Abstract
A combination of representational difference analysis and comparative DNA sequencing revealed that four type I (sheep) isolates of Mycobacterium avium subsp. paratuberculosis were differentiated from nine type II (bovine) isolates by the presence of an 11-bp insertion in a novel M. avium subsp. paratuberculosis-specific region of genomic DNA. Further, our studies show that M. avium subsp. paratuberculosis type I isolates contain three type-specific loci that are missing in M. avium subsp. paratuberculosis type II but are present in M. avium subsp. avium. Taken together, the results are consistent with the hypothesis that M. avium subsp. paratuberculosis type I strains are an evolutionary intermediate between M. avium subsp. avium and M. avium subsp. paratuberculosis type II isolates or share a subset of M. avium subsp. avium type-specific loci through horizontal transfer.
Mycobacterium avium subsp. paratuberculosis is the causative agent of paratuberculosis (Johne's diseases), a chronic granulomatous enteritis in ruminants. The disease is prevalent in domestic and wild animals worldwide (15, 16, 24, 31) and has a considerable economic impact on the livestock industry (13). In humans, M. avium subsp. paratuberculosis has been suggested to be involved in Crohn's disease (6, 11, 20). M. avium subsp. paratuberculosis shows a DNA homology of more than 90% to M. avium subsp. avium (14, 26). It differs from other subspecies of M. avium by its dependence on the iron chelator mycobactin for growth in culture (9, 30) and by the presence of multiple copies of the insertion sequences IS900 (12) and ISMav2 (29).
By using analyses by pulsed-field electrophoresis (PFGE) and IS900-restriction fragment length polymorphisms, M. avium subsp. paratuberculosis isolates have been divided into two distinct types, with type I comprising very slow-growing, mostly pigmented isolates forming smooth and uniform colonies mainly obtained from sheep and other small ruminants and type II comprising slow-growing, nonpigmented isolates forming rough and nonuniform colonies exhibiting a very broad host range (27). Both types can also be differentiated by IS1311-PCR (19) and by a specific multiplex PCR (8). However, the molecular differences between M. avium subsp. paratuberculosis type I and type II strains which might give important clues with respect to (i) the evolutionary relationship between the two M. avium subsp. paratuberculosis types, (ii) the different phenotypes, and (iii) the differences in host preference have not as yet been elucidated.
In this study we investigated the differences between M. avium subsp. paratuberculosis type I and type II strains by applying the technique of representational difference analysis (RDA) (18) to isolate M. avium subsp. paratuberculosis subspecies as well as type-specific DNA fragments. The isolation was followed by PCR, restriction endonuclease digest, and nucleotide sequence analyses of the respective fragments obtained from M. avium subsp. paratuberculosis isolated from various hosts.
The bacterial strains and plasmids used in this study are listed in Table 1. Mycobacteria were grown on Middlebrook 7H10 agar supplemented with a solution of oleic acid-albumin-dextrose-catalase enrichment (DIFCO, Augsburg, Germany), Tween 80 (0.05%), and mycobactin J (2 μg ml−1; Synbiotics, Lyon, France). Escherichia coli transformants were grown in Luria-Bertani medium supplemented with ampicillin (100 μg ml−1). The M. avium subsp. paratuberculosis isolates were classified as type I and type II by PFGE (27) and two M. avium subsp. paratuberculosis PCR tests (8, 19); three pigmented ovine isolates (M189, 213G, 21P) were classified as type I by all three tests, and one nonpigmented caprine isolate CAM42 was classified as type I by both PCR tests but exhibited a PFGE profile intermediate between type I and type II. All other isolates were classified as type II strains. All type I and type II isolates were independent and originated from different farms.
TABLE 1.
Strains and plasmids used in this study
| Isolate or primer | Origin or descriptiona |
|---|---|
| M. avium subsp. paratuberculosis | |
| ATCC 19698, bovine | American Type Culture Collection, Rockville, Md. |
| Isolate 6783 (DSM 44135), bovine | Laboratory reference strain |
| Isolate M189, pigmented ovine | MRI |
| Isolate 99PW, bovine | MRI |
| Isolate 51/91, nonpigmented ovine | MRI |
| Isolate 502038-191, nonpigmented ovine | MRI |
| Isolate JD143, nonpigmented ovine | MRI |
| Isolate 21P, pigmented ovine | MRI |
| Isolate 213G, pigmented ovine | MRI |
| Isolate M100/C, caprine | MRI |
| Isolate 176P, caprine | MRI |
| Isolate CAM42, caprine | MRI |
| Isolate 52291V.Veen-HPC, human | MRI |
| M. avium subsp. avium | |
| ATCC 35712 | DSMZ |
| ATCC 25291 (DSM 44156) | DSMZ |
| DSM 44158 | DSMZ |
| MAA-S4 (M. avium subsp. avium serotype 4) | J. Magee, Newcastle Public Health Laboratory |
| NCTC 8559 | NCTC |
| Other Mycobacterium isolates | |
| M. intracellulare NCTC 10425 | NCTC |
| M. scrofulaceum NCTC 10803 | NCTC |
| M. smegmatis NCTC 8159 | NCTC |
| M. phlei NCTC 8151 | NCTC |
| M. tuberculosis H37Rv NCTC 7416 | NCTC |
| M. bovis NCTC 10772 | NCTC |
| M. microti NCTC 8710 | NCTC |
| M. terrae NCTC 10856 | NCTC |
| M. kansasii NCTC 10268 | NCTC |
| M. xenopi NCTC 10042 | NCTC |
| M. malmoense NCTC 11298 | NCTC |
| M. gordonae NCTC 10267 | NCTC |
| M. szulgai NCTC 10831 | NCTC |
| M. flavescens NCTC 10271 | NCTC |
| M. marinum NCTC 2275 | NCTC |
| M. chelonae subsp. chelonae NCTC 946 | NCTC |
| M. chelonae subsp. abscessus NCTC 10882 | NCTC |
| M. fortuitum NCTC 10394 | NCTC |
| M. peregrinum NCTC 10264 | NCTC |
| M. haemophilum NCTC 11185 | NCTC |
| E. coli TOP 10F′ | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 rec A1 deoR araD139 Δ(araleu)7697 galU galK rpsL (StrR) endA1 nupG; TOPO TA cloning vector (Invitrogen) |
| Plasmid pCR 2.1-TOPO | E. coli cloning vector carrying an ampicillin and kanamycin resistance determinant (Invitrogen) |
MRI, strain collection at the Moredun Research Institute; DSMZ, Deutsche Sammlung fuer Mikroorganismen und Zellkulturen; NCTC, National Collection of Type Cultures.
The RDA was performed as previously described (29). For the identification of M. avium subsp. paratuberculosis-specific DNA regions the genome of M. avium subsp. avium strain ATCC 25291 (driver) was subtracted from that of the bovine M. avium subsp. paratuberculosis strain 6783 (tester). For the identification of M. avium subsp. paratuberculosis type-specific DNA regions the nonpigmented bovine strain 6783 was used as the tester and the pigmented ovine M. avium subsp. paratuberculosis strain M189 was used as the driver, and vice versa. Briefly, tester and driver DNA were digested simultaneously with BamHI, BglII, and BclI (New England Biolabs, Bad Schwalbach, Germany). Tester fragments were ligated to the oligonucleotide adapter RBam12/RBam24 (Table 2). Ligation products were diluted 10- to 1,000-fold, and a constant amount of digested driver DNA was added, resulting in 4-, 40-, and 400-fold excesses of driver DNA. DNA was ethanol precipitated, dried, resuspended in 5 μl of Taq polymerase buffer (Invitrogen, Groningen, The Netherlands) containing 1 M NaCl, overlaid with paraffin oil, and hybridized for 20 h at 67°C. The hybridized DNA was diluted with 15 μl of H2O, 5 μl of each hybridization reaction mixture was removed, and the overhangs were filled in by using Taq polymerase in the recommended buffer (Invitrogen, Groningen, The Netherlands) containing deoxynucleoside triphosphates (0.2 mM) at 72°C for 20 min in a 20-μl volume. Primer RBam24 (25 pmol in a 5-μl volume) was added, and tester-specific DNA was exponentially amplified in a 25-μl volume with an initial denaturation step at 94°C for 3 min, 35 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 3 min, and a final extension at 72°C for 10 min. PCR products were analyzed on a 1.5% agarose gel and were cloned by using a TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands); E. coli transformants were tested for inserts in a PCR with RBam24 as the primer. PCR products were analyzed on a 1.5% agarose gel, and their specificity was tested by differential Southern blot analyses using chromosomal DNA of the tester and driver as probes. Agarose gel electrophoresis, Southern blot analyses, PCR, DNA cloning, and transformation of E. coli were done by following standard procedures (25). Primers were purchased from Invitrogen, and DNA sequencing reactions were done by SeqLab (Göttingen, Germany). GenBank database searches were performed by using BLASTX and BLASTN (1). In addition, BLASTX alignments of DNA sequences were performed by using GenBank/EMBL as well as the M. avium subsp. avium (The Institute for Genome Research [TIGR] strain 104) and M. avium subsp. paratuberculosis (UMN strain K-10) unfinished genome sequences of TIGR (http://www.tigr.org) and the University of Minnesota M. avium subsp. paratuberculosis database (http://genome.cm.umn.edu/cgi-bin/blast/web_blast.cgi).DNA-modifying enzymes were purchased from New England Biolabs (Karlsruhe, Germany). Primers were designed for the PCR analyses of M. avium subsp. paratuberculosis subspecies- and type I-specific DNA regions (Table 2). PCR was carried out in 25-μl reaction volumes containing standard PCR buffer (Invitrogen) according to the manufacturer's instructions at annealing temperatures optimal for each primer pair (Table 2), and PCR products were analyzed on a 1.5% agarose gel.
TABLE 2.
Primers used in this study
| Primer and/or fragment | Sequence |
|---|---|
| RDA primersa | |
| RBam12 | 5′-GAT CCT CGG TGA-3′ |
| RBam24 | 5′-AGC ACT CTC CAG CCT CTC ACC GAG-3′ |
| ISMav2 primersb | |
| ISMav1 | 5′-GTA TCA GGC CGT GAT GGC GG-3′ |
| ISMav2 | 5′-CGC GAC CAG CGC TCG ATA CA-3′ |
| IS900 primersc | |
| MK5 | 5′-TTC TTG AAG GGT GTT CGG GGC C-3′ |
| MK6 | 5′-GCG ATG ATC GCA GCG TCT TTG G-3′ |
| IS901 primersd | |
| MK7 | 5′-GTC TGG GAT TGG ATG TCC TG-3′ |
| MK8 | 5′-CAC CAC GTG GTT AGC AAT CC-3′ |
| RDA fragments specific for M. avium subsp. paratuberculosise | |
| RD I130 | |
| I 130A | 5′-TGT GAG GAC ATT CGG TCG GTC-3′ |
| I 131A | 5′-TCT ACC TGC ACC CAC GAT GAG-3′ |
| RD II60 | |
| II 60A | 5′-TGC CGA CGT GTA CGA ATC AG-3′ |
| II 61A | 5′-TCG TTC CGG TCT CTG CGC TA-3′ |
| RD III10 | |
| III 10B | 5′-TGC ACG CCC GTT ACA TCA TCC-3′ |
| III 11B | 5′-TAG CGG CAG TCA CGA TCG AG-3′ |
| RDA fragments specific for M. avium subsp. paratuberculosis type I strainsf | |
| pig-RDA10 | |
| p19 | 5′-TAG CGG TCC CGC AGT TTG GC-3′ |
| p20 | 5′-TCA AGC CGA ACG AGG TGG TCG-3′ |
| pig-RDA20 | |
| p21 | 5′-TCG TCC CGT CCC GAT GCT GT-3′ |
| p22 | 5′-TGA GTC CTG TCG TGC ATG CG-3′ |
| pig-RDA30 | |
| p23 | 5′-TGA AGA GCC CGG ACA AGG GG-3′ |
| p24 | 5′-TAG GTC TCA GTG GTC CAC CAG C-3′ |
The RDA with M. avium subsp. paratuberculosis as tester and M. avium subsp. avium as driver revealed three fragments specific for M. avium subsp. paratuberculosis, designated RDI130, RDII60, and RDIII10, ranging from 456 to 652 bp (Table 3) with 54 to 58% homology to the M. avium subsp. avium and 100% homology to the M. avium subsp. paratuberculosis genome (Table 3). The specificity of these fragments was confirmed by dot blots of genomic DNA from 22 different species of mycobacteria (Table 1). Fragment RDI130 is located inside a 15-kb M. avium subsp. paratuberculosis-specific region containing the previously described F57 fragment (23) and fragments Mpt52.16 and Mpt54.16 (21). Fragment RDII60 is located on a 6-kb M. avium subsp. paratuberculosis-specific region containing fragment Mpt62.12 (21). Fragment RDIII10 overlaps a pks oxidoreductase-like gene adjacent to IS900 locus 6 described by Bull and coworkers (5). Using specific primers and stringent annealing temperatures (Table 2), PCR for the fragments RDII60 and RDIII10 was positive for all 13 M. avium subsp. paratuberculosis isolates tested but was negative for all 4 isolates of M. avium subsp. avium. In addition, the nucleotide sequence was identical in all 13 strains, indicating a high degree of genetic stability among strains isolated from different hosts.
TABLE 3.
Molecular data for the M. avium subsp. paratuberculosis subspecies- and type I-specific fragments
| RDA fragment | Length (bp) | Accession no. | Identity with the M. avium subsp. avium genome (%) | Position in M. avium subsp. paratuberculosis genome (UMN strain K-10, contig 16) | Position in M. avium subsp. avium genome (TIGR strain 104, contig 3294) |
|---|---|---|---|---|---|
| M. avium subsp. paratuberculosis specific | |||||
| RDA I130 | 631 | AY 254383 | 54 | 2,637,820-2,638,450 | |
| RDA I130 (pigmented) | 642 | AY 254384 | 54 | ||
| RDA II60 | 652 | AY 254385 | 56 | 1,188,511-1,187,860 | |
| RDA III10 | 456 | AY 254386 | 58 | 3,987,903-3,988,358 | |
| M. avium subsp. paratuberculosis type I specific | |||||
| pig-RDA10 | 233 | AY 266300 | 99 | 5,394,822-5,395,054 | |
| pig-RDA20 | 197 | AY 266301 | 98 | 1,986,505-1,986,701 | |
| pig-RDA30 | 548 | AY 266302 | 99 | 3,021,830-3,022,376 |
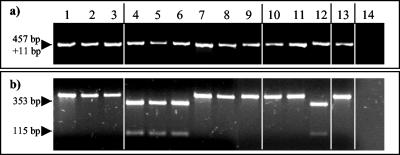
Fragment RDI130 contained identical sequences in all nine M. avium subsp. paratuberculosis type II isolates tested but included 11 additional nucleotides (AGTGACGGCTG) in all M. avium subsp. paratuberculosis type I isolates. This difference was verified by restriction digests of RDI130-specific PCR products with the endonuclease Tsp45I (Fig. 1). Analysis of the sequence adjacent to fragment RDI130 on the M. avium subsp. paratuberculosis genome revealed that the fragment is part of a putative polycistronic operon carrying a set of five genes. The 11-bp insertion is located in the second open reading frame (ORF) and contains an in-frame stop codon. The fourth ORF of this cluster encodes a protein with FtsK motifs that may be involved in cell division (2, 3, 4). The fifth ORF of this operon has already been identified as a M. avium subsp. paratuberculosis-specific phage integrase (GenBank accession number L39071).
FIG. 1.
PCR analysis of M. avium subsp. paratuberculosis isolates of different host groupings by using the internal primer pair I130A/131A and Tsp45I restriction of the PCR product RD I130. The DNA template used was derived from three nonpigmented bovine (lanes 1 to 3), three pigmented ovine (lanes 4 to 6), three nonpigmented ovine (lanes 7 to 9), three nonpigmented caprine (lanes 10 to 12), and one nonpigmented human M. avium subsp. paratuberculosis isolate (lane 13) as well as the M. avium subsp. avium ATCC 25291 isolate (lane 14). (a) The PCR with the primers I130A/I131A resulted in products of different sizes for the M. avium subsp. paratuberculosis type I and type II isolates. (b) The PCR product RD I130 of the M. avium subsp. paratuberculosis type I isolates was cut into two fragments by digestion with Tsp45I. The arrows to the left indicate the size of the corresponding PCR products.
To investigate M. avium subsp. paratuberculosis type II-specific DNA regions, the bovine M. avium subsp. paratuberculosis type II strain 6783 was used as tester and the pigmented ovine type I strain M189 as driver. This RDA did not result in any specific product, suggesting that M. avium subsp. paratuberculosis type II strains, unlike type I strains (see below), do not contain extended loci of type-specific DNA or that the sensitivity of the assay was not sufficient to detect these fragments.
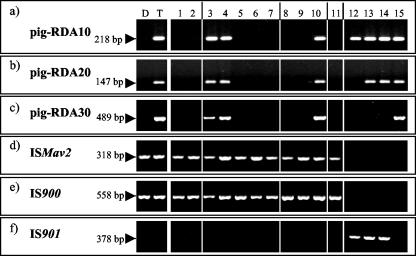
To investigate M. avium subsp. paratuberculosis type I-specific DNA regions the RDA was repeated upon reversing tester and driver strains. This RDA resulted in three M. avium subsp. paratuberculosis type I-specific DNA fragments, designated pig-RDA10, pig-RDA20, and pig-RDA30, that showed no homology to the M. avium subsp. paratuberculosis K-10 genome (a type II strain) but contained 98 to 99% homology to M. avium subsp. avium sequences (BLAST search in the TIGR database) (Table 3). The PCR with specific primers (Table 2) resulted in specific products in all M. avium subsp. paratuberculosis type I strains, whereas all type II strains remained negative (Fig. 2a to c). Furthermore, three M. avium subsp. avium serotype 2 isolates (ATCC 35712, ATCC 25291, and DSM 44158) showed a specific PCR product with the primer pair p19/p20 (specific for pig-RDA10), and two of them (ATCC 25291 and DSM 44158) showed a PCR product with p21/p22 specific for pig-RDA20 (Fig. 2a and b and 3a) but did not show a PCR product with the primer pair p23/p24 specific for pig-RDA30 (Fig. 2c and 3b). The M. avium subsp. avium serotype 4 strain (MAA S4) was positive in all three PCRs. The four M. avium subsp. paratuberculosis type I isolates, like all other nonpigmented isolates, were positive in an M. avium subsp. paratuberculosis-specific IS900- and ISMav2-PCR (Fig. 2d and e, Table 2) but were negative in an IS901-PCR with primers MK7 and MK8 (Fig. 2f, Table 2).
FIG. 2.
PCR analyses of M. avium subsp. paratuberculosis and M. avium subsp. avium isolates with the primer pair p19/p20, specific for pig-RDA10 (a); p21/p22, specific for pig-RDA20 (b); p23/p24, specific for pig-RDA30 (c); ISMav2 I/ISMav2 II, specific for M. avium subsp. paratuberculosis (d); MK5/MK6, specific for IS900 of M. avium subsp. paratuberculosis (e); and (f) MK7/MK8, specific for IS901 of M. avium subsp. avium. The template DNA used was derived from the nonpigmented type II driver strain (lane D) and the pigmented type I tester strain (lane T). Other template DNA used was from two nonpigmented bovine (lanes 1 and 2), two pigmented (lanes 3 and 4), three nonpigmented ovine (lanes 5 to 7), three nonpigmented caprine (lanes 8 to 10), and one nonpigmented human (lane 11) M. avium subsp. paratuberculosis isolate as well as four M. avium subsp. avium isolates (ATCC 35712, ATCC 25291, DSM 44158, and MAA S4; lanes 12 to 15).
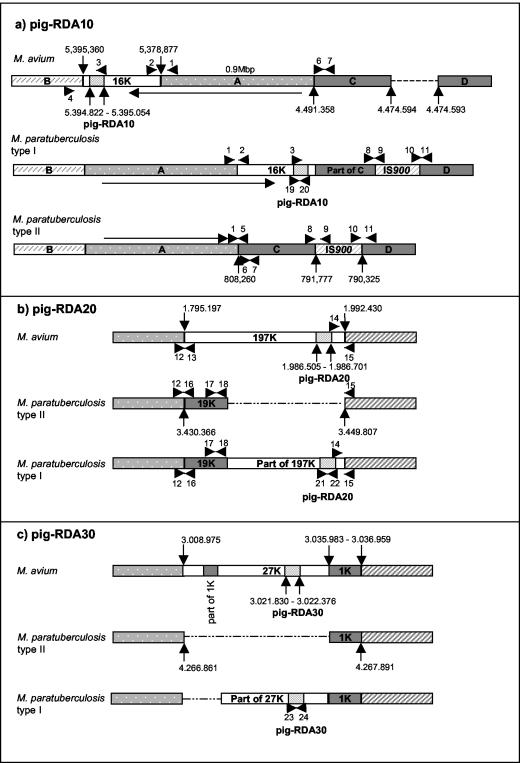
FIG. 3.
Genetic organization of the loci pig-RDA10, pig-RDA20, and pig-RDA30 specific for M. avium subsp. paratuberculosis type I isolates. The numbers given in the maps relate to the positions on the M. avium subsp. avium (TIGR strain 104, contig 3294) and M. avium subsp. paratuberculosis (UMN strain K-10, contig 16) genomes. (a) pig-RDA10 locus. The sequence identified is also present in the 16K locus present in M. avium subsp. avium (top) and M. avium subsp. paratuberculosis type I (middle) but is absent from M. avium subsp. paratuberculosis type II (bottom). The 16K locus is flanked by a 0.9-Mbp sequence (designated A) present in M. avium subsp. avium and partially present in M. avium subsp. paratuberculosis but is oriented in the opposite direction (indicated by an arrow). These sequences are bounded by regions (designated B, C, and D) which are interrupted by IS900 locus 15 in both M. avium subsp. paratuberculosis type I and type II isolates. A putative organization of the locus in M. avium subsp. paratuberculosis type I strains is depicted in the middle map. (b) pig-RDA20 locus. The sequence identified is also present in the 197K locus specific for M. avium subsp. avium (top). In M. avium subsp. paratuberculosis the same flanking regions embrace the 19Klocus specific for M. avium subsp. paratuberculosis (middle). The probable organization of the locus in M. avium subsp. paratuberculosis type I strains is depicted in the bottom map. (c) pig-RDA30 locus. The sequence identified is also present in the 27K locus specific for M. avium subsp. avium (top) flanked by a transposase (designated 1K) and containing an additional truncated 1K region. In M. avium subsp. paratuberculosis the 27K locus is missing (middle). The probable organization of the locus in M. avium subsp. paratuberculosis type I strains is depicted in the bottom map. The arrows are numbered to indicate the positions of the primers used (Table 4).
Further analysis using partial alignments of the M. avium subsp. avium TIGR 104 and M. avium subsp. paratuberculosis K-10 genome sequences revealed that pig-RDA10 maps to an ORF near the end of a 16,483-bp sequence found in M. avium subsp. avium (designated 16K region) (Fig. 3a) containing genes encoding reducing and hydrolyzing enzymes. This sequence is flanked in M. avium subsp. avium by a large 0.9-Mbp region (A region) which is 98% homologous to the corresponding M. avium subsp. paratuberculosis region. However, the 16K region along with the immediately adjacent A region are inverted in M. avium subsp. paratuberculosis type I, while in M. avium subsp. paratuberculosis type II the A region is inverted but the 16K region is deleted. Flanking these inversions in both M. avium subsp. paratuberculosis type I and type II are regions homologous to M. avium subsp. avium (C region) (Fig. 3a; 4,491,358 to 4,474,594) but are interrupted by a copy of IS900 (IS900 in locus 15) exactly 16,483 bp away from the A region. The identical length of this C region with that of the 16K region and the proximity to a copy of IS900 suggests that this element may have been involved in this very large genomic inversion. Our PCR analyses suggest that the entire M. avium subsp. avium 16K locus is present in M. avium subsp. paratuberculosis type I strains but is absent in type II strains (Fig. 3a, Table 4), while the IS900 in locus 15 is present in both M. avium subsp. paratuberculosis type I and type II strains but is absent in M. avium subsp. avium. No product could be obtained with primers p1/p5 and primers p3/p5 (Fig. 3a, Table 4). This suggests that M. avium subsp. paratuberculosis type I isolates contain only part of the M. avium subsp. paratuberculosis type II C locus in this chromosomal position. However, PCR with primers p6/p7 amplifying the left-hand end of the C locus was positive (Table 4), suggesting that it may be present but is translocated to a different genomic location in these strains.
TABLE 4.
Primers for the PCR analyses of the M. avium subsp. avium 16K locus and the M. avium subsp. paratuberculosis C locus
| Primer location and sequence | Position in M. avium subsp. avium genome (TIGR strain 104, contig 3294) | Position in the M. avium subsp. paratuberculosis genome (UMN strain K-10, contig 16) | PCR results for:
|
||
|---|---|---|---|---|---|
| M. avium subsp. avium S2/S4 | M. avium subsp. paratuber- culosis type I | M. avium subsp. paratuber- culosis type II | |||
| Left transition side of M. avium subsp. avium 16K | |||||
| p1 (5′-TCC GCA GCC AGA ACG GCG AA-3′) | 5,378,644-5,378,663 | 808,474-808,469 | +/+ | + | − |
| p2 (5′-TCA CCG AGA CCA TGA CGC TGC-3′) | 5,379,037-5,379,057 | ||||
| Right transition side of M. avium subsp. avium 16K | |||||
| p3 (5′-TGT GCG AGC GCG AAA ACC GC-3′) | 5,394,672-5,394,691 | +/+ | − | − | |
| p4 (5′-TGA TCA GCG CGA CCT TGC CC-3′) | 5,395,421-5,395,440 | ||||
| Left transition side of M. avium subsp. avium subsp. paratuberculosis C locus | |||||
| p1 (5′-TCC GCA GCC AGA ACG GCG AA-3′) | 5,378,644-5,378,663 | 808,474-808,469 | −/− | − | + |
| p5 (5′-TCA GCT CCT CGA TCC CCG TC-3′) | 4,491,296-4,491,315 | 808,191-808,210 | |||
| Left-hand end of M. avium subsp. paratuberculosis C locus | |||||
| p6 (5′-TGT TCG CGC CCA TTC TGC GG-3′) | 4,491,321-4,491,340 | 808,235-808,216 | +/+ | + | + |
| p7 (5′-TCA GGA TCC GCT CCA CCT CG-3′) | 4,491,245-4,491,264 | 808,140-808,159 | |||
| Right-hand end of M. avium subsp. avium 16K plus left-hand end of M. avium subsp. paratuberculosis C locus | |||||
| p3 (5′-TGT GCG AGC GCG AAA ACC GC-3′) | 5,394,672-5,394,691 | +/+ | − | − | |
| p5 (5′-TCA GCT CCT CGA TCC CCG TC-3′) | 4,491,296-4,491,315 | 808,191-808,210 | |||
| M. avium subsp. paratuberculosis IS900 in locus 15, left-hand end | |||||
| p8 (5′-TCG ATG GTG CCG CTG GCG TT-3′) | 792,007-792,026 | −/− | + | + | |
| p9 (5′-TAC CTG TCG GCC TTG GTC AGC-3′) | 791,573-791,593 (and many other hits) | ||||
| M. avium subsp. paratuberculosis IS900 locus 15, right-hand end | |||||
| p10 (5′-GCG ATG ATC GCA GCG TCT TTG G-3′) | 790,893-790,872 (and many other hits) | −/− | + | + | |
| p11 (5′-TCG AAC AGC AGG GGT TCG GTC-3′) | 4,474,535-4,474,555 | 790,267-790,287 | |||
RDA fragment pig-RDA20 maps to a large 197,233-bp M. avium subsp. avium-specific region (designated 197K locus) (Fig. 3b). Sequence analyses of this region indicate that it encodes several systems involved in metabolic pathways, with fragment pig-RDA20 positioned within an ORF predicted to encode a peptide synthase. Sequences flanking the 197K locus are 98% homologous in M. avium subsp. avium and M. avium subsp. paratuberculosis, but in M. avium subsp. paratuberculosis the 197K locus is replaced by a 19,441-bp sequence (19K locus) carrying a gene cassette containing close homologues to mce genes involved in mycobacterial entry into mammalian cells (7, 22). Primers specific for the left- and right-hand end of the M. avium subsp. avium 197K locus and the M. avium subsp. paratuberculosis 19K locus were designed (Table 5). PCRs with primers specific for the left- and right-hand end of the M. avium subsp. paratuberculosis 19K locus suggest that M. avium subsp. paratuberculosis type I isolates contain this entire locus (Fig. 3b, Table 5). Additional PCRs showed that the left-hand end of the M. avium subsp. avium 197K locus was absent, whereas the right-hand end was present in all M. avium subsp. paratuberculosis type I isolates (Fig. 3b, Table 5).
TABLE 5.
Primers for the PCR analyses of the M. avium subsp. avium 197K locus and the M. avium subsp. paratuberculosis 19K locus
| Primer location and sequence | Position in M. avium subsp. avium genome (TIGR strain 104, contig 3294) | Position in M. avium subsp. paratuberculosis genome (UMN strain K-10, contig 16) | PCR results for:
|
||
|---|---|---|---|---|---|
| M. avium subsp. avium S2/S4 | M. avium subsp. paratuber- culosis type I | M. avium subsp. paratuber- culosis type II | |||
| Left transition side of M. avium subsp. avium 197K | |||||
| p12 (5′-TGA TCC GGG CGA CGA TCT GG-3′) | 1,794,970-1,794,989 and 2,073,405-2,073,422 | 3,430,140-3,430,159 | −/+ | − | − |
| p13 (5′-TGG CGT TGA TAT CGC GAC TGG-3′) | 1,795,236-1,795,256 | ||||
| Right-hand end of M. avium subsp. avium 197K | |||||
| p14 (5′-TCG TCC AGG TAG CCG TTC AAC TC-3′) | 1,991,916-1,991,938 | +/+ | + | − | |
| p15 (5′-TGC AGC AGG TGT TCG GGA TGG-3′) | 1,992,400-1,992,420 and 2 other hits, 2,449,453-2,449,472 and 4,813,981-4,814,000 | ||||
| Left transition side of M. avium subsp. paratuberculosis 19K | |||||
| p12 (5′-TGA TCC GGG CGA CGA TCT GG-3′) | 1,794,970-1,794,989 | 3,430,140-3,430,159 | −/− | + | + |
| p16 (5′-TGT CGT CAA CGC GGT TGG CG-3′) | 3,430,532-3,430,551 | ||||
| Right-hand end of M. avium subsp. paratuberculosis 19K | |||||
| p17 (5′-TAC TAC GCC TAC GCA CCG CG-3′) | 3,448,574-3,448,593 | −/− | + | + | |
| p18 (5′-TAG ATC AGG ATG TGC CCG GCC-3′) | 3,448,995-3,448,975 | ||||
| Right-hand end of M. avium subsp. paratuberculosis 19K plus left-hand end of M. avium subsp. avium 197K | |||||
| p17 (5′-TAC TAC GCC TAC GCA CCG CG-3′) | 3,448,574-3,448,593 | −/− | − | − | |
| p13 (5′-TGG CGT TGA TAT CGC GAC TGG-3′) | 1,795,236-1,795,256 | ||||
RDA fragment pig-RDA30 maps to a 27,008-bp M. avium subsp. avium-specific region (27K region) (Fig. 3c) positioned within the carboxy-terminal end of a tetR regulation gene that is absent in M. avium subsp. paratuberculosis type II isolates. The sequences flanking the 27K region are 98% homologous in M. avium subsp. avium and M. avium subsp. paratuberculosis. A 1,030-bp (1K region) sequence encoding a transposase is located on the right-hand side (Fig. 3c) as well as in several other locations in the M. avium subsp. paratuberculosis and the M. avium subsp. avium genomes. The presence of several copies of different insertion sequences within the left-hand end of the M. avium subsp. avium 27K region prevented the design of primers specific for this region. We were therefore unable to determine the extent to which the 27K region is present in M. avium subsp. paratuberculosis type I strains (Fig. 3c).
Our study characterizes, for the first time, genomic differences between M. avium subsp. paratuberculosis type I and type II strains by using four and nine independent isolates, respectively. It suggests that, while neither M. avium subsp. paratuberculosis type I nor type II isolates contain unique type-specific loci, type II strains appear to have undergone more deletions and rearrangements of regions than type I strains that have corresponding loci in M. avium subsp. avium. The presence of the 0.9-Mbp reversed region containing pig-RDA10 in both M. avium subsp. paratuberculosis type I and type II isolates suggests that both types of M. avium subsp. paratuberculosis strains originate from a common progenitor and that the two types are not derived from divergent M. avium subsp. avium strains that have similarly acquired IS900. These findings are also consistent with the hypothesis that M. avium subsp. paratuberculosis type I isolates are an evolutionary intermediate between M. avium subsp. avium and M. avium subsp. paratuberculosis type II strains. Alternatively, since many of the fragments map to regions that are likely to be carried by phage or to be adjacent to mobile genetic elements, the results may be indicative of the fact that type I isolates and some isolates of M. avium subsp. avium are more likely to share some of these elements through horizontal transfer. Further studies based on large-scale comparative sequence analysis as well as a larger number of isolates will be needed to reconstruct the evolutionary history of this closely related group of animal-pathogenic mycobacteria.
Acknowledgments
This study was funded by the EU project Assess Mptb Risk QLK2-CT-2001-01420, by Action Research, United Kingdom, by the Scottish Executive Environment Rural Affairs Department, and by a mobility grant jointly financed by the Academic Research Council (ARC) and the Deutscher Akademischer Austauschdienst (DAAD). M. avium subsp. paratuberculosis genome sequencing in the laboratory of V.K. is supported by competitive grants from the USDA-CSREES.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barre, F. X., M. Aroyo, S. D. Colloms, A. Helfrich, F. Cornet, and D. J. Sherratt. 2000. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 14:2976-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, D. S., D. Grant, G. C. Draper, and W. D. Donachie. 2000. All major regions of FtsK are required for resolution of chromosome dimers. J. Bacteriol. 182:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146:2185-2197. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlin, W., D. Y. Graham, K. Hulten, H. M. El Zimaity, M. R. Schwartz, S. Naser, I. Shafran, and F. A. El Zaatari. 2001. Review article: Mycobacterium avium subsp. paratuberculosis as one cause of Crohn's disease. Aliment. Pharmacol. Ther. 15:337-346. [DOI] [PubMed] [Google Scholar]
- 7.Chitale, S., S. Ehrt, I. Kawamura, T. Fujimura, N. Shimono, N. Anand, S. W. Lu, L. Cohen-Gould, and L. W. Riley. 2001. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 3:247-254. [DOI] [PubMed] [Google Scholar]
- 8.Collins, D. M., M. De Zoete, and S. M. Cavaignac. 2002. Mycobacterium avium subsp. paratuberculosis strains from cattle and sheep can be distinguished by a PCR test based on a novel DNA sequence difference. J. Clin. Microbiol. 40:4760-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Voss, J. J., K. Rutter, B. G. Schroeder, and C. E. Barry III. 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doran, T. J., J. K. Davies, A. J. Radford, and A. L. Hodgson. 1994. Putative functional domain within ORF2 on the Mycobacterium insertion sequences IS900 and IS902. Immunol. Cell Biol. 72:427-434. [DOI] [PubMed] [Google Scholar]
- 11.El Zaatari, F. A., M. S. Osato, and D. Y. Graham. 2001. Etiology of Crohn's disease: the role of Mycobacterium avium subsp. paratuberculosis. Trends Mol. Med. 7:247-252. [DOI] [PubMed] [Google Scholar]
- 12.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imaeda, T., G. Broslawski, and S. Imaeda. 1988. Genomic relatedness among mycobacterial species by nonisotopic blot hybridization. Int. J. Syst. Bacteriol. 38:151-156. [Google Scholar]
- 15.Johnston, W. S., and G. K. MacLachlan. 1986. Johne's disease in sheep. Vet. Rec. 7:75. [DOI] [PubMed] [Google Scholar]
- 16.Kreeger, J. M. 1991. Ruminant paratuberculosis - a century of progress and frustration. J. Vet. Diagn. Investig. 3:373-382. [DOI] [PubMed] [Google Scholar]
- 17.Kunze, Z. M., S. Wall, R. Appelberg, M. T. Silva, F. Portaels, and J. J. McFadden. 1991. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol. Microbiol. 5:2265-2272. [DOI] [PubMed] [Google Scholar]
- 18.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 19.Marsh, I., R. Whittington, and D. Cousins. 1999. PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol. Cell Probes 13:115-126. [DOI] [PubMed] [Google Scholar]
- 20.McFadden, J. J., P. D. Butcher, R. Chiodini, and J. Hermon-Taylor. 1987. Crohn's disease-isolated mycobacteria are identical to Mycobacterium paratuberculosis, as determined by DNA probes that distinguish between mycobacterial species. J. Clin. Microbiol. 25:796-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen, K., and P. Ahrens. 2002. Putative in vitro expressed gene fragments unique to Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 214:199-203. [DOI] [PubMed] [Google Scholar]
- 22.Parker, S. L., Y. L. Tsai, and C. J. Palmer. 1995. Comparison of PCR-generated fragments of the mce gene from Mycobacterium tuberculosis, Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum. Clin. Diagn. Lab. Immunol. 2:770-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poupart, P., M. Coene, H. Van Heuverswyn, and C. Cocito. 1993. Preparation of a specific RNA probe for detection of Mycobacterium paratuberculosis and diagnosis of Johne's disease. J. Clin. Microbiol. 31:1601-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Power, S. B., J. Haagsma, and D. P. Smyth. 1993. Paratuberculosis in farmed red deer (Cervus elaphus) in Ireland. Vet. Rec. 132:213-216. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Saxegaard, F., I. Baess, and E. Jantzen. 1988. Characterization of clinical isolates of Mycobacterium paratuberculosis by DNA-DNA hybridization and cellular fatty acid analysis. Acta Pathol. Microbiol. Immunol. Scand. 96:497-502. [PubMed] [Google Scholar]
- 27.Stevenson, K., V. M. Hughes, L. de Juan, N. F. Inglis, F. Wright, and J. M. Sharp. 2002. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 40:1798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stratmann, J., B. Strommenger, K. Stevenson, and G. F. Gerlach. 2002. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J. Clin. Microbiol. 40:4244-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strommenger, B., K. Stevenson, and G. F. Gerlach. 2001. Isolation and diagnostic potential of ISMav2, a novel insertion sequence-like element from Mycobacterium avium ssp. paratuberculosis. FEMS Microbiol. Lett. 196:31-37. [DOI] [PubMed] [Google Scholar]
- 30.Thorel, M. F., M. Krichevsky, and V. V. Levy-Frebault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40:254-260. [DOI] [PubMed] [Google Scholar]
- 31.Weber, A., R. Gürke, K. Bauer, and K. Schreyer. 1992. Bacterial studies on the occurrence of Mycobacterium paratuberculosis in fecal samples of zoo ruminants. Berl. Munch. Tierarztl. Wochenschr. 105:161-164. [PubMed] [Google Scholar]