Abstract
Among 100 patients with group G beta-hemolytic streptococcal bacteremia in a 6-year period (1997 to 2002), seven had bacteremia caused by erythromycin-resistant strains. Five of the seven patients had cellulitis and/or abscesses. The two isolates resistant to erythromycin and clindamycin possessed erm genes, one ermTR and the other ermB. The five isolates resistant to erythromycin but sensitive to clindamycin and one of those resistant to both erythromycin and clindamycin possessed mef genes.
Macrolides constitute an important group of drugs because of their antimicrobial and immunomodulatory activities (3, 21, 22, 25, 27). As for the treatment and prophylaxis of infections caused by streptococci, the main role of macrolides is their use in patients with beta-lactam hypersensitivity. Macrolide resistance in streptococci has been increasingly reported (8, 9, 14, 18, 26) and is mediated through two major mechanisms, target site modification (mediated through various erythromycin resistance methylase [erm] genes) and the use of efflux pumps (mediated through mef genes). Different types of erm genes that mediate the MLSB phenotype and resistance to macrolides, lincomycin, and streptogramin B have been described in streptococci (1, 2, 5, 6, 8, 10, 16, 18, 20). Recently it was noticed that 65% of Streptococcus bovis strains isolated from patients with S. bovis bacteremia were erythromycin resistant (10). These erythromycin-resistant isolates possessed either ermB or ermT genes. In another study, it was noticed that 36.5% of the isolates associated with invasive Streptococcus pyogenes infections in Hong Kong were resistant to erythromycin and the resistance was mediated through the possession of the ermTR gene, the mef gene, both the ermTR and mef genes, or the ermB gene (5). As for Streptococcus pneumoniae, 27 and 73% of erythromycin-resistant isolates possessed erm and mef genes, respectively (8).
Although beta-hemolytic group G streptococci are the most common cause of beta-hemolytic streptococcal bacteremia in many parts of the world (17, 23), no study describing the clinical and molecular epidemiology of macrolide resistance in this group of streptococci was found in the literature. In this study, we report phenotypic and genotypic characterization of the erythromycin resistance in beta-hemolytic group G streptococci recovered from blood cultures of patients over a 6-year period. Risk factors for bacteremia caused by erythromycin-resistant, beta-hemolytic group G streptococci were also analyzed.
The 100 patients in this study were hospitalized at the Queen Mary Hospital in Hong Kong during a 6-year period (1997 to 2002). All suspect colonies were identified by standard conventional biochemical methods (13), and streptococci were further identified using the API system (20 STREP; Biomerieux Vitek, Hazelwood, Mo.). Lancefield serogrouping was performed using Streptex (Murex Biotech Ltd., Dartford, United Kingdom) according to the manufacturer's instructions. Antimicrobial susceptibility was tested by the Kirby Bauer disk diffusion method, and results were interpreted according to the NCCLS criteria. MICs of penicillin, erythromycin, clindamycin, and vancomycin for the seven strains of erythromycin-resistant beta-hemolytic group G streptococci were determined using the macrodilution broth method (13).
Bacterial DNA extraction from the seven erythromycin-resistant beta-hemolytic streptococcus isolates was performed using a previously published protocol (24). PCR amplification and DNA sequencing of the ermTR, ermB, and mef genes were performed as described in previous publications (5, 10, 16, 18). The PCR mixture (50 μl) contained bacterial DNA, primers (LPW511, 5′-GGTTATAATGAAMCAGAAAAACCC-3′, and LPW512, 5′-CKATACTTTTTGTAGTCCTTC-3′, for ermTR; LPW509, 5′-AACGARTGAAAARGTACTCAACC-3′, and LPW510, 5′-AGAATTATTTCCTCCCGTTAAATA-3′, for ermB; and LPW707, 5′-ATGGAAAAATACAACAATTGG-3′, and LPW708, 5′-TTATTTTAAATCTAATTTTCT-3′, for mef; Gibco BRL, Rockville, Md.), PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2 mM MgCl2, and 0.01% gelatin), 200 μM (each) deoxynucleoside triphosphates, and 1.0 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). The mixtures were amplified in 40 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min, in an automated thermal cycler (Perkin-Elmer Cetus, Gouda, The Netherlands). Distilled water was used as the negative control.
The PCR products were gel purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI 377 automated sequencer according to the instructions of the manufacturer (Perkin-Elmer, Foster City, Calif.) by using the corresponding PCR primers. The sequences of the PCR products were compared with known erm and mef gene sequences from GenBank by using multiple sequence alignment with the CLUSTAL W program (19), and phylogenetic tree construction was performed using the PileUp method with GrowTree (Genetics Computer Group, Inc.).
A comparison was made of characteristics of patients with bacteremia due to resistant beta-hemolytic group G streptococci and those of patients with bacteremia due to streptococci that were sensitive to erythromycin. The chi-square test was used for categorical variables, and Student's t test was used for age. P values of <0.05 were regarded as statistically significant.
During the 6-year period, 100 patients had beta-hemolytic group G streptococcal bacteremia. All 100 isolates were identified as Streptococcus dysgalactiae subsp. equisimilis. For the seven patients with bacteremia caused by erythromycin-resistant beta-hemolytic group G streptococci, the median age was 64 (range, 45 to 97). The male-female ratio was 4:3. No source of the bacteremia was identified in two patients, whereas four and two patients had cellulitis and abscess formation, respectively (with one having both cellulitis and abscess formation). All seven patients had community-acquired monomicrobial bacteremia, with the beta-hemolytic group G streptococci isolated from single blood cultures. None of the seven patients died.
The characteristics of patients with bacteremia due to resistant beta-hemolytic group G streptococci and those of patients with bacteremia due to streptococci that were sensitive to erythromycin were compared and summarized in Table 1. Erythromycin resistance was associated with cellulitis or abscess formation (P of <0.05 and <0.005, respectively).
TABLE 1.
Comparison of characteristics of patients with bacteremia due to resistant beta-hemolytic group G streptococci and those of patients with bacteremia due to streptococci that were sensitive to erythromycin
| Characteristic | No. of patients (%) or value for group with streptococci found to bea:
|
|
|---|---|---|
| Resistant (n = 7) | Sensitive (n = 93) | |
| Sex | ||
| Male | 4 (57) | 49 (53) |
| Female | 3 (43) | 44 (47) |
| Mean age ± SEM (yr) | 67 ± 22.9 | 73 ± 16.1 |
| Underlying condition | ||
| Hypertension | 3 (43) | 26 (28) |
| Malignancy | 2 (29) | 27 (29) |
| Immobilization and/or bed sore | 2 (29) | 25 (27) |
| Diabetes mellitus | 2 (29) | 18 (19) |
| Chronic renal failure | 2 (29) | 9 (10) |
| Dementia | 1 (14) | 17 (18) |
| Cirrhosis | 1 (14) | 6 (6) |
| Cerebrovascular accident | 0 (0) | 27 (29) |
| Lymphedema | 0 (0) | 10 (11) |
| Intravenous drug abuse | 0 (0) | 8 (9) |
| Diagnosis | ||
| Primary bacteremia | 2 (29) | 52 (56) |
| Cellulitis | 4 (57) | 20 (22) |
| Abscess | 2 (29) | 2 (2) |
| Bed sore/wound infection | 0 (0) | 8 (9) |
| Infective endocarditis | 0 (0) | 4 (4) |
| Septic arthritis | 0 (0) | 3 (3) |
| Pneumonia | 0 (0) | 2 (2) |
| Disseminated infection | 0 (0) | 2 (2) |
| Toxic shock syndrome | 0 (0) | 1 (2) |
| Aquisition source | ||
| Community | 7 (100) | 83 (89) |
| Hospital | 0 (0) | 10 (11) |
| No. of positive blood cultures | ||
| Single | 7 (100) | 86 (92) |
| Multiple | 0 (0) | 7 (8) |
| Nature of infection | ||
| Monomicrobial | 7 (100) | 84 (90) |
| Polymicrobial | 0 (0) | 9 (10) |
| Positive culture at other sites | 1 (14) | 12 (13) |
| Outcome | ||
| Cured | 7 (100) | 80 (86) |
| Died | 0 (0) | 13 (14) |
P values were not significant except for those associated with the diagnoses of cellutitis and abscess. These values were < 0.05 and < 0.005, respectively.
The beta-hemolytic group G streptococcal isolates recovered from 93 patients were sensitive to penicillin, erythromycin, clindamycin, and vancomycin. Two isolates were resistant to both erythromycin and clindamycin, and five were resistant to erythromycin but sensitive to clindamycin. The MICs of penicillin for all seven isolates were <0.016 μg/ml. The MICs of vancomycin for five and two isolates were 0.5 and 0.25 μg/ml, respectively. The median MIC of erythromycin was 4 μg/ml (range, 0.5 to >256 μg/ml). The MICs of clindamycin for the two isolates resistant to clindamycin were 0.5 μg/ml, and those for the five isolates sensitive to clindamycin were 0.125 μg/ml.
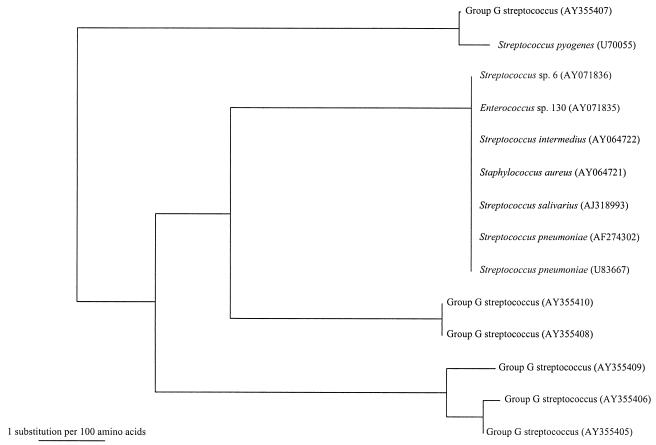
The two beta-hemolytic group G streptococcal isolates resistant to both erythromycin and clindamycin possessed erm genes; one possessed the ermTR gene (encoding ErmA protein) (15), and the other possessed the ermB gene (encoding ErmB protein). The five isolates resistant to erythromycin but sensitive to clindamycin and one of those resistant to both erythromycin and clindamycin possessed mef genes (Fig. 1).
FIG. 1.
Phylogenetic tree showing the relationship of the amino acid sequences corresponding to the mef genes of the six erythromycin-resistant beta-hemolytic group G streptococcus isolates that possess mef genes to those corresponding to other mef genes. The tree was inferred from data on 392 amino acid sequences by the neighbor-joining method. The scale bar indicates the estimated number of substitutions per 100 amino acids by using the Kimura correction. Names and accession numbers are given as cited in the GenBank database.
There are major geographical differences associated with the mechanisms of macrolide resistance. In one study, it was noted that 31 (97%) of 32 erythromycin-resistant beta-hemolytic group G streptococcus isolates recovered in Finland possessed erm genes, of which 30 (94%) possessed the ermTR gene and only one possessed the ermB gene (9). On the other hand, in the present study, six (86%) of seven beta-hemolytic group G streptococcus isolates recovered from blood cultures possessed mef genes, but only two (29%) of seven possessed erm genes, although the two erm genes recovered from our patients were also ermTR and ermB genes. The present observation is in line with that reported previously, which showed that 9 (50%) of 18 erythromycin-resistant group C and group G beta-hemolytic streptococcus isolates possessed mef genes, but only 5 (28%) of 18 erythromycin-resistant group C and group G beta-hemolytic streptococcus isolates possessed erm genes, although in that study the authors did not mention the different types of genes possessed by the group C and group G beta-hemolytic streptococci (7).
The type of erm gene in beta-hemolytic group G streptococci may be related to the source of the streptococcal infections. Recently, we found that the ermB genes in the S. bovis isolates were highly homologous to the ermB genes in Clostridium difficile, Clostridium perfringens, Lactobacillus reuteri, Streptococcus agalactiae, Enterococcus faecalis, Enterococcus faecium, Lactobacillus fermentum, and Escherichia coli, whereas the ermT genes in the S. bovis isolates were highly homologous to the ermT genes in L. reuteri and another Lactobacillus species, which are bacteria from the gastrointestinal tract (10). We speculated that these genes in S. bovis were acquired through horizontal gene transfer from the other bacteria that reside in the gastrointestinal tract. In the present study, the ermTR gene recovered from the beta-hemolytic group G streptococcus strain isolated from a patient with cellulitis complicating glossectomy, selective neck dissection, and radiotherapy for his carcinoma of the tongue showed high homology to the ermTR genes of S. pyogenes. We speculate that the skin or pharynx, which is the source of S. pyogenes, was also the source of this beta-hemolytic group G streptococcus isolate. On the other hand, the ermB gene recovered from the beta-hemolytic group G streptococcus strain isolated from a patient with a leg abscess complicating alcoholic cirrhosis with recurrent spontaneous bacterial peritonitis showed high homologies with the ermB genes of other bacteria of the gastrointestinal tract. It is well known that intestinal mucosal edema and local immunosuppression secondary to portal venous congestion vasculopathy are very common in patients with decompensated liver cirrhosis with portal hypertension and ascites, and infections in these patients often originate from bacteria in the gastrointestinal tract (4). Therefore, we speculate that the source of the beta-hemolytic group G streptococcus isolate from this patient was the gastrointestinal tract and that the ermB gene in this bacterium was also acquired from horizontal gene transfer from other bacteria of the gastrointestinal tract.
Macrolide resistance in beta-hemolytic group G streptococci has resulted from horizontal transfer of mef genes among different species of streptococci, staphylococci, and enterococci, as well as among the beta-hemolytic group G streptococci themselves. Efflux of macrolides, mediated by mef genes, has been described mainly in various gram-positive cocci (5, 7, 8, 11, 12). From the available sequence information, it can be observed that the amino acid sequence corresponding to the mef gene (AY355407) of the beta-hemolytic group G streptococcus strain isolated from one patient shared more than 99% identity with that corresponding to a mef gene (U70055) from a strain of S. pyogenes (Fig. 1). Moreover, the amino acid sequences corresponding to the mef genes (AY355408 and AY355410) of the beta-hemolytic group G streptococci isolated from two patients were identical, and those corresponding to the mef genes (AY355405, AY355406, and AY355409) of the beta-hemolytic group G streptococci isolated from three other patients also showed very high sequence identity. These findings implied that there was horizontal transfer of mef genes among the various gram-positive cocci. This is in line with the evidence from a study which showed that it is possible to move the mef genes from all 11 erythromycin-resistant S. pneumoniae isolates tested to erythromycin-susceptible S. pneumoniae and/or E. faecalis recipients (11).
Nucleotide sequence accession numbers.
The erm and mef gene sequences of the seven erythromycin-resistant beta-hemolytic group G streptococcus isolates have been deposited with the GenBank sequence database under accession numbers AY355403, AY355404, AY355405, AY355406, AY355407, AY355408, AY355409, and AY355410.
REFERENCES
- 1.Brantl, S., C. Kummer, and D. Behnke. 1994. Complete nucleotide sequence of plasmid pGB3631, a derivative of the Streptococcus agalactiae plasmid pIP501. Gene 142:155-156. [DOI] [PubMed] [Google Scholar]
- 2.Ceglowski, P., and J. C. Alonzo. 1994. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the orfŋ-orfS region. Gene 145:33-39. [DOI] [PubMed] [Google Scholar]
- 3.Chow, L. W. C., K. Y. Yuen, P. C. Y. Woo, and W. I. Wei. 2000. Clarithromycin attenuates mastectomy-induced acute inflammatory response. Clin. Diagn. Lab. Immunol. 7:925-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corredoira, J. M., J. Ariza, R. Pallares, J. Carratala, P. F. Viladrich, G. Rufi, R. Verdaguer, and F. Gudiol. 1994. Gram-negative bacillary cellulitis in patients with hepatitic cirrhosis. Eur. J. Clin. Microbiol. Infect. Dis. 13:19-24. [DOI] [PubMed] [Google Scholar]
- 5.Ho, P. L., D. R. Johnson, A. W. Y. Yue, D. N. C. Tsang, T. L. Que, B. Beall, and E. L. Kaplan. 2003. Epidemiologic analysis of invasive and noninvasive group A streptococcal isolates in Hong Kong. J. Clin. Microbiol. 41:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horinouchi, S., W. Byeon, and B. A. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ip, M., D. J. Lyon, T. Leung, and A. F. B. Cheng. 2002. Macrolide resistance and distribution of erm and mef genes among beta-haemolytic streptococci in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 21:238-240. [DOI] [PubMed] [Google Scholar]
- 8.Ip, M., D. J. Lyon, R. W. H. Yung, C. Chan, and A. F. B. Cheng. 2001. Macrolide resistance in Streptococcus pneumoniae in Hong Kong. Antimicrob. Agents Chemother. 45:1578-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kataja, J., H. Seppälä, M. Skurnik, H. Sarkkinen, and P. Huovinen. 1998. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob. Agents Chemother. 42:1493-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, R. A., P. C. Y. Woo, A. P. C. To, S. K. P. Lau, S. S. Y. Wong, and K. Y. Yuen. 2003. Geographical difference of disease association in Streptococcus bovis bacteremia. J. Med. Microbiol. 52:903-908. [DOI] [PubMed] [Google Scholar]
- 11.Luna, V. A., P. Coates, E. A. Eady, J. H. Cove, T. T. H. Nguyen, and M. C. Roberts. 1999. A variety of Gram-positive bacteria carry mobile mef genes. J. Antimicrob. Chemother. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 12.Luna, V. A., S. Cousin, Jr., W. L. H. Whittington, and M. C. Roberts. 2000. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob. Agents Chemother. 44:2503-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray, P. R., E. J. Baro, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 14.Reinert, R. R., R. Lutticken, A. Bryskier, and A. Al-Lahham. 2003. Macrolide-resistant Streptococcus pneumoniae and Streptococcus pyogenes in the pediatric population in Germany during 2000-2001. Antimicrob. Agents Chemother. 47:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppälä. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinant. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skogberg, K., H. Simonen, O. V. Renkonen, and V. V. Valtonen. 1988. Beta-haemolytic group A, B, C and G streptococcal septicaemia: a clinical study. Scand. J. Infect. Dis. 20:119-125. [DOI] [PubMed] [Google Scholar]
- 18.Teng, L. J., P. R. Hsueh, S. W. Ho, and K. T. Luh. 2001. High prevalence of inducible erythromycin resistance among Streptococcus bovis isolates in Taiwan. Antimicrob. Agents Chemother. 45:3362-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Coirralin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed]
- 21.Woo, P. C. Y., L. W. C. Chow, E. S. K. Ma, and K. Y. Yuen. 1999. Clarithromycin attenuates the inflammatory response induced by surgical trauma in a guinea pig model. Pharmacol. Res. 39:49-54. [DOI] [PubMed] [Google Scholar]
- 22.Woo, P. C. Y., W. F. Ng, H. C. H. Leung, H. W. Tsoi, and K. Y. Yuen. 2000. Clarithromycin attenuates cyclophosphamide-induced mucositis in mice. Pharmacol. Res. 41:527-532. [DOI] [PubMed] [Google Scholar]
- 23.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, S. S. Y. Wong, and K. Y. Yuen. 2001. Group G beta-hemolytic streptococcal bacteremia characterized by 16S ribosomal RNA gene sequencing. J. Clin. Microbiol. 39:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo, P. C. Y., D. M. W. Tam, K. W. Leung, S. K. P. Lau, J. L. L. Teng, M. K. M. Wong, and K. Y. Yuen. 2002. Streptococcus sinensis sp. nov., a novel Streptococcus species isolated from a patient with infective endocarditis. J. Clin. Microbiol. 40:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo, P. C. Y., S. K. P. Lau, and K. Y. Yuen. 2002. Macrolides as immunomodulatory agents. Curr. Med. Chem. Anti-Inflamm. Anti-Allergy Agents 1:131-141. [Google Scholar]
- 26.Wu, J. J., K. Y. Lin, P. R. Hsueh, J. W. Liu, H. I. Pan, and S. M. Sheu. 1997. High incidence of erythromycin-resistant streptococci in Taiwan. Antimicrob. Agents Chemother. 41:844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuen, K. Y., P. C. Y. Woo, J. W. M. Tai, A. K. W. Lie, J. Luk, and R. Liang. 2001. Effects of clarithromycin on oral mucositis in bone marrow transplant recipients. Haematologica 86:554-555. [PubMed] [Google Scholar]