, Stack HF, Waters MD, Lohman PH. Heritable and cancer risks of exposures to anticancer drugs: inter-species comparisons of covalent deoxyribonucleic acid-binding agents. Mutat Res. 1998;400:509–40. doi: 10.1016/s0027-5107(98)00060-8. [DOI] [PubMed] [Google Scholar]
- (8).O’Connor PM, Kohn KW. Comparative pharmacokinetics of DNA lesion formation and removal following treatment of L1210 cells with nitrogen mustards. Cancer Commun. 1990;2:387–94. doi: 10.3727/095535490820873949. [DOI] [PubMed] [Google Scholar]
- (9).Noronha AM, Wilds CJ, Miller PS. N(4)C-alkyl-N(4)C cross-linked DNA: bending deformations in duplexes that contain a -CNG-interstrand cross-link. Biochemistry. 2002;41:8605–12. doi: 10.1021/bi025671j. [DOI] [PubMed] [Google Scholar]
- (10).Noll DM, Noronha AM, Miller PS. Synthesis and characterization of DNA duplexes containing an N(4)C-ethyl-N(4)C interstrand cross-link. J Am Chem Soc. 2001;123:3405–11. doi: 10.1021/ja003340t. [DOI] [PubMed] [Google Scholar]
- (11).Noronha AM, Noll DM, Miller PS. Syntheses of DNA duplexes containing a C-C interstrand cross -link. Nucleosides Nucleotides Nucleic Acids. 2001;20:1303–7. doi: 10.1081/NCN-100002542. [DOI] [PubMed] [Google Scholar]
- (12).Noronha AM, Noll DM, Wilds CJ, Miller PS. N(4)C-ethyl-N(4)C cross-linked DNA: synthesis and characterization of duplexes with interstrand cross-links of different orientations. Biochemistry. 2002;41:760–71. doi: 10.1021/bi011610u. [DOI] [PubMed] [Google Scholar]
- (13).Noll DM, Webba da Silva M, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Structure, flexibility, and repair of two different orientations of the same alkyl interstrand DNA cross-link. Biochemistry. 2005;44:6764–75. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- (14).Chiu TK, Dickerson RE. 1 A crystal structures of B-DNA reveal sequence-specific binding and groove-specific bending of DNA by magnesium and calcium. J Mol Biol. 2000;301:915–45. doi: 10.1006/jmbi.2000.4012. [DOI] [PubMed] [Google Scholar]
- (15).Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- (16).Kleygwegt GJ, Zou JY, Kjeldgaard M, Jones TA. Around O. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography, Volume F. Crystallography of Biological Macromolecules. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2001. pp. 353–356.pp. 366–367. [Google Scholar]
- (17).Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- (18).Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- (19).Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- (20).Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. [Google Scholar]
- (21).Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KMJ, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- (22).Mackerell AD, Wiorkiewiczkuczera J, Karplus M. An all-atom empirical energy function for the simulation of nucleic acids. J Am Chem Soc. 1995;117:11946–11975. [Google Scholar]
- (23).Collaborative Computational Project The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst D50. 1994:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- (24).Jeffrey GA. An Introduction to Hydrogen Bonding. Oxford University Press; New York: 1997. [Google Scholar]
- (25).Garbuzynskiy SO, Melnik BS, Lobanov MY, Finkelstein AV, Galzitskaya OV. Comparison of X-ray and NMR structures: is there a systematic difference in residue contacts between X-ray- and NMR-resolved protein structures? Proteins. 2005;60:139–47. doi: 10.1002/prot.20491. [DOI] [PubMed] [Google Scholar]
- (26).Prive GG, Yanagi K, Dickerson RE. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J Mol Biol. 1991;217:177–99. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- (27).Gold B, Marky LM, Stone MP, Williams LD. A review of the role of the sequence-dependent electrostatic landscape in DNA alkylation patterns. Chem Res Toxicol. 2006;19:1402–14. doi: 10.1021/tx060127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Minasov G, Tereshko V, Egli M. Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing. J Mol Biol. 1999;291:83–99. doi: 10.1006/jmbi.1999.2934. [DOI] [PubMed] [Google Scholar]
- (29).Katz AK, Glusker JP, Beebe SA, Bock CW. Calcium ion coodination: A comparison with that of beryllium, magnesium, and zinc. J Am Chem Soc. 1996;118:5752–5763. [Google Scholar]
- (30).Vitali J, Ding J, Jiang J, Zhang Y, Krainer AR, Xu RM. Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1. Nucleic Acids Res. 2002;30:1531–8. doi: 10.1093/nar/30.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Thickman KR, Sickmier EA, Kielkopf CL. Alternative conformations at the RNA binding surface of the N-terminal U2AF65 RNA recognition motif. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.11.077. in press, doi: 10.1016/j.jmb.2006.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Mukherjee S, Majumdar S, Bhattacharyya D. Role of hydrogen bonds in protein-DNA recognition: Effect of nonplanar amino groups. J Phys Chem B. 2005;109:10484–10492. doi: 10.1021/jp0446231. [DOI] [PubMed] [Google Scholar]
- (33).Luisi B, Orozco M, Sponer J, Luque FJ, Shakked Z. On the potential role of the amino nitrogen atom as a hydrogen bond acceptor in macromolecules. J Mol Biol. 1998;279:1123–36. doi: 10.1006/jmbi.1998.1833. [DOI] [PubMed] [Google Scholar]
- (34).Bludsky O, Sponer J, Leszczynski J, Spirka V, Hobza P. Amino groups in nucleic acid bases, aniline, aminopyridines, and aminotriazine are nonplanar: Results of correlated ab initio quantum chemical calculations and anharmonic analysis of the aniline inversion motion. J Chem Phys. 1996;105:11042–11050. [Google Scholar]
- (35).Arai S, Chatake T, Ohhara T, Kurihara K, Tanaka I, Suzuki N, Fujimoto Z, Mizuno H, Niimura N. Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements. Nucleic Acids Res. 2005;33:3017–24. doi: 10.1093/nar/gki616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Rink SM, Hopkins PB. A mechlorethamine-induced DNA interstrand cross-link bends duplex DNA. Biochemistry. 1995;34:1439–45. doi: 10.1021/bi00004a039. [DOI] [PubMed] [Google Scholar]
- (37).Cohen SM, Lippard SJ. Cisplatin: from DNA damage to cancer chemotherapy. Prog Nucleic Acid Res Mol Biol. 2001;67:93–130. doi: 10.1016/s0079-6603(01)67026-0. [DOI] [PubMed] [Google Scholar]
- (38).Kartalou M, Essigmann JM. Recognition of cisplatin adducts by cellular proteins. Mutat Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- (39).Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 A resolution: hydration at the platinated site. Nucleic Acids Res. 1999;27:1837–46. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Paquet F, Perez C, Leng M, Lancelot G, Malinge JM. NMR solution structure of a DNA decamer containing an interstrand cross-link of the antitumor drug cis-diamminedichloroplatinum (II) J Biomol Struct Dyn. 1996;14:67–77. doi: 10.1080/07391102.1996.10508930. [DOI] [PubMed] [Google Scholar]
- (41).Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB. Solution structure of a cisplatin-induced DNA interstrand cross-link. Science. 1995;270:1842–5. doi: 10.1126/science.270.5243.1842. [DOI] [PubMed] [Google Scholar]
- (42).Malinge JM, Perez C, Leng M. Base sequence-independent distorsions induced by interstrand cross-links in cis-diamminedichloroplatinum (II)-modified DNA. Nucleic Acids Res. 1994;22:3834–9. doi: 10.1093/nar/22.19.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Brenner M, Herzinger T, Berking C, Plewig G, Degitz K. Phototherapy and photochemotherapy of sclerosing skin diseases. Photodermatol Photoimmunol Photomed. 2005;21:157–65. doi: 10.1111/j.1600-0781.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- (44).Hearst JE. Psoralen photochemistry. Annu Rev Biophys Bioeng. 1981;10:69–86. doi: 10.1146/annurev.bb.10.060181.000441. [DOI] [PubMed] [Google Scholar]
- (45).Spielmann HP, Dwyer TJ, Sastry SS, Hearst JE, Wemmer DE. DNA structural reorganization upon conversion of a psoralen furan-side monoadduct to an interstrand cross-link: implications for DNA repair. Proc Natl Acad Sci U S A. 1995;92:2345–9. doi: 10.1073/pnas.92.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Spielmann HP, Dwyer TJ, Hearst JE, Wemmer DE. Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics. Biochemistry. 1995;34:12937–53. [PubMed] [Google Scholar]
- (47).Hwang GS, Kim JK, Choi BS. The solution structure of a psoralen cross-linked DNA duplex by NMR and relaxation matrix refinement. Biochem Biophys Res Commun. 1996;219:191–7. doi: 10.1006/bbrc.1996.0204. [DOI] [PubMed] [Google Scholar]
- (48).Chalasani R, Giblin M, Conway RM. Role of topical chemotherapy for primary acquired melanosis and malignant melanoma of the conjunctiva and cornea: review of the evidence and recommendations for treatment. Clin Experiment Ophthalmol. 2006;34:708–14. doi: 10.1111/j.1442-9071.2006.01356.x. [DOI] [PubMed] [Google Scholar]
- (49).Eifel PJ. Chemoradiotherapy in the treatment of cervical cancer. Semin Radiat Oncol. 2006;16:177–85. doi: 10.1016/j.semradonc.2006.02.007. [DOI] [PubMed] [Google Scholar]
- (50).Bolenz C, Cao Y, Arancibia MF, Trojan L, Alken P, Michel MS. Intravesical mitomycin C for superficial transitional cell carcinoma. Expert Rev Anticancer Ther. 2006;6:1273–82. doi: 10.1586/14737140.6.8.1273. [DOI] [PubMed] [Google Scholar]
- (51).Norman D, Live D, Sastry M, Lipman R, Hingerty BE, Tomasz M, Broyde S, Patel DJ. NMR and computational characterization of mitomycin cross-linked to adjacent deoxyguanosines in the minor groove of the d(T-A-C-G-T-A).d(T-A-C-G-T-A) duplex. Biochemistry. 1990;29:2861–75. doi: 10.1021/bi00463a032. [DOI] [PubMed] [Google Scholar]
- (52).Rink SM, Lipman R, Alley SC, Hopkins PB, Tomasz M. Bending of DNA by the mitomycin C-induced, GpG intrastrand cross-link. Chem Res Toxicol. 1996;9:382–9. doi: 10.1021/tx950156q. [DOI] [PubMed] [Google Scholar]
- (53).Iyer VN, Szybalski W. Mitomycins and Porfiromycin: Chemical Mechanism of Activation and Cross-Linking of DNA. Science. 1964;145:55–8. doi: 10.1126/science.145.3627.55. [DOI] [PubMed] [Google Scholar]
- (54).Iyer VN, Szybalski W. A Molecular Mechanism of Mitomycin Action: Linking of Complementary DNA Strands. Proc Natl Acad Sci U S A. 1963;50:355–62. doi: 10.1073/pnas.50.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Pu WT, Kahn R, Munn MM, Rupp WD. UvrABC incision of N-methylmitomycin A-DNA monoadducts and cross-links. J Biol Chem. 1989;264:20697–704. [PubMed] [Google Scholar]
- (56).Vidal LS, Santos LB, Lage C, Leitao AC. Enhanced sensitivity of Escherichia coli uvrB mutants to mitomycin C points to a UV-C distinct repair for DNA adducts. Chem Res Toxicol. 2006;19:1351–6. doi: 10.1021/tx060035y. [DOI] [PubMed] [Google Scholar]
- (57).Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.